Preparation, Optimization, and In-Vitro Evaluation of Brusatol- and Docetaxel-Loaded Nanoparticles for the Treatment of Prostate Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Preparation and Optimization of Stealth Blank Nanoparticles
2.2.2. Initial Screening Using the One-Factor-at-a-Time Method
| Factors | Variables |
|---|---|
| Solvent composition (Organic phase) | Ethyl acetate (EA):Dimethylformamide (DMF) |
| Ethyl acetate (EA):Dimethyl sulfoxide (DMSO) | |
| Ethyl acetate (EA):Acetonitrile | |
| Ethyl acetate (EA):Acetone | |
| Ethyl acetate (EA):DMSO:Acetone | |
| Solvent ratio (EA: other solvent) (Total volume = 2 mL) | 1.2:0.8 |
| 1.4:0.6 | |
| 1.6:0.4 | |
| 1.8:0.2 | |
| mPEG-PLGA concentration (mg/2 mL) | 20, 50, 75, 100, 150 |
| Organic: Aqueous ratio | 2:8 |
| 2:10 | |
| 2:12 | |
| 2:14 | |
| 2:16 | |
| Polyvinyl alcohol concentration (%) | 0.25, 0.5, 1.0, 2.0, 3.0 |
| Sonication pulse rate (on:off) | 7:3, 8:2; 9:1 |
| Amplitude (%) | 25, 30, 35, 40 |
| Sonication time (minutes) | 2.5, 5, 7.5, 10 |
2.2.3. Full Factorial Statistical Experimental Design
2.3. Preparation of Stealth Brusatol- and Docetaxel-Loaded Nanoparticles
2.3.1. Evaluation of Particle and Zeta Potential
2.3.2. Morphological Studies
2.3.3. Infrared Spectroscopy Analysis
2.3.4. Drug Content Determination
2.3.5. Release Profile of Docetaxel- and Brusatol-Loaded Nanoparticles
2.4. Cell Culture Experiments
2.4.1. Combination Index Determination
2.4.2. Viable Cell Count and In Vitro Cytotoxicity
2.4.3. Flow Cytometry Studies
Cell Cycle Arrest Analysis
Caspase 3/7 Activity Assay
Immunoblot Analysis
2.5. Statistical Analysis
3. Results and Discussion
3.1. Screening of Formulation and Process Variables Using the One-Factor-at-a-Time Approach
3.2. Design of Experiments—Full Factorial Statistical Experimental Design
3.3. Regression Equation
3.4. Nanoparticle Morphology, Particle Size, and Zeta Potential Characterization
3.5. Drug-Loading Studies
3.6. FT-IR Spectroscopy Evaluation
3.7. Drug Release Studies
3.8. Evaluation of Cytotoxicity of Pure Drugs and Determination of Combination Index
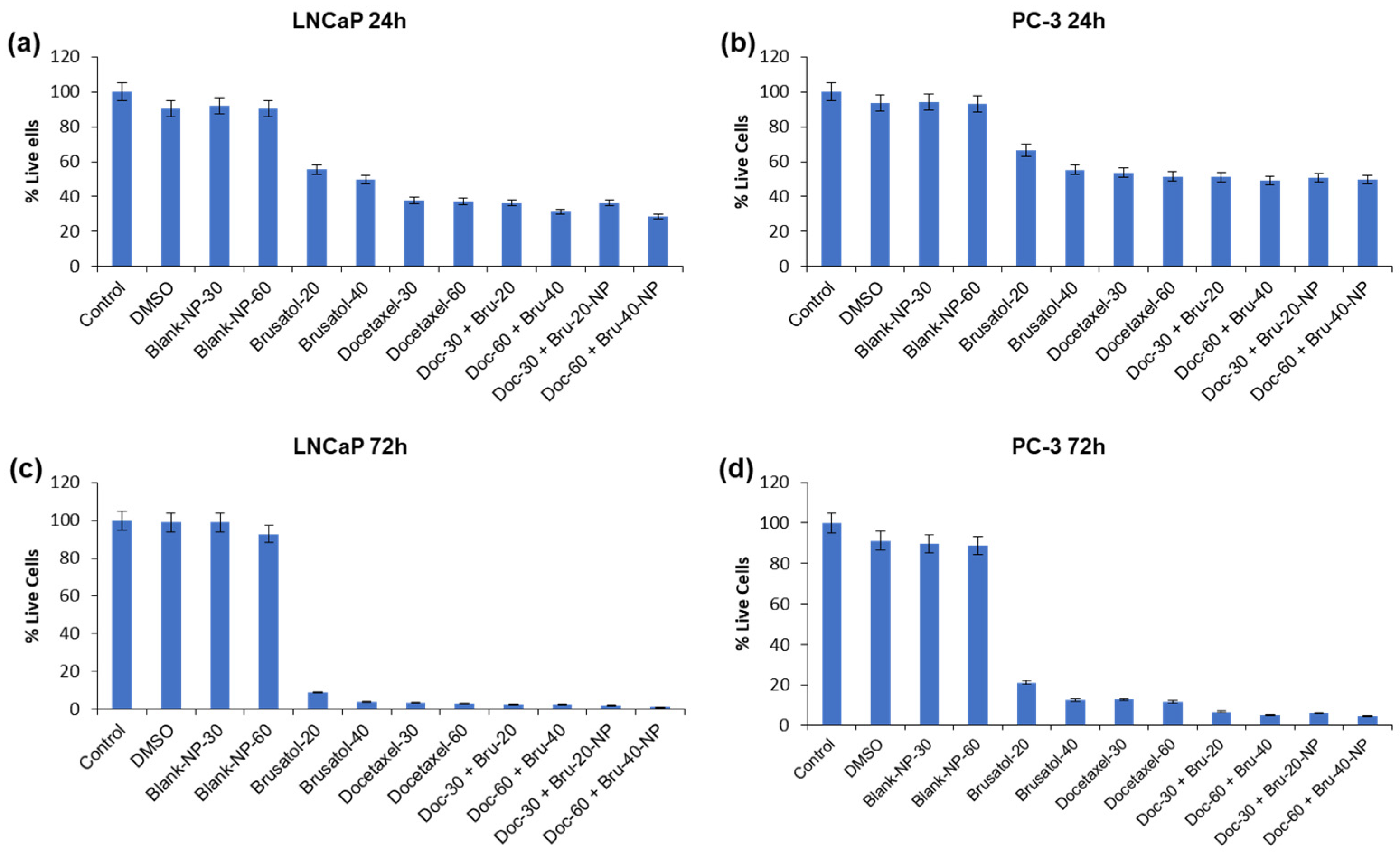
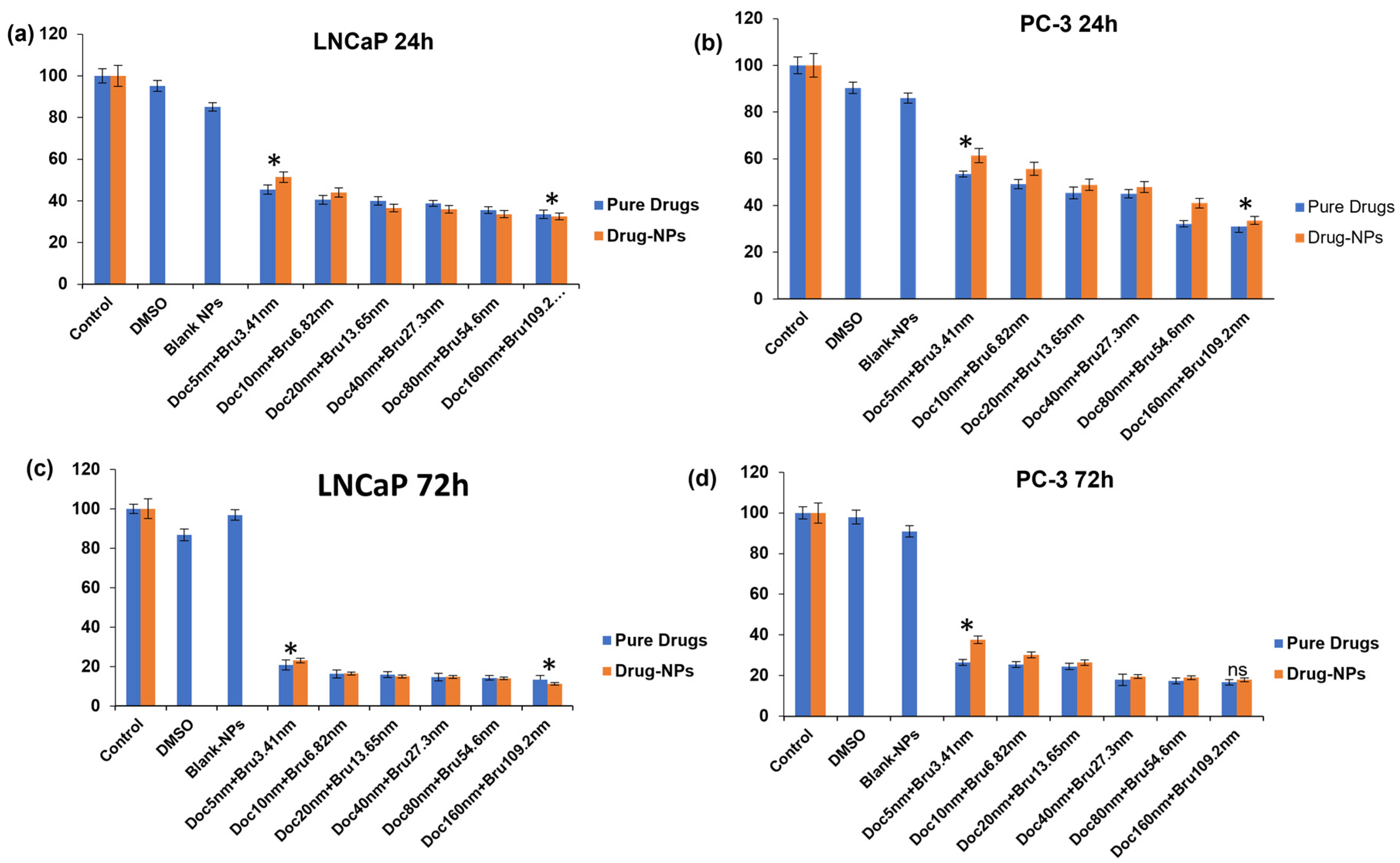
3.9. Cytotoxicity Evaluations of Brusatol- and Docetaxel-Loaded Nanoparticle Formulations
3.9.1. Viable Cell Count and In Vitro Cytotoxicity
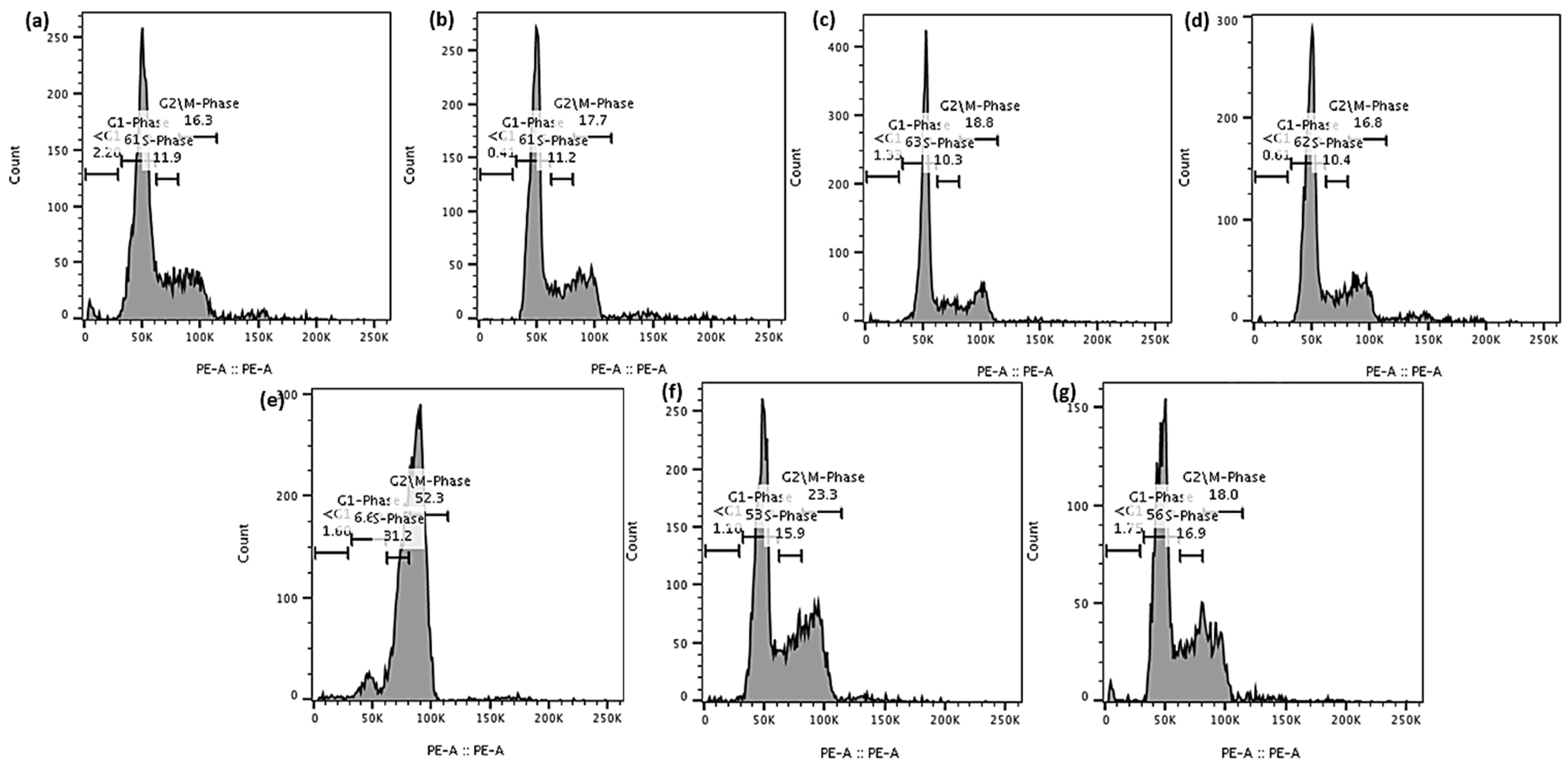
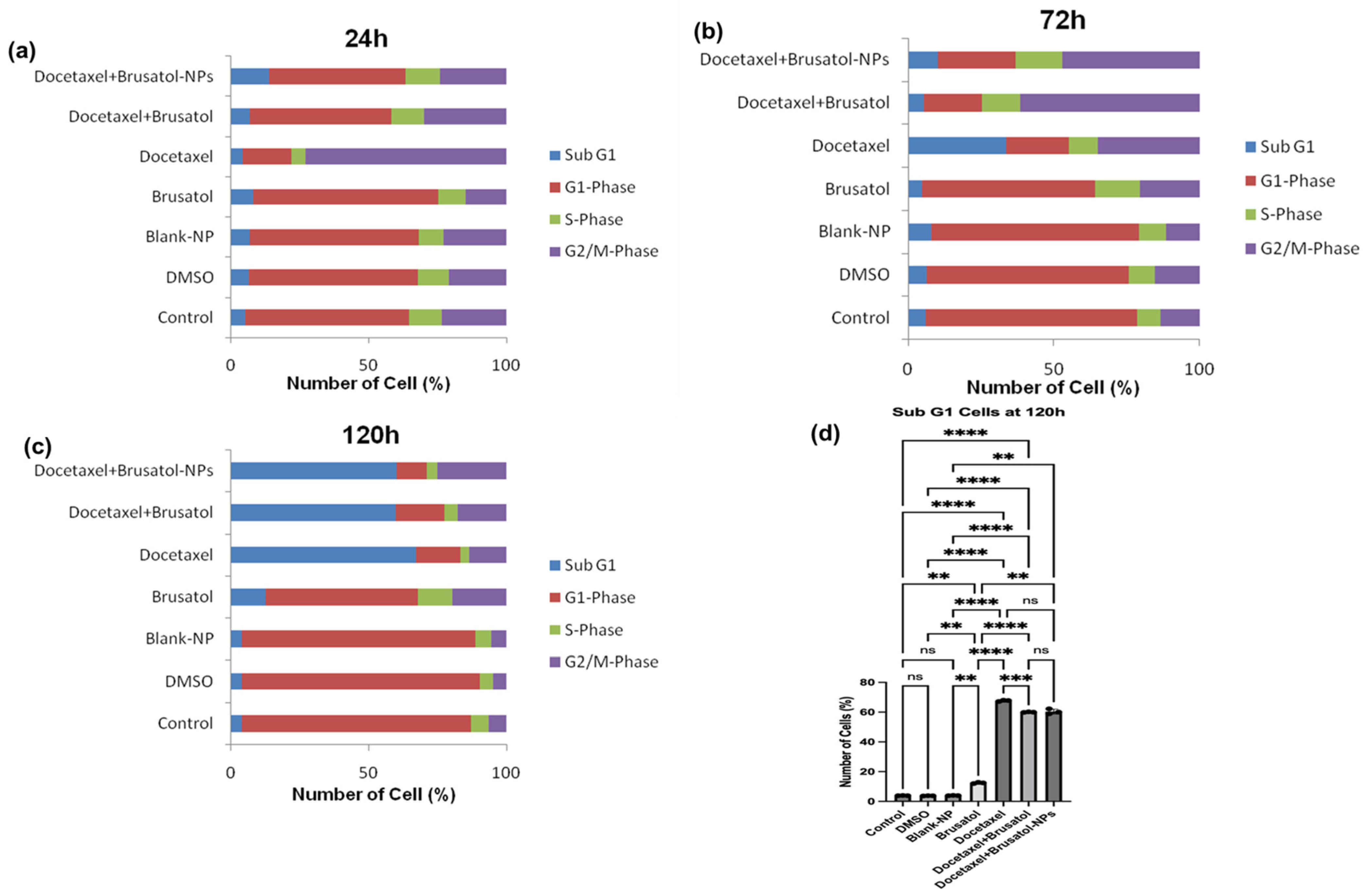
3.9.2. Cell Cycle Analysis
3.9.3. Caspase-3/7 Activity
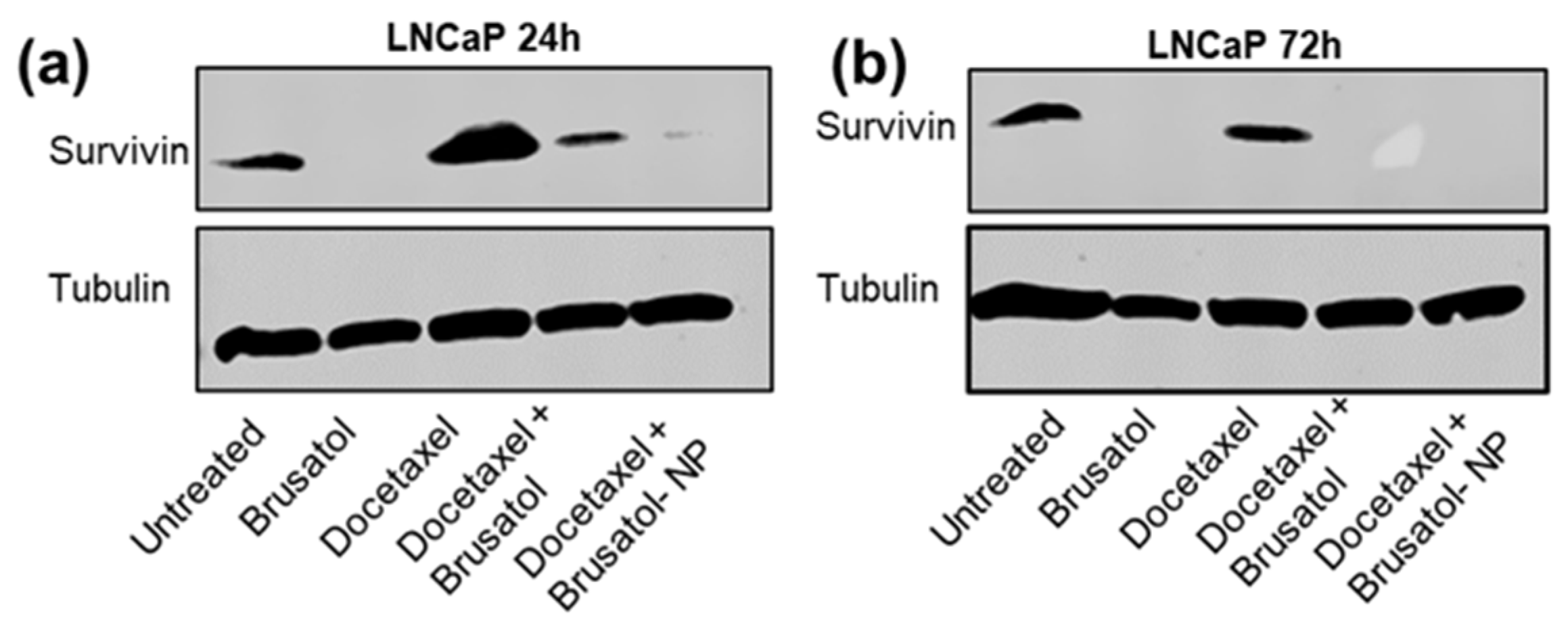
3.9.4. Immunoblotting Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Cancer Society. Cancer Facts & Figures 2023; American Cancer Society: Atlanta, GA, USA, 2023. [Google Scholar]
- National Cancer Institute. SEER Cancer Stat Facts: Prostate Cancer. Available online: https://seer.cancer.gov/statfacts/html/prost.html (accessed on 3 May 2023).
- Liang, Y.; Rong, E.; Qian, J.; Ma, C.; Hu, J. Transcriptome subtyping of metastatic Castration Resistance Prostate Cancer (mCRPC) for the precision therapeutics: An in silico analysis. Prostate Cancer Prostatic Dis. 2022, 25, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Henríquez, I.; Roach, M., III; Morgan, T.M.; Bossi, A.; Gómez, J.A.; Abuchaibe, O.; Couñago, F. Current and Emerging Therapies for Metastatic Castration-Resistant Prostate Cancer (mCRPC). Biomedicines 2021, 9, 1247. [Google Scholar] [CrossRef]
