Gut Microbiota-Mediated Pharmacokinetic Drug–Drug Interactions between Mycophenolic Acid and Trimethoprim-Sulfamethoxazole in Humans
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Drug Administration and Sample Collection
2.3. Determination of MPA and MPAG
2.4. Pharmacokinetic Analysis
2.5. Bacterial DNA Extraction, PCR Amplification, and Sequencing
2.6. Gut Microbiome Analysis
3. Results
3.1. Demographics of Study Participants
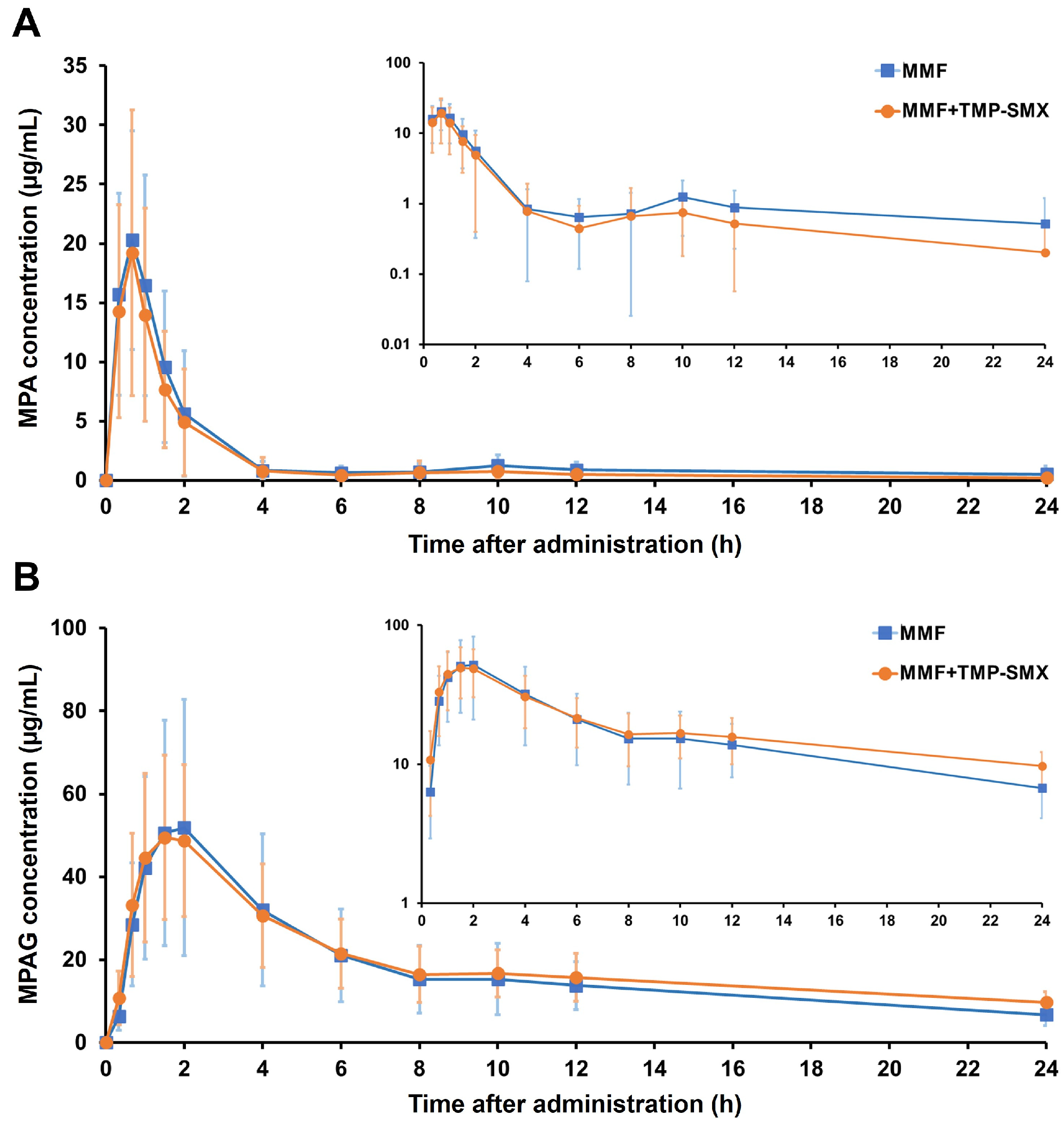
3.2. Effect of TMP-SMX on the Pharmacokinetic Parameters of MPA and MPAG
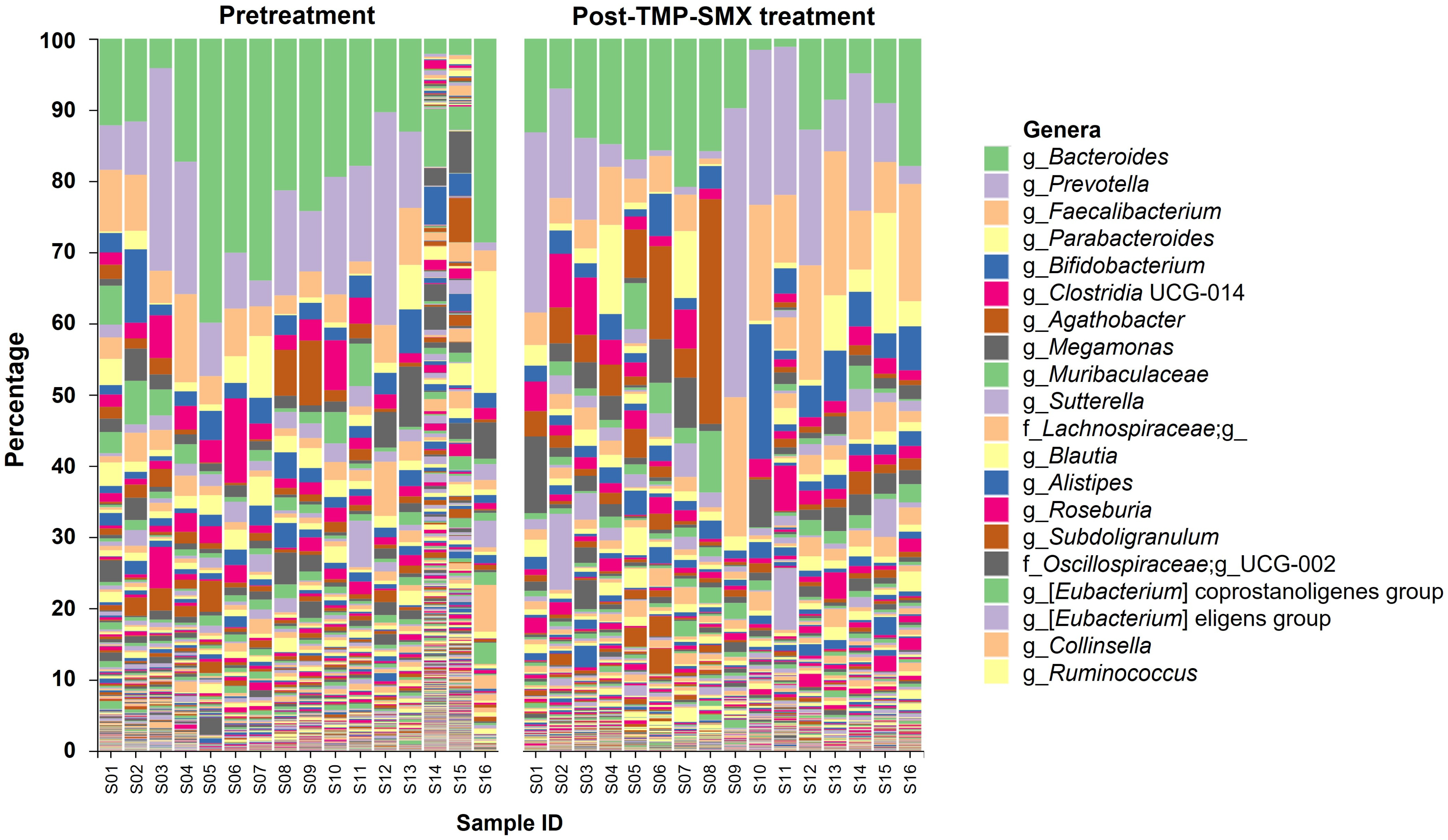
3.3. Gut Microbiota Composition before and after TMP-SMX Treatment
3.4. Co-Occurrence Network of the Highly Abundant Genera in Gut Microbiota
3.5. Correlation between Gut Microbiota and the Pharmacokinetic Parameters of MPA and MPAG
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Enderby, C.; Keller, C.A. An overview of immunosuppression in solid organ transplantation. Am. J. Manag. Care 2015, 21, s12–s23. [Google Scholar] [PubMed]
