Novel Human/Non-Human Primate Cross-Reactive Anti-Transferrin Receptor Nanobodies for Brain Delivery of Biologics
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Immunization and Nanobody Library Preparation
2.3. Cell Line Generation
2.4. Nanobody Selection, Expression and Purification
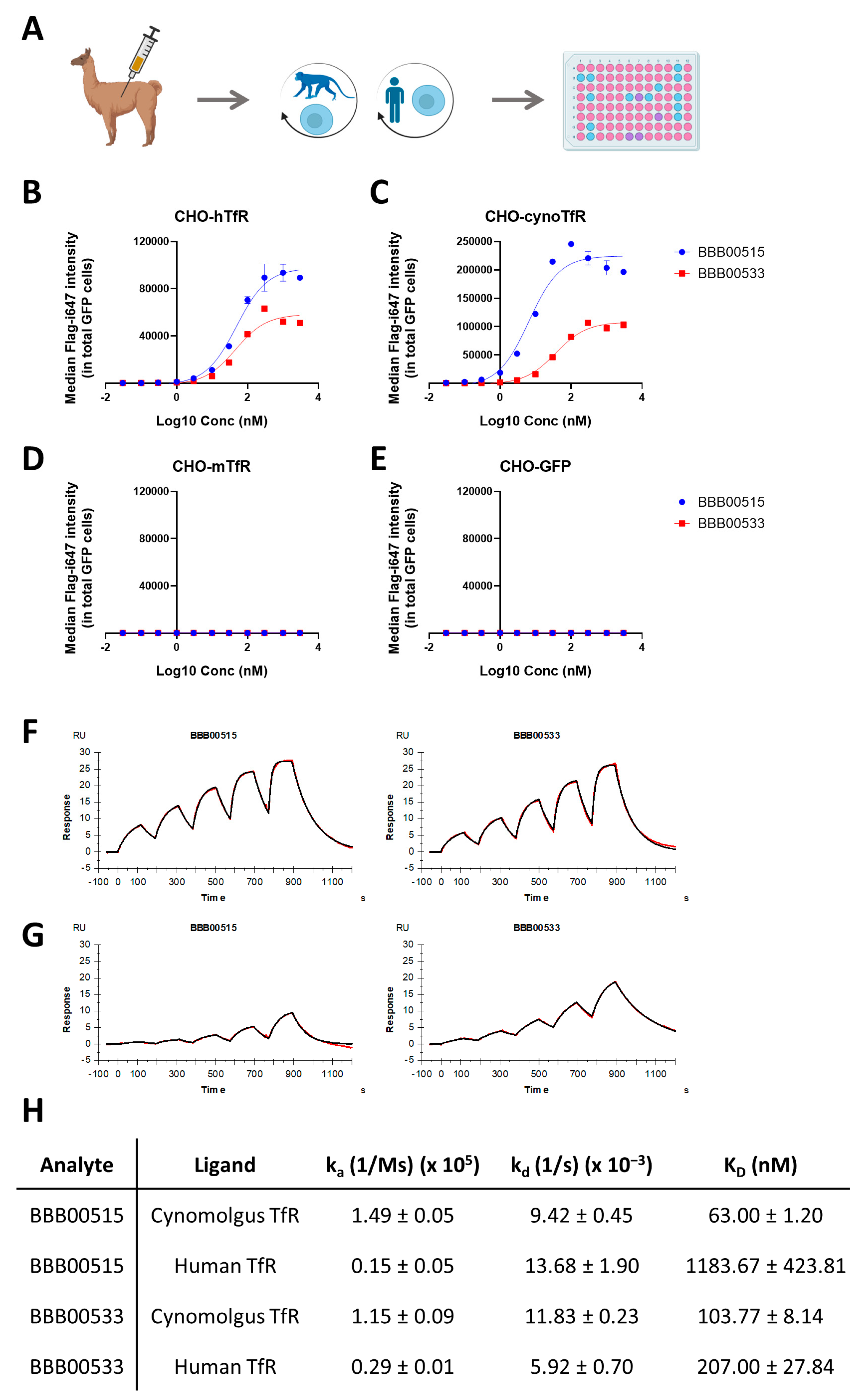
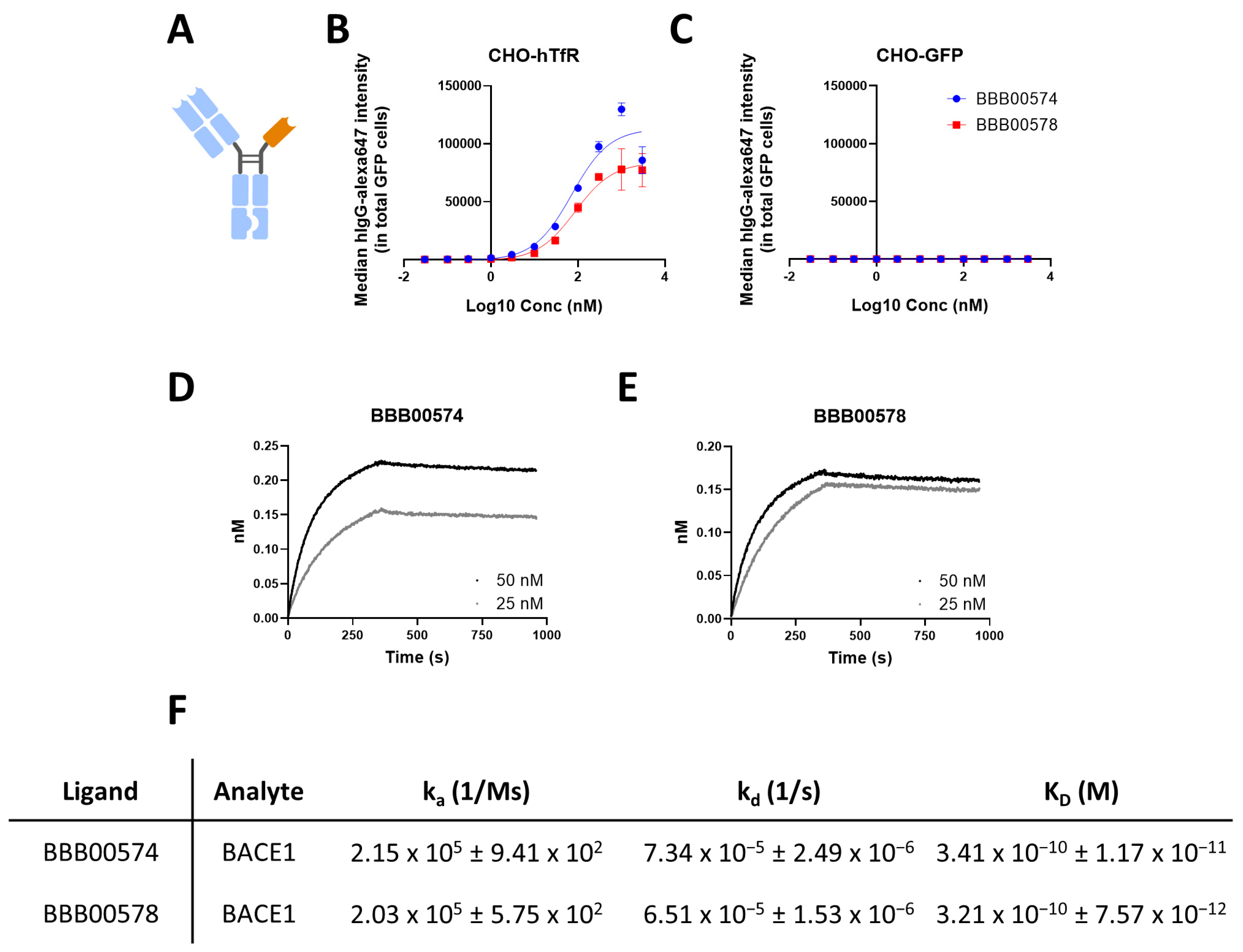
2.5. Flow Cytometry-Based Binder Screening and Validation
2.6. Surface Plasmon Resonance
2.7. Bio-Layer Interferometry (BLI)
2.8. Bispecific Antibodies Engineering and Expression
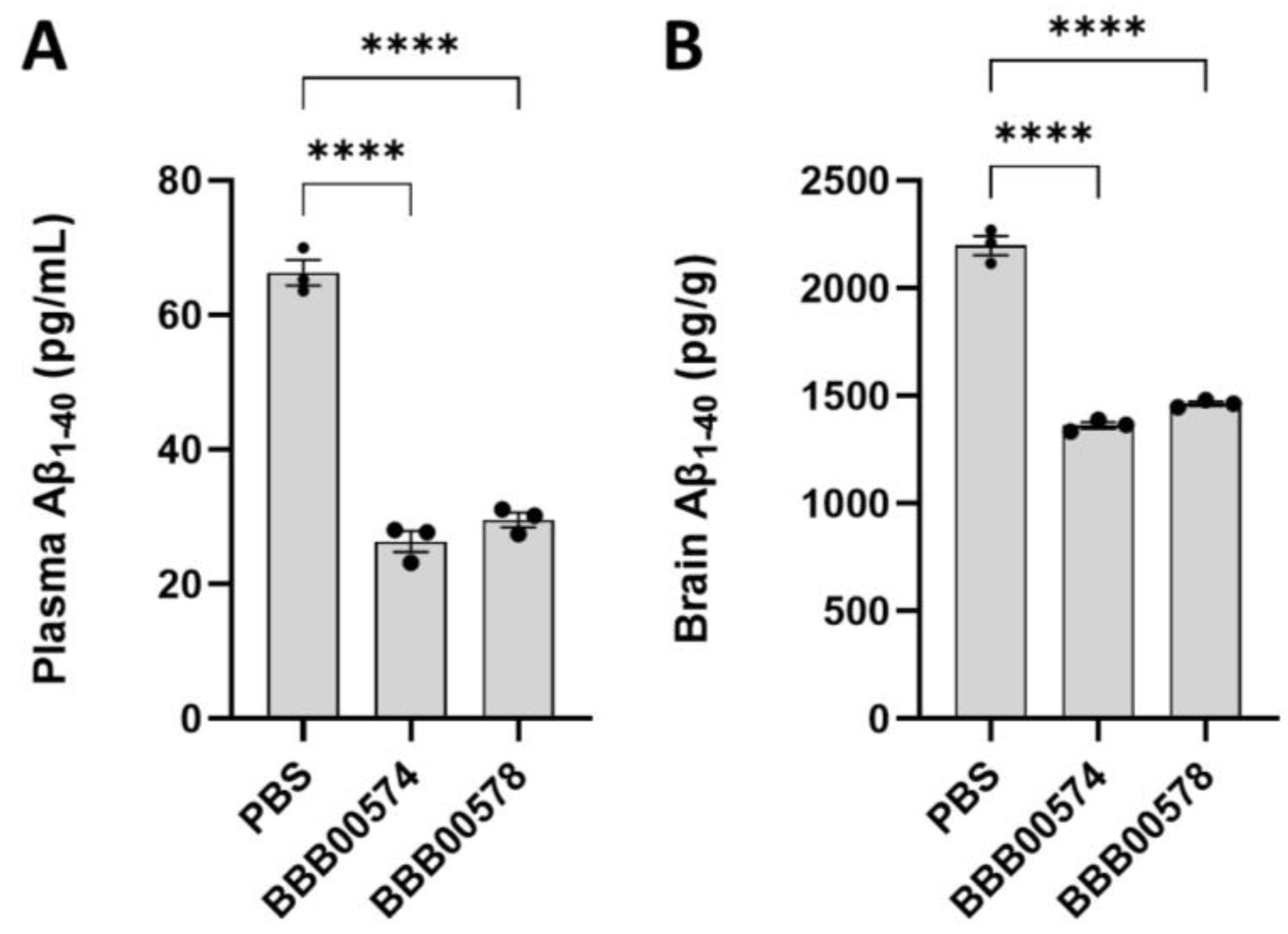
2.9. Sample Collection, Aβ Extraction and Enzyme-Linked Immunosorbent Assay (ELISA)
2.10. Statistical Analysis
3. Results
3.1. Identification of Human/Cynomolgus TfR Binders
3.2. Anti-Human/Cynomolgus TfR Nanobodies Shuttle Anti-BACE1 mAb into the Brain
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daneman, R.; Prat, A. The Blood–Brain Barrier. Cold Spring Harb. Perspect. Biol. 2015, 7, a020412. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J.; Patabendige, A.A.K.; Dolman, D.E.M.; Yusof, S.R.; Begley, D.J. Structure and Function of the Blood–Brain Barrier. Neurobiol. Dis. 2010, 37, 13–25. [Google Scholar] [CrossRef]
- Freskgård, P.-O.; Urich, E. Antibody Therapies in CNS Diseases. Neuropharmacology 2017, 120, 38–55. [Google Scholar] [CrossRef] [PubMed]
- St-Amour, I.; Paré, I.; Alata, W.; Coulombe, K.; Ringuette-Goulet, C.; Drouin-Ouellet, J.; Vandal, M.; Soulet, D.; Bazin, R.; Calon, F. Brain Bioavailability of Human Intravenous Immunoglobulin and Its Transport through the Murine Blood–Brain Barrier. J. Cereb Blood Flow Metab. 2013, 33, 1983–1992. [Google Scholar] [CrossRef]