- Autio, K.A.; Dreicer, R.; Anderson, J.; Garcia, J.A.; Alva, A.; Hart, L.L.; Milowsky, M.I.; Posadas, E.M.; Ryan, C.J.; Graf, R.P.; et al. Safety and Efficacy of BIND-014, a Docetaxel Nanoparticle Targeting Prostate-Specific Membrane Antigen for Patients with Metastatic Castration-Resistant Prostate Cancer A Phase 2 Clinical Trial. JAMA Oncol. 2018, 4, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Baochen, Z.; Yue, Z.; Cheng, W.; Yali, W.; Zongxi, S.; Wantong, Z.; Yang, L.; Shouying, D. Network Pharmacology-Based identification of pharmacological mechanism of SQFZ injection in combination with Docetaxel on lung cancer. Sci. Rep. 2019, 9, 4533. [Google Scholar] [CrossRef]
- Sekino, Y.; Teishima, J. Molecular mechanisms of docetaxel resistance in prostate cancer. Cancer Drug Resist. 2020, 3, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Shuai, W.; Wang, G.; Zhang, Y.; Bu, F.; Zhang, S.; Miller, D.D.; Li, W.; Ouyang, L.; Wang, Y. Recent Progress on Tubulin Inhibitors with Dual Targeting Capabilities for Cancer Therapy. J. Med. Chem. 2021, 64, 7963–7990. [Google Scholar] [CrossRef]
- Plana, D.; Palmer, A.C.; Sorger, P.K. Independent Drug Action in Combination Therapy: Implications for Precision Oncology. Cancer Discov. 2022, 12, 606–624. [Google Scholar] [CrossRef]
- Hu, C.J.; Aryal, S.; Zhang, L. Nanoparticle-assisted combination therapies for effective cancer treatment. Ther. Deliv. 2010, 1, 323–334. [Google Scholar] [CrossRef]
- Wang, H.; Huang, Y. Combination therapy based on nano codelivery for overcoming cancer drug resistance. Med. Drug Disc. 2020, 6, 100024. [Google Scholar] [CrossRef]
- Jaaks, P.; Coker, E.A.; Vis, D.J.; Edwards, O.; Carpenter, E.F.; Leto, S.M.; Dwane, L.; Sassi, F.; Lightfoot, H.; Barthorpe, S.; et al. Effective drug combinations in breast, colon and pancreatic cancer cells. Nature 2022, 603, 166–173. [Google Scholar] [CrossRef]
- Ren, D.; Villeneuve, N.F.; Jiang, T.; Wu, T.; Lau, A.; Toppin, H.A.; Zhang, D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. USA 2011, 108, 1433–1438. [Google Scholar] [CrossRef] [PubMed]
- Olayanju, A.; Copple, I.M.; Bryan, H.K.; Edge, G.T.; Sison, R.L. Brustaol provokes a rapid and transient inhibition of Nrf2 signaling and sensitizes mammalian cells to chemical toxicity-implications for therapeutic targeting of Nrf2. Free Radic. Biol. Med. 2015, 78, 202–212. [Google Scholar]
- Hayes, J.D.; McMahon, M. NRF2 and KEAP1 mutations: Permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 2008, 34, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Dai, N.; He, Q.; Guo, P.; Xiang, Y.; Zhang, Q.; Hong, Z.; Zhang, Q. Comprehensive anti-tumor effect of Brusatol through inhibition of cell viability and promotion of apoptosis caused by autophagy via the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Biomed. Pharmacother. 2018, 105, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Tian, Z.; Guo, R.; Ren, F. Nrf2 inhibitor, brusatol in combination with trastuzumab exerts synergistic antitumor activity in HER2-positive cancers by inhibiting Nrf2/HO-1 and HER2-AKT/ERK1/2 pathways. Oxid. Med. Cell. Longev. 2020, 2020, 9867595. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Lai, Z.Q.; Leung, A.W.; Leung, P.S.; Li, Z.S.; Lin, Z.X. Exploring brusatol as a new anti-pancreatic cancer adjuvant: Biological evaluation and mechanistic studies. Oncotarget 2017, 8, 84974. [Google Scholar] [CrossRef]
- Xiang, Y.; Ye, W.; Huang, C.; Yu, D.; Chen, H.; Deng, T.; Zhang, F.; Lou, B.; Zhang, J.; Shi, K.; et al. Brusatol enhances the chemotherapy efficacy of gemcitabine in pancreatic cancer via the Nrf2 signalling pathway. Oxid. Med. Cell. Longev. 2018, 2018, 2360427. [Google Scholar] [CrossRef]
- Adesina, S.K.; Reid, T.E. Nanoparticle formulation of brusatol: A novel therapeutic option for cancers. J. Pharm. Drug Deliv. Res. 2018, 7, 1. [Google Scholar] [CrossRef]
- Bovilla, V.R.; Kuruburu, M.G.; Bettada, V.G.; Krishnamurthy, J.; Sukocheva, O.A.; Thimmulappa, R.K.; Shivananju, N.S.; Balakrishna, J.P.; Madhunapantula, S.V. Targeted inhibition of anti-inflammatory regulator Nrf2 results in breast cancer retardation in vitro and in vivo. Biomedicines 2021, 9, 1119. [Google Scholar] [CrossRef]
- Xing, S.; Nong, F.; Wang, Y.; Huang, D.; Qin, J.; Chen, Y.F.; He, D.H.; Wu, P.E.; Huang, H.; Zhan, R.; et al. Brusatol has therapeutic efficacy in non-small cell lung cancer by targeting Skp1 to inhibit cancer growth and metastasis. Pharmacol. Res. 2022, 176, 106059. [Google Scholar] [CrossRef]
- Yao, Y.; Zhou, Y.; Liu, L.; Xu, Y.; Chen, Q.; Wang, Y.; Wu, S.; Deng, Y.; Zhang, J.; Shao, A. Nanoparticle-based drug delivery in cancer therapy and its role in overcoming drug resistance. Front. Mol. Biosci. 2020, 7, 193. [Google Scholar] [CrossRef] [PubMed]
- Hines, D.J.; Kaplan, D.L. Poly (lactic-co-glycolic) acid– controlled-release systems: Experimental and modeling insights. Crit. Rev. Ther. Drug Carr. Syst. 2013, 30, 257–276. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cheng, D.; Niu, B.; Wang, X.; Wu, X.; Wang, A. Properties of Poly (Lactic-co-Glycolic Acid) and Progress of Poly (Lactic-co-Glycolic Acid)-Based Biodegradable Materials in Biomedical Research. Pharmaceuticals 2023, 16, 454. [Google Scholar] [CrossRef] [PubMed]
- Ejigah, V.; Owoseni, O.; Bataille-Backer, P.; Ogundipe, O.D.; Fisusi, F.A.; Adesina, S.K. Approaches to Improve Macromolecule and Nanoparticle Accumulation in the Tumor Microenvironment by the Enhanced Permeability and Retention Effect. Polymers 2022, 14, 2601. [Google Scholar] [CrossRef] [PubMed]
- Adesina, S.K.; Wight, S.A.; Akala, E.O. Optimization of the fabrication of novel stealth PLA-based nanoparticles by dispersion polymerization using D-optimal mixture design. Drug Dev. Ind. Pharm. 2014, 40, 1547–1556. [Google Scholar] [CrossRef] [PubMed]
- Amin, H.; Osman, S.K.; Mohammed, A.M.; Zayed, G. Gefitinib-loaded starch nanoparticles for battling lung cancer: Optimization by full factorial design and in vitro cytotoxicity evaluation. Saudi Pharm. J. 2023, 31, 29–54. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, M.J.; Loureiro, J.A.; Coelho, M.A.N.; Pereira, M.C. Factorial Design as a Tool for the Optimization of PLGA Nanoparticles for the Co-Delivery of Temozolomide and O6-Benzylguanine. Pharmaceutics 2019, 11, 401. [Google Scholar] [CrossRef]
- Hernández-Giottonini, K.Y.; Rodríguez-Córdova, R.J.; Gutiérrez-Valenzuela, C.A.; Peñuñuri-Miranda, O.; Zavala-Rivera, P.; Guerrero-Germán, P.; Lucero-Acuña, A. PLGA nanoparticle preparations by emulsification and nanoprecipitation techniques: Effects of formulation parameters. RSC Adv. 2020, 10, 4218–4231. [Google Scholar] [CrossRef]
- Berko, Y.A.; Funmilola, A.F.; Akala, E.O. Fabrication of Paclitaxel and 17AAG-loaded Poly-ε-Caprolactone Nanoparticles for Breast Cancer Treatment. J. Pharm. Drug Deliv. Res. 2021, 10, 196. [Google Scholar]