- Poduslo, J.F.; Curran, G.L.; Berg, C.T. Macromolecular Permeability across the Blood-Nerve and Blood-Brain Barriers. Proc. Natl. Acad. Sci. USA 1994, 91, 5705–5709. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J.; Rönnbäck, L.; Hansson, E. Astrocyte–Endothelial Interactions at the Blood–Brain Barrier. Nat. Rev. Neurosci. 2006, 7, 41–53. [Google Scholar] [CrossRef]
- Pardridge, W. Targeted Delivery of Protein and Gene Medicines through the Blood-Brain Barrier. Clin. Pharmacol. Ther. 2015, 97, 347–361. [Google Scholar] [CrossRef]
- Pardridge, W.M. Receptor-Mediated Peptide Transport through the Blood-Brain Barrier. Endocr. Rev. 1986, 7, 314–330. [Google Scholar] [CrossRef]
- Sehlin, D.; Stocki, P.; Gustavsson, T.; Hultqvist, G.; Walsh, F.S.; Rutkowski, J.L.; Syvänen, S. Brain Delivery of Biologics Using a Cross-Species Reactive Transferrin Receptor 1 VNAR Shuttle. FASEB J. 2020, 34, 13272–13283. [Google Scholar] [CrossRef]
- Stocki, P.; Szary, J.; Rasmussen, C.L.M.; Demydchuk, M.; Northall, L.; Logan, D.B.; Gauhar, A.; Thei, L.; Moos, T.; Walsh, F.S.; et al. Blood-Brain Barrier Transport Using a High Affinity, Brain-Selective VNAR Antibody Targeting Transferrin Receptor 1. FASEB J. 2021, 35, e21172. [Google Scholar] [CrossRef]
- Su, S.; Esparza, T.J.; Brody, D.L. Selection of Single Domain Anti-Transferrin Receptor Antibodies for Blood-Brain Barrier Transcytosis Using a Neurotensin Based Assay and Histological Assessment of Target Engagement in a Mouse Model of Alzheimer’s Related Amyloid-Beta Pathology. PLoS ONE 2022, 17, e0276107. [Google Scholar] [CrossRef] [PubMed]