- Almuzaini, N.; Moore, M.; Robert-Guroff, M.; Thomas, M.A. Disruption of NBS1/MRN Complex Formation by E4orf3 Supports NF-κB That Licenses E1B55K-Deleted Adenovirus-Infected Cells to Accumulate DNA> 4n. Microbiol. Spectr. 2022, 10, e01881-21. [Google Scholar] [CrossRef]
- Seredick, B.; Archer, C.; Bradford, J.; Olszowy, M.W. Monitor Caspase 3/7 Activity without Cell Fixation: A Novel Apoptosis Reagent from Molecular Probes. 2013. Available online: https://assets.thermofisher.com/TFS-Assets/BID/posters/seredick-cellevent-cyto-2013.pdf (accessed on 5 September 2023).
- AAT Bioquest, Inc. Quest Graph™ IC50 Calculator. Available online: https://www.aatbio.com/tools/ic50-calculator (accessed on 21 January 2023).
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Baranov, M.V.; Kumar, M.; Sacanna, S.; Thutupalli, S.; Van den Bogaart, G. Modulation of immune responses by particle size and shape. Front. Immunol. 2021, 11, 3854. [Google Scholar] [CrossRef] [PubMed]
- Di, J.; Gao, X.; Du, Y.; Zhang, H.; Gao, J.; Zheng, A. Size, shape, charge and “stealthy” surface: Carrier properties affect the drug circulation time in vivo. Asian J. Pharm. Sci. 2021, 16, 444–458. [Google Scholar] [CrossRef]
- Sindhwani, S.; Syed, A.M.; Ngai, J.; Kingston, B.R.; Maiorino, L.; Rothschild, J.; MacMillan, P.; Zhang, Y.; Rajesh, N.U.; Hoang, T.; et al. The entry of nanoparticles into solid tumours. Nat. Mater. 2020, 19, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Pandit, S.; Dutta, D.; Nie, S. Active transcytosis and new opportunities for cancer nanomedicine. Nat. Mater. 2020, 19, 478–480. [Google Scholar] [CrossRef] [PubMed]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; HasanzadehDavarani, F.; Javanmard, R.; Dokhani, A.; Mozafari, M.R. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef]
- Cheng, J.; Teply, B.A.; Sherifi, I.; Sung, J.; Luther, G.; Gu, F.X.; Levy-Nissenbaum, E.; Radovic-Moreno, A.F.; Langer, R.; Farokhzad, O.C. Formulation of functionalized PLGA–PEG nanoparticles for in vivo targeted drug delivery. Biomaterials 2007, 28, 869–876. [Google Scholar] [CrossRef]
- Sharma, N.; Madan, P.; Lin, S. Effect of process and formulation variables on the preparation of parenteral paclitaxel-loaded biodegradable polymeric nanoparticles: A co-surfactant study. Asian J. Pharm. Sci. 2016, 11, 404–416. [Google Scholar] [CrossRef]
- Soomherun, N.; Kreua-Ongarjnukool, N.; Chumnanvej, S.; Thumsing, S. Encapsulation of Nicardipine Hydrochloride and Release from Biodegradable Poly(D,L-lactic-co-glycolic acid) Microparticles by Double Emulsion Process: Effect of Emulsion Stability and Different Parameters on Drug Entrapment. Int. J. Biomater. 2017, 2017, 1743765. [Google Scholar] [CrossRef]
- Gutema, E.M.; Gopal, M.; Lemu, H. Temperature Optimization by Using Response Surface Methodology and Desirability Analysis of Aluminium 6061. Materials 2022, 15, 5892. [Google Scholar] [CrossRef]
- Makita-Chingombe, F.; Kutscher, H.L.; DiTursi, S.L.; Morse, G.D.; Maponga, C.C. Poly (lactic-co-glycolic) acid-chitosan dual loaded nanoparticles for antiretroviral nanoformulations. J. Drug Deliv. 2016, 2016, 3810175. [Google Scholar] [CrossRef] [PubMed]
- Çalış, S.; Atar, K.Ö.; Arslan, F.B.; Eroğlu, H.; Çapan, Y. Nanopharmaceuticals as Drug-Delivery Systems: For, Against, and Current Applications. In Nanocarriers for Drug Delivery; Elsevier: Amsterdam, The Netherlands, 2019; pp. 133–154. [Google Scholar]