- Kariolis, M.S.; Wells, R.C.; Getz, J.A.; Kwan, W.; Mahon, C.S.; Tong, R.; Kim, D.J.; Srivastava, A.; Bedard, C.; Henne, K.R.; et al. Brain Delivery of Therapeutic Proteins Using an Fc Fragment Blood-Brain Barrier Transport Vehicle in Mice and Monkeys. Sci Transl. Med. 2020, 12, eaay1359. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.J.; Atwal, J.K.; Zhang, Y.; Tong, R.K.; Wildsmith, K.R.; Tan, C.; Bien-Ly, N.; Hersom, M.; Maloney, J.A.; Meilandt, W.J.; et al. Therapeutic Bispecific Antibodies Cross the Blood-Brain Barrier in Nonhuman Primates. Sci. Transl. Med. 2014, 6, 261ra154. [Google Scholar] [CrossRef] [PubMed]
- Ullman, J.C.; Arguello, A.; Getz, J.A.; Bhalla, A.; Mahon, C.S.; Wang, J.; Giese, T.; Bedard, C.; Kim, D.J.; Blumenfeld, J.R.; et al. Brain Delivery and Activity of a Lysosomal Enzyme Using a Blood-Brain Barrier Transport Vehicle in Mice. Sci. Transl. Med. 2020, 12, eaay1163. [Google Scholar] [CrossRef]
- Wouters, Y.; Jaspers, T.; De Strooper, B.; Dewilde, M. Identification and in Vivo Characterization of a Brain-Penetrating Nanobody. Fluids Barriers CNS 2020, 17, 62. [Google Scholar] [CrossRef]
- Wouters, Y.; Jaspers, T.; Rué, L.; Serneels, L.; De Strooper, B.; Dewilde, M. VHHs as Tools for Therapeutic Protein Delivery to the Central Nervous System. Fluids Barriers CNS 2022, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Niewoehner, J.; Bohrmann, B.; Collin, L.; Urich, E.; Sade, H.; Maier, P.; Rueger, P.; Stracke, J.O.; Lau, W.; Tissot, A.C.; et al. Increased Brain Penetration and Potency of a Therapeutic Antibody Using a Monovalent Molecular Shuttle. Neuron 2014, 81, 49–60. [Google Scholar] [CrossRef]
- Edavettal, S.; Cejudo-Martin, P.; Dasgupta, B.; Yang, D.; Buschman, M.D.; Domingo, D.; Van Kolen, K.; Jaiprasat, P.; Gordon, R.; Schutsky, K.; et al. Enhanced Delivery of Antibodies across the Blood-Brain Barrier via TEMs with Inherent Receptor-Mediated Phagocytosis. Med 2022, 3, 860–882.e15. [Google Scholar] [CrossRef]