- Sohail, M.F.; Rehman, M.; Sarwar, H.S.; Naveed, S.; Salman, O.; Bukhari, N.I.; Hussain, I.; Webster, T.J.; Shahnaz, G. Advancements in the oral delivery of Docetaxel: Challenges, current state-of-the-art and future trends. Intl. J. Nanomed. 2018, 13, 3145. [Google Scholar] [CrossRef]
- Zhang, J.; Fang, X.; Li, Z.; Chan, H.F.; Lin, Z.; Wang, Y.; Chen, M. Redox-sensitive micelles composed of disulfide-linked Pluronic-linoleic acid for enhanced anticancer efficiency of brusatol. Int. J. Nanomed. 2018, 13, 939. [Google Scholar] [CrossRef] [PubMed]
- Maderuelo, C.; Zarzuelo, A.; Lanao, J.M. Critical factors in the release of drugs from sustained release hydrophilic matrices. J. Control. Release 2011, 154, 2–19. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, W.; Wang, J.; Chen, P.; Jin, J. Effect of docetaxel on the regulation of proliferation and apoptosis of human prostate cancer cells. Mol. Med. Rep. 2019, 19, 3864–3870. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C.; Paul, T. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzym. Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Chou, T.C. The combination index (CI < 1) as the definition of synergism and of synergy claims. Synergy 2018, 7, 49–50. [Google Scholar]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method synergy quantification method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef]
- Cheng, C.; Yuan, F.; Chen, X.P.; Zhang, W.; Zhao, X.L.; Jiang, Z.P.; Zhou, H.H.; Zhou, G.; Cao, S. Inhibition of Nrf2-mediated glucose metabolism by brusatol synergistically sensitizes acute myeloid leukemia to Ara-C. Biomed. Pharm. 2021, 142, 111652. [Google Scholar] [CrossRef]
- Jorge, J.; Magalhães, N.; Alves, R.; Lapa, B.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B. Antitumor Effect of Brusatol in Acute Lymphoblastic Leukemia Models Is Triggered by Reactive Oxygen Species Accumulation. Biomedicines 2022, 10, 2207. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Jing, H.; Cui, L. Highly Efficient Labeling of Human Lung Cancer Cells Using Cationic Poly-l-lysine-Assisted Magnetic Iron Oxide Nanoparticles. Nano-Micro Lett. 2015, 7, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Hwang, N.; Lang, F.; Zhou, L.; Wong, J.H.Y.; Singh, R.K.; Jha, H.C.; El-Deiry, W.S.; Du, Y.; Robertson, E.S. Quassinoidanalogs with enhanced efficacy for treatment of hematologic malignancies target the PI3Kγ isoform. Commun. Biol. 2020, 3, 267. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Chung, J.Y.; Lee, S.G.; Kim, Y.J.; Park, J.E.; Yun, J.; Park, Y.C.; Kim, B.G.; Yoo, Y.H.; Kim, J.M. Interferes with microtubule-stabilizing agent-induced apoptosis in prostate and colorectal cancer cells. Int. J. Mol. Med. 2013, 31, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Srivastava, S.K.; Singh, S.; Arora, S.; Tyagi, N.; Andrews, J.; McClellan, S.; Carter, J.E.; Singh, A.P. CXCL12/CXCR4 signaling counteracts docetaxel-induced microtubule stabilization via p21-activated kinase 4-dependent activation of LIM domain kinase 1. Oncotarget 2014, 5, 11490. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Lin, Z.Q.; Liu, B.; Wei, Y.Q. Caspase-mediated programmed cell death pathways as potential therapeutic targets in cancer. Cell Prolif. 2012, 45, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Kesavardhana, S.; Malireddi, R.S.; Kanneganti, T.D. Caspases in cell death, inflammation, and pyroptosis. Ann. Rev. Immunol. 2020, 38, 567–595. [Google Scholar] [CrossRef] [PubMed]
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef]
- Mhaidat, N.M.; Wang, Y.; Kiejda, K.A.; Zhang, X.D.; Hersey, P. Docetaxel-induced apoptosis in melanoma cells is dependent on activation of caspase-2. Mol. Cancer Ther. 2007, 6, 752–761. [Google Scholar] [CrossRef]
- Mediavilla-Varela, M.; Pacheco, F.J.; Almaguel, F.; Perez, J.; Sahakian, E.; Daniels, T.R.; Leoh, L.S.; Padilla, A.; Wall, N.R.; Lilly, M.B.; et al. Docetaxel-induced prostate cancer cell death involves concomitant activation of caspase and lysosomal pathways and is attenuated by LEDGF/p75. Mol. Cancer 2009, 8, 68. [Google Scholar] [CrossRef]
- Liu, X.; Yang, W.; Guan, Z.; Yu, W.; Fan, B.; Xu, N.; Liao, D.J. There are only four basic modes of cell death, although there are many ad-hoc variants adapted to different situations. Cell Biosci. 2018, 8, 6. [Google Scholar] [CrossRef]
- Tait, S.W.; Ichim, G.; Green, D.R. Die another way–non-apoptotic mechanisms of cell death. J. Cell Sci. 2014, 127, 2135–2144. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Strasser, A.; McDunn, J.E.; Swanson, P.E. Cell death. N. Engl. J. Med. 2009, 361, 1570–1583. [Google Scholar] [CrossRef] [PubMed]
- Garg, H.; Suri, P.; Gupta, J.C.; Talwar, G.P.; Dubey, S. Survivin: A unique target for tumor therapy. Cancer Cell Int. 2016, 16, 49. [Google Scholar] [CrossRef] [PubMed]
- Mita, A.C.; Mita, M.M.; Nawrocki, S.T.; Giles, F.J. Survivin: Key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin. Cancer Res. 2008, 14, 5000–5005. [Google Scholar] [CrossRef] [PubMed]
- Han, T.L.; Sha, H.; Ji, J.; Li, Y.T.; Wu, D.S.; Lin, H.; Hu, B.; Jiang, Z.X. Depletion of Survivin suppresses docetaxel-induced apoptosis in HeLa cells by facilitating mitotic slippage. Sci. Rep. 2021, 11, 2283. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365. [Google Scholar] [PubMed]
- Liang, H.; Zhang, L.; Xu, R.; Ju, X.L. Silencing of survivin using YM155 induces apoptosis and chemosensitization in neuroblastomas cells. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 2909–2915. [Google Scholar]
- Nakahara, T.; Takeuchi, M.; Kinoyama, I.; Minematsu, T.; Shirasuna, K.; Matsuhisa, A.; Kita, A.; Tominaga, F.; Yamanaka, K.; Kudoh, M.; et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007, 67, 8014–8021. [Google Scholar] [CrossRef]
- Altieri, D.C. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene 2003, 22, 8581–8589. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, H.; Li, R.; Guo, Y.; Zheng, R. High expression of survivin predicts poor prognosis in cervical squamous cell carcinoma treated with paclitaxel and carboplatin. Medicine 2019, 98, e15607. [Google Scholar] [CrossRef]
- Jaiswal, P.K.; Goel, A.; Mittal, R.D. Survivin: A molecular biomarker in cancer. Indian J. Med. Res. 2015, 141, 389. [Google Scholar] [PubMed]
- Fenstermaker, R.A.; Figel, S.A.; Qiu, J.; Barone, T.A.; Dharma, S.S.; Winograd, E.K.; Galbo, P.M.; Wiltsie, L.M.; Ciesielski, M.J. Survivin Monoclonal Antibodies Detect Survivin Cell Surface Expression and Inhibit Tumor Growth In Vivo. Clin. Cancer Res. 2018, 24, 2642–2652. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, S.; Ma, T.P.; Lee, J.; Haverty, P.M.; Kirkpatrick, D.S.; Yu, K.; Stokoe, D. Application of Mass Spectrometry Profiling to Establish Brusatol as an Inhibitor of Global Protein Synthesis. Mol. Cell. Proteom. 2016, 15, 1220–1231. [Google Scholar] [CrossRef] [PubMed]