- Giugliani, R.; Martins, A.M.; So, S.; Yamamoto, T.; Yamaoka, M.; Ikeda, T.; Tanizawa, K.; Sonoda, H.; Schmidt, M.; Sato, Y. Iduronate-2-Sulfatase Fused with Anti-HTfR Antibody, Pabinafusp Alfa, for MPS-II: A Phase 2 Trial in Brazil. Mol. Ther. 2021, 29, 2378–2386. [Google Scholar] [CrossRef]
- Muyldermans, S. Applications of Nanobodies. Annu. Rev. Anim. Biosci. 2021, 9, 401–421. [Google Scholar] [CrossRef]
- Yogi, A.; Hussack, G.; van Faassen, H.; Haqqani, A.S.; Delaney, C.E.; Brunette, E.; Sandhu, J.K.; Hewitt, M.; Sulea, T.; Kemmerich, K.; et al. Brain Delivery of IGF1R5, a Single-Domain Antibody Targeting Insulin-like Growth Factor-1 Receptor. Pharmaceutics 2022, 14, 1452. [Google Scholar] [CrossRef]
- Farrington, G.K.; Caram-Salas, N.; Haqqani, A.S.; Brunette, E.; Eldredge, J.; Pepinsky, B.; Antognetti, G.; Baumann, E.; Ding, W.; Garber, E.; et al. A Novel Platform for Engineering Blood-brain Barrier-crossing Bispecific Biologics. FASEB J. 2014, 28, 4764–4778. [Google Scholar] [CrossRef] [PubMed]
- Marque, T.; Leach, M.W. Nonclinical Toxicology Testing Strategies and Applicable International Regulatory Guidelines for Using Nonhuman Primates in the Development of Biotherapeutics. In The Nonhuman Primate in Nonclinical Drug Development and Safety Assessment; Elsevier: Amsterdam, The Netherlands, 2015; pp. 315–336. ISBN 978-0-12-417144-2. [Google Scholar]
- Pardon, E.; Laeremans, T.; Triest, S.; Rasmussen, S.G.F.; Wohlkönig, A.; Ruf, A.; Muyldermans, S.; Hol, W.G.J.; Kobilka, B.K.; Steyaert, J. A General Protocol for the Generation of Nanobodies for Structural Biology. Nat. Protoc. 2014, 9, 674–693. [Google Scholar] [CrossRef] [PubMed]