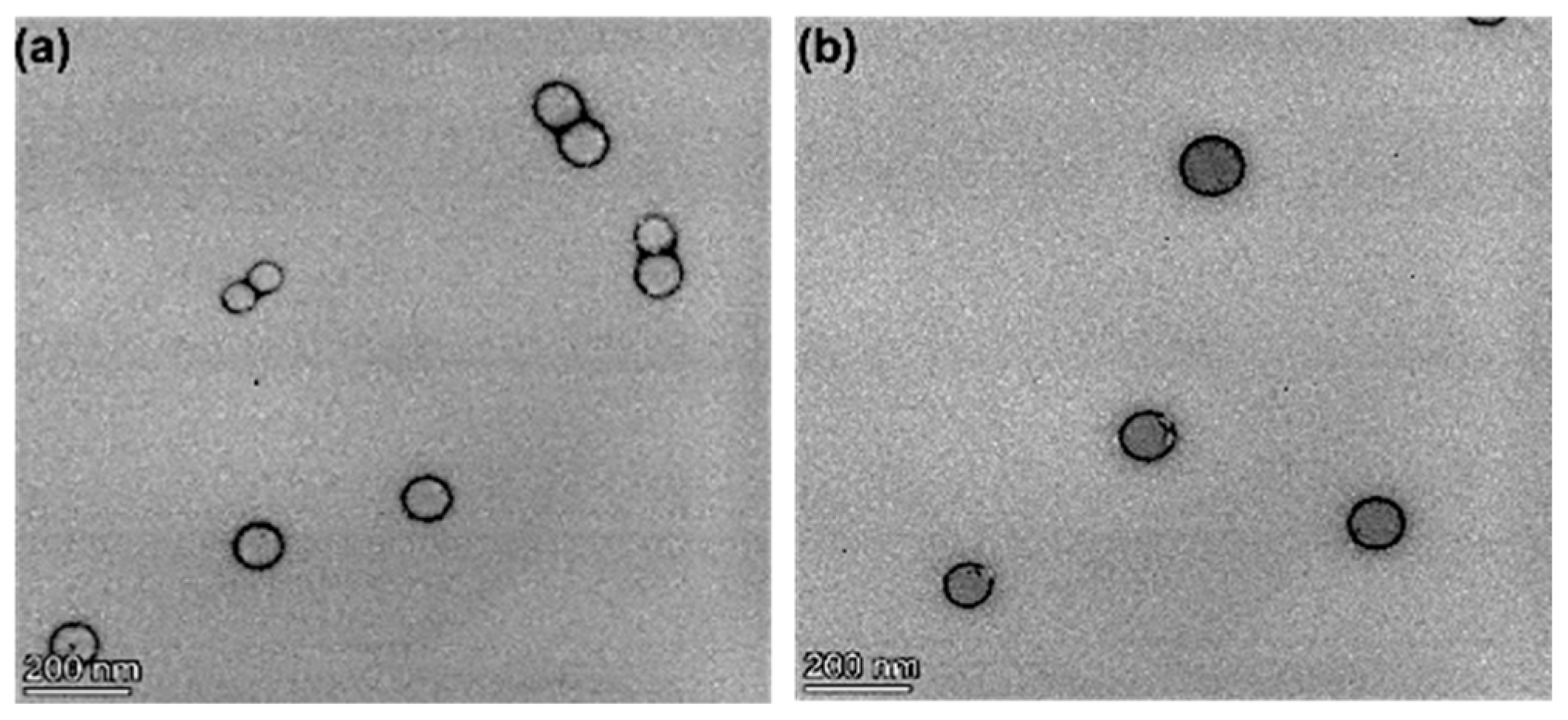
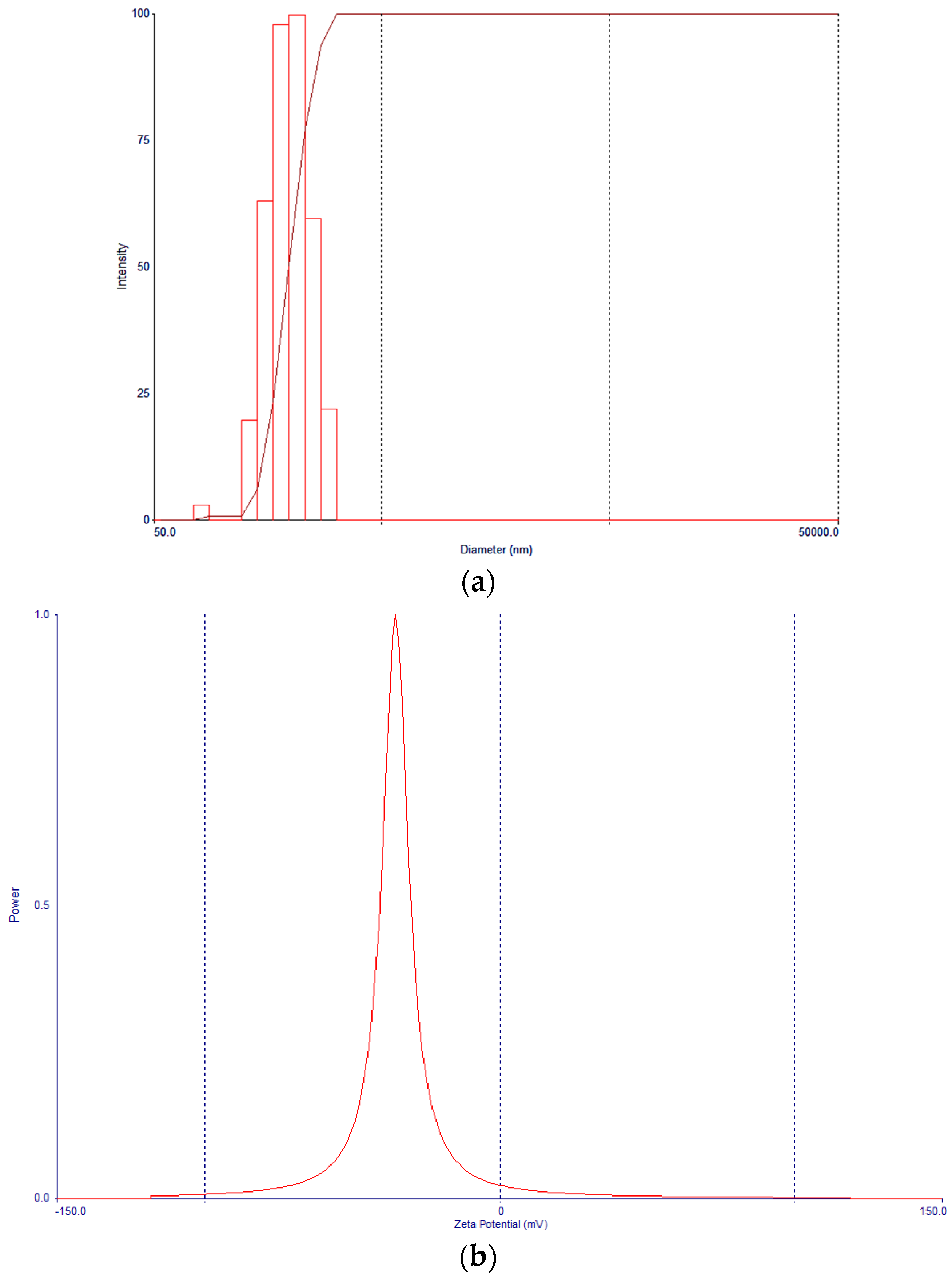

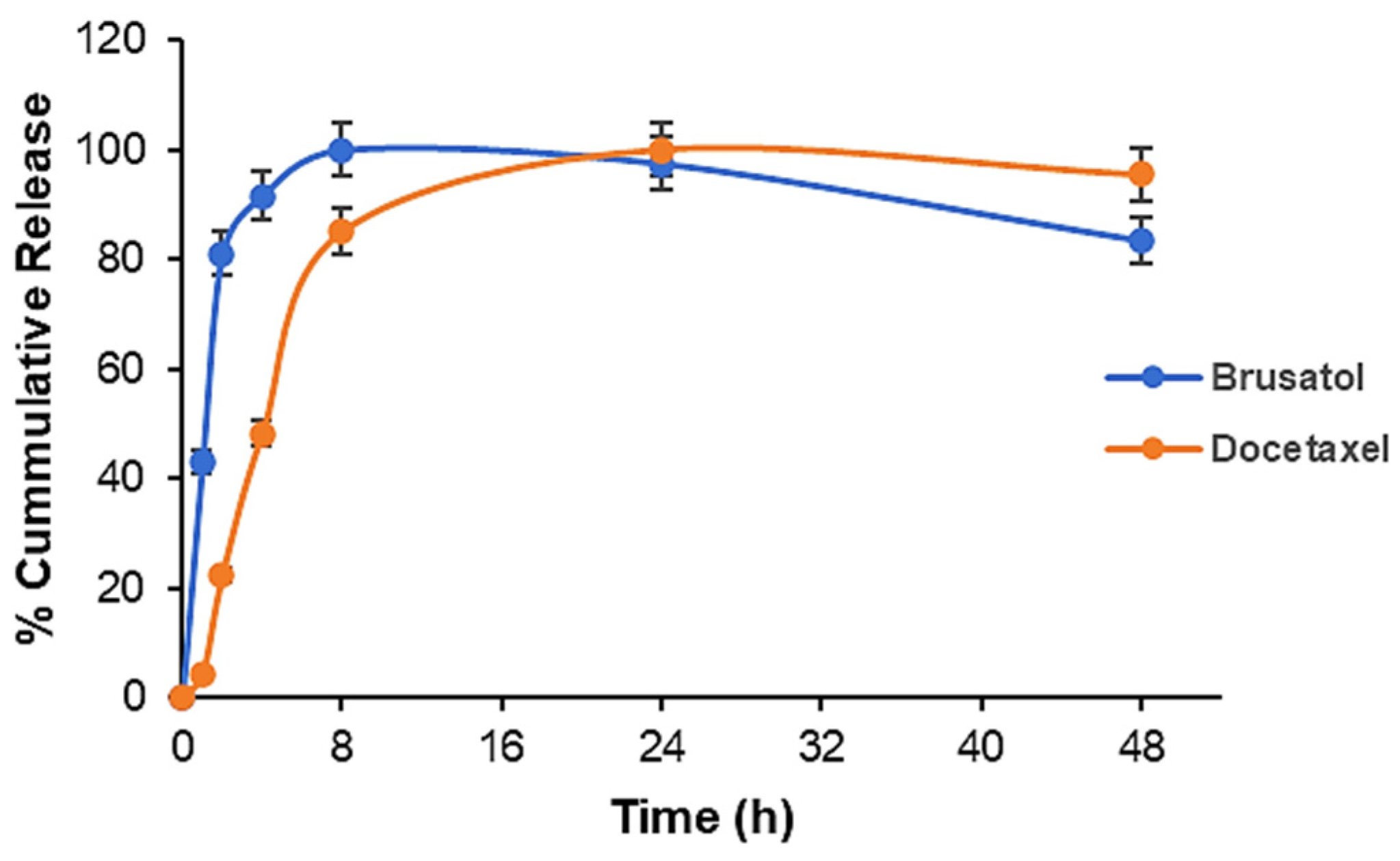
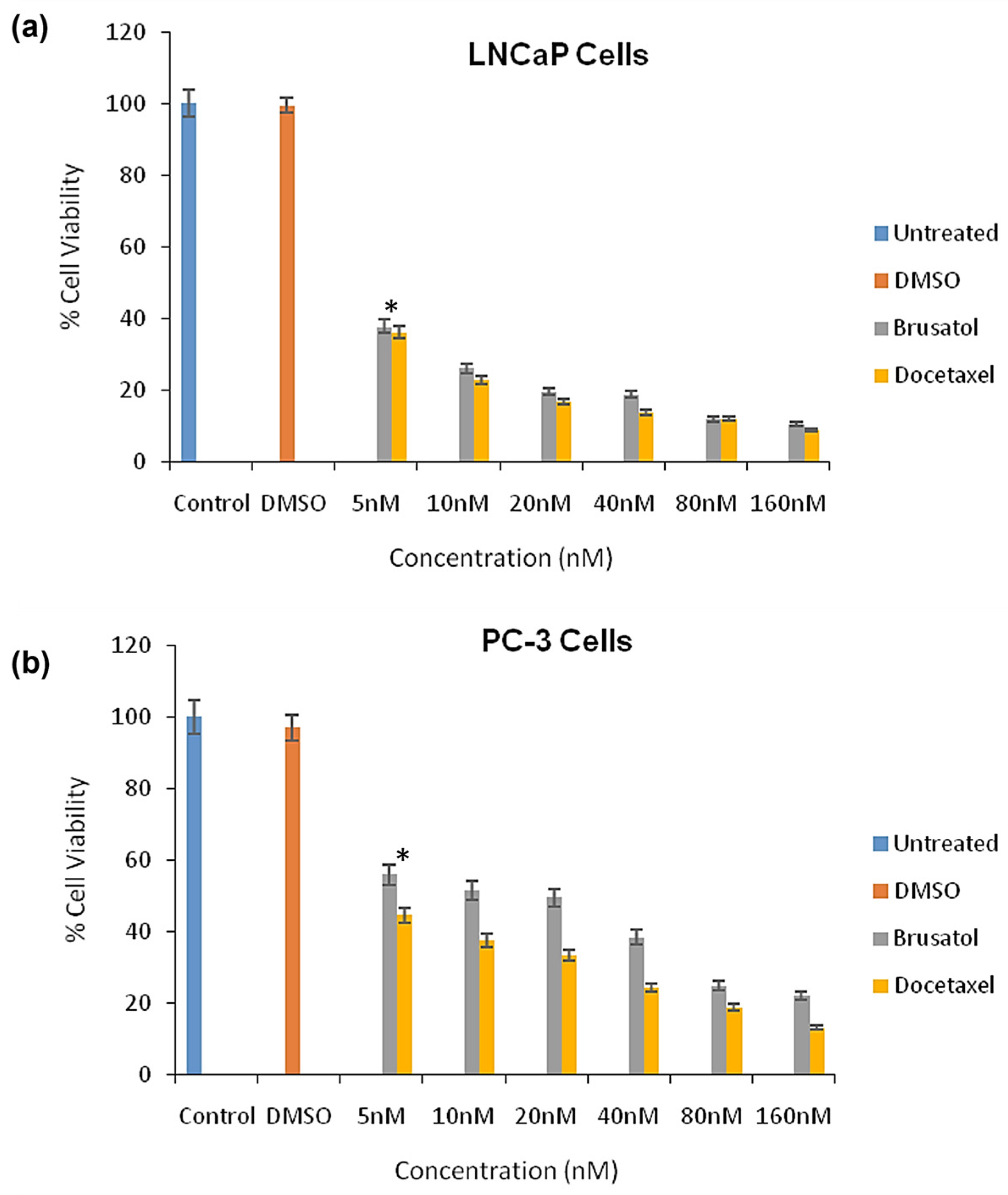




| Formulation | Docetaxel (mg) | Brusatol (mg) | Drug:Polymer (%) |
|---|---|---|---|
| 1 | 7 | 3 | 20% |
| 2 | 6 | 4 | 20% |
| 3 | 5 | 5 | 20% |
| 4 | 4 | 6 | 20% |
| 5 | 2.5 | 7.5 | 20% |
| 6 | 3 | 7 | 20% |
| 7 | 2 | 8 | 20% |
| 8 | 1 | 9 | 20% |
| 9 | 4.5 | 10.5 | 30% |
| 10 | 3 | 12 | 30% |
| Formulation Factor | Variation | Size (nm) | Process Factor | Variation | Size (nm) |
|---|---|---|---|---|---|
| Solvent composition | Ethyl acetate:DMF | 202.8 | Pulse (on/off, sec) | No pulse | 188.7 |
| Ethyl acetate:acetone | 206.9 | 7:3 | 230.8 | ||
| Ethyl acetate:DMSO | 212.8 | 8:2 | 178.1 | ||
| Ethyl acetate:ACN | 207.6 | 9:1 | 175.9 | ||
| Solvent ratio | 1.2:0.8 | 371.6 | Amplitude (%) | 25 | 176.9 |
| 1.4:0.6 | 391 | 30 | 196.1 | ||
| 1.6:0.4 | 179.2 | 35 | 193.3 | ||
| 1.8:0.2 | 200.4 | 40 | 203.8 | ||
| Polymer conc. | 20 mg/2 mL | 207.1 | Sonic. time (min) | 2.5 | 199.3 |
| 50 mg/2 mL | 189.6 | 5 | 188.8 | ||
| 75 mg/2 mL | 185.7 | 7.5 | 186.8 | ||
| 100 mg/2 mL | 194.5 | 10 | 186.1 | ||
| 150 mg/2 mL | 355.1 | ||||
| Organic:Aqueous | 2:8 | 203 | |||
| 2:10 | 193.4 | ||||
| 2:12 | 192.1 | ||||
| 2:14 | 318.5 | ||||
| 2:16 | 399.4 | ||||
| PVA conc. (%) | 0.25 | 186.9 | |||
| 0.5 | 182.1 | ||||
| 1 | 184.5 | ||||
| 2 | 195 | ||||
| 3 | 195 |
| Formulation | Polymer Conc. (mg/2 mL) | Organic:Aqu | PVA Conc. (%) | Amplitude (%) | Size (nm) | Polydispersity |
|---|---|---|---|---|---|---|
| 1 | 25 | 2|8 | 1.5 | 30 | 225.2 | 0.149 |
| 2 | 50 | 2|8 | 0.5 | 30 | 212.5 | 0.048 |
| 3 | 25 | 2|8 | 1.5 | 26 | 230.4 | 0.134 |
| 4 | 50 | 2|8 | 1.5 | 26 | 201.7 | 0.104 |
| 5 | 50 | 2|8 | 1.5 | 30 | 206.7 | 0.049 |
| 6 | 37.5 | 2|10 | 1 | 28 | 195 | 0.081 |
| 7 | 25 | 2|8 | 0.5 | 30 | 220.5 | 0.064 |
| 8 | 50 | 2|8 | 1.5 | 30 | 212.9 | 0.058 |
| 9 | 50 | 2|12 | 0.5 | 30 | 203.3 | 0.067 |
| 10 | 25 | 2|12 | 1.5 | 26 | 186.7 | 0.128 |
| 11 | 25 | 2|8 | 1.5 | 26 | 223.8 | 0.105 |
| 12 | 37.5 | 2|10 | 1 | 28 | 201.8 | 0.088 |
| 13 | 50 | 2|12 | 1.5 | 26 | 185.6 | 0.032 |
| 14 | 37.5 | 2|10 | 1 | 28 | 190.6 | 0.116 |
| 15 | 50 | 2|8 | 0.5 | 26 | 197.8 | 0.089 |
| 16 | 25 | 2|12 | 1.5 | 30 | 177.3 | 0.102 |
| 17 | 50 | 2|12 | 0.5 | 30 | 181.6 | 0.059 |
| 18 | 25 | 2|8 | 1.5 | 30 | 210.5 | 0.098 |
| 19 | 25 | 2|12 | 1.5 | 26 | 158.4 | 0.08 |
| 20 | 25 | 2|12 | 0.5 | 30 | 173.3 | 0.114 |
| 21 | 37.5 | 2|10 | 1 | 28 | 184.9 | 0.046 |
| 22 | 50 | 2|12 | 1.5 | 30 | 181 | 0.085 |
| 23 | 25 | 2|12 | 0.5 | 26 | 173.7 | 0.042 |
| 24 | 25 | 2|8 | 0.5 | 26 | 203.3 | 0.079 |
| 25 | 50 | 2|8 | 0.5 | 30 | 192.3 | 0.089 |
| 26 | 25 | 2|12 | 1.5 | 30 | 170.4 | 0.093 |
| 27 | 37.5 | 2|10 | 1 | 28 | 175.1 | 0.147 |
| 28 | 25 | 2|8 | 0.5 | 30 | 186 | 0.075 |
| 29 | 25 | 2|12 | 0.5 | 30 | 167.7 | 0.119 |
| 30 | 25 | 2|8 | 0.5 | 26 | 181.9 | 0.079 |
| 31 | 50 | 2|12 | 0.5 | 26 | 173.1 | 0.069 |
| 32 | 50 | 2|12 | 1.5 | 26 | 172.7 | 0.077 |
| 33 | 50 | 2|8 | 1.5 | 26 | 184.1 | 0.106 |
| 34 | 50 | 2|12 | 1.5 | 30 | 176 | 0.061 |
| 35 | 25 | 2|12 | 0.5 | 26 | 171.7 | 0.105 |
| 36 | 50 | 2|8 | 0.5 | 26 | 183.8 | 0.138 |
| 37 | 50 | 2|12 | 0.5 | 26 | 159.6 | 0.071 |
| Analysis of Variance | |||||
|---|---|---|---|---|---|