- Nesspor, T.C.; Kinealy, K.; Mazzanti, N.; Diem, M.D.; Boye, K.; Hoffman, H.; Springer, C.; Sprenkle, J.; Powers, G.; Jiang, H.; et al. High-Throughput Generation of Bipod (Fab × ScFv) Bispecific Antibodies Exploits Differential Chain Expression and Affinity Capture. Sci. Rep. 2020, 10, 7557. [Google Scholar] [CrossRef] [PubMed]
- Serneels, L.; T’Syen, D.; Perez-Benito, L.; Theys, T.; Holt, M.G.; De Strooper, B. Modeling the β-Secretase Cleavage Site and Humanizing Amyloid-Beta Precursor Protein in Rat and Mouse to Study Alzheimer’s Disease. Mol. Neurodegener 2020, 15, 60. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Anderson, J.P.; Barbour, R.; Basi, G.S.; Caccavello, R.; Davis, D.; Doan, M.; Dovey, H.F.; Frigon, N.; Hong, J.; et al. Purification and Cloning of Amyloid Precursor Protein β-Secretase from Human Brain. Nature 1999, 402, 537–540. [Google Scholar] [CrossRef]
- Zhou, L.; Chávez-Gutiérrez, L.; Bockstael, K.; Sannerud, R.; Annaert, W.; May, P.C.; Karran, E.; De Strooper, B. Inhibition of Beta-Secretase in Vivo via Antibody Binding to Unique Loops (D and F) of BACE1. J. Biol. Chem. 2011, 286, 8677–8687. [Google Scholar] [CrossRef]
- Sonoda, H.; Morimoto, H.; Yoden, E.; Koshimura, Y.; Kinoshita, M.; Golovina, G.; Takagi, H.; Yamamoto, R.; Minami, K.; Mizoguchi, A.; et al. A Blood-Brain-Barrier-Penetrating Anti-Human Transferrin Receptor Antibody Fusion Protein for Neuronopathic Mucopolysaccharidosis II. Mol. Ther. 2018, 26, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Atwal, J.K.; Chen, Y.; Chiu, C.; Mortensen, D.L.; Meilandt, W.J.; Liu, Y.; Heise, C.E.; Hoyte, K.; Luk, W.; Lu, Y.; et al. A Therapeutic Antibody Targeting BACE1 Inhibits Amyloid-β Production in Vivo. Sci. Transl. Med. 2011, 3, 84ra43. [Google Scholar] [CrossRef]