| Source | DF | Adj SS | Adj MS | F-Value | p-Value |
| Model | 6 | 9113.7 | 1518.94 | 10.91 | 0.000 |
| Linear | 4 | 7926.1 | 1981.54 | 14.23 | 0.000 |
| Pol. Conc. (mg/2 mL) | 1 | 62.7 | 62.72 | 0.45 | 0.507 |
| Org:Aqueous | 1 | 6903.1 | 6903.12 | 49.59 | 0.000 |
| PVA Conc. (%) | 1 | 528.1 | 528.12 | 3.79 | 0.061 |
| Amp (%) | 1 | 432.2 | 432.18 | 3.10 | 0.088 |
| 2-Way Interactions | 2 | 1187.5 | 593.75 | 4.27 | 0.023 |
| Pol. Conc. (mg/2 mL) * Org:Aqueous | 1 | 723.9 | 723.90 | 5.20 | 0.030 |
| Org:Aqueous * PVA Conc. (%) | 1 | 463.6 | 463.60 | 3.33 | 0.078 |
| Error | 30 | 4176.4 | 139.21 | ||
| Curvature | 1 | 4.4 | 4.36 | 0.03 | 0.863 |
| Lack-of-Fit | 9 | 1301.2 | 144.58 | 1.01 | 0.466 |
| Pure Error | 20 | 2870.8 | 143.54 | ||
| Total | 36 | 13,290.1 | |||
| Solution | Pol. Conc. (mg/2 mL) | Org:Aqu | PVA Conc. (%) | Amp (%) | Predicted Particle Size (nm) | Composite Desirability | Experimental Mean Particle Size (nm) |
|---|---|---|---|---|---|---|---|
| 1 | 25 | 0.167 | 0.5 | 26 | 168.333 | 0.868436 | 169.09 |
| 2 | 50 | 0.1670 | 0.5 | 26 | 175.046 | 0.77953 | 175.15 |
| Formulation | Doc (mg) | Bru (mg) | Size (nm) | Doc Loading (%) | Bru Loading (%) | Drug:Polymer (%) |
|---|---|---|---|---|---|---|
| 1 | 7 | 3 | 201.87 ± 1.40 | 7.84 ± 0.17 | 0.87 ± 0.17 | 20% |
| 2 | 6 | 4 | 195.70 ± 1.15 | 6.37 ± 0.11 | 0.79 ± 0.07 | 20% |
| 3 | 5 | 5 | 198.27 ± 2.78 | 6.32 ± 0.11 | 1.14 ± 0.03 | 20% |
| 4 | 4 | 6 | 182.57 ± 1.90 | 3.73 ± 0.25 | 0.82 ± 0.09 | 20% |
| 5 | 3 | 7 | 194.97 ± 0.75 | 2.49 ± 0.13 | 0.79 ± 0.08 | 20% |
| 6 | 2.5 | 7.5 | 184.80 ± 1.30 | 2.04 ± 0.04 | 0.89 ± 0.02 | 20% |
| 7 | 2 | 8 | 188.80 ± 0.80 | 1.41 ± 0.17 | 0.99 ± 0.15 | 20% |
| 8 | 1 | 9 | 193.83 ± 1.59 | 0.70 ± 0.06 | 1.48 ± 0.28 | 20% |
| 9 | 4.5 | 10.5 | 185.07 ± 3.55 | 3.33 ± 0.96 | 0.89 ± 0.19 | 30% |
| 10 | 3 | 12 | 183.03 ± 5.66 | 1.88 ± 0.57 | 0.92 ± 0.30 | 30% |
| Total Dose (nM) | CI in LNCaP Cells | CI in PC-3 Cells |
|---|---|---|
| 5.0 | 0.33061 | 0.10535 |
| 10.0 | 0.59098 | 0.18948 |
| 20.0 | 0.64905 | 0.33766 |
| 40.0 | 0.76064 | 0.29589 |
| 80.0 | 0.79381 | 0.55168 |
| 160.0 | 0.95830 | 0.99662 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adekiya, T.A.; Moore, M.; Thomas, M.; Lake, G.; Hudson, T.; Adesina, S.K. Preparation, Optimization, and In-Vitro Evaluation of Brusatol- and Docetaxel-Loaded Nanoparticles for the Treatment of Prostate Cancer. Pharmaceutics 2024, 16, 114. https://doi.org/10.3390/pharmaceutics16010114
Adekiya TA, Moore M, Thomas M, Lake G, Hudson T, Adesina SK. Preparation, Optimization, and In-Vitro Evaluation of Brusatol- and Docetaxel-Loaded Nanoparticles for the Treatment of Prostate Cancer. Pharmaceutics. 2024; 16(1):114. https://doi.org/10.3390/pharmaceutics16010114
Chicago/Turabian StyleAdekiya, Tayo Alex, Madison Moore, Michael Thomas, Gabriel Lake, Tamaro Hudson, and Simeon K. Adesina. 2024. "Preparation, Optimization, and In-Vitro Evaluation of Brusatol- and Docetaxel-Loaded Nanoparticles for the Treatment of Prostate Cancer" Pharmaceutics 16, no. 1: 114. https://doi.org/10.3390/pharmaceutics16010114
APA StyleAdekiya, T. A., Moore, M., Thomas, M., Lake, G., Hudson, T., & Adesina, S. K. (2024). Preparation, Optimization, and In-Vitro Evaluation of Brusatol- and Docetaxel-Loaded Nanoparticles for the Treatment of Prostate Cancer. Pharmaceutics, 16(1), 114. https://doi.org/10.3390/pharmaceutics16010114