- Zhang, Y.; Lee, D.H.S. Sink Hypothesis and Therapeutic Strategies for Attenuating Aβ Levels. Neuroscientist 2011, 17, 163–173. [Google Scholar] [CrossRef]
- Georgievska, B.; Gustavsson, S.; Lundkvist, J.; Neelissen, J.; Eketjäll, S.; Ramberg, V.; Bueters, T.; Agerman, K.; Juréus, A.; Svensson, S.; et al. Revisiting the Peripheral Sink Hypothesis: Inhibiting BACE1 Activity in the Periphery Does Not Alter β-Amyloid Levels in the CNS. J. Neurochem. 2015, 132, 477–486. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rué, L.; Jaspers, T.; Degors, I.M.S.; Noppen, S.; Schols, D.; De Strooper, B.; Dewilde, M. Novel Human/Non-Human Primate Cross-Reactive Anti-Transferrin Receptor Nanobodies for Brain Delivery of Biologics. Pharmaceutics 2023, 15, 1748. https://doi.org/10.3390/pharmaceutics15061748
Rué L, Jaspers T, Degors IMS, Noppen S, Schols D, De Strooper B, Dewilde M. Novel Human/Non-Human Primate Cross-Reactive Anti-Transferrin Receptor Nanobodies for Brain Delivery of Biologics. Pharmaceutics. 2023; 15(6):1748. https://doi.org/10.3390/pharmaceutics15061748
Chicago/Turabian StyleRué, Laura, Tom Jaspers, Isabelle M. S. Degors, Sam Noppen, Dominique Schols, Bart De Strooper, and Maarten Dewilde. 2023. "Novel Human/Non-Human Primate Cross-Reactive Anti-Transferrin Receptor Nanobodies for Brain Delivery of Biologics" Pharmaceutics 15, no. 6: 1748. https://doi.org/10.3390/pharmaceutics15061748
APA StyleRué, L., Jaspers, T., Degors, I. M. S., Noppen, S., Schols, D., De Strooper, B., & Dewilde, M. (2023). Novel Human/Non-Human Primate Cross-Reactive Anti-Transferrin Receptor Nanobodies for Brain Delivery of Biologics. Pharmaceutics, 15(6), 1748. https://doi.org/10.3390/pharmaceutics15061748