- Ritter, M.L.; Pirofski, L. Mycophenolate mofetil: Effects on cellular immune subsets, infectious complications, and antimicrobial activity. Transpl. Infect. Dis. 2009, 11, 290–297. [Google Scholar] [CrossRef]
- Mok, C.C.; Tse, S.M.; Chan, K.L.; Ho, L.Y. Effect of immunosuppressive therapies on survival of systemic lupus erythematosus: A propensity score analysis of a longitudinal cohort. Lupus 2018, 27, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Davulcu, E.A.; Vural, F. Immunosuppressive agents in hematopoietic stem cell transplantation. Trends Transplant. 2017, 10, 1–3. [Google Scholar] [CrossRef]
- Holt, C.D. Overview of immunosuppressive therapy in solid organ transplantation. Anesthesiol. Clin. 2017, 35, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Staatz, C.E.; Tett, S.E. Clinical Pharmacokinetics and Pharmacodynamics of Mycophenolate in Solid Organ Transplant Recipients. Clin. Pharmacokinet. 2007, 46, 13–58. [Google Scholar] [CrossRef]
- Lamba, V.; Sangkuhl, K.; Sanghavi, K.; Fish, A.; Altman, R.B.; Klein, T.E. Pharm GKB Summary: Mycophenolic acid pathway. Pharm. Genom. 2014, 24, 73–79. [Google Scholar] [CrossRef]
- Kiang, T.K.L.; Ensom, M.H.H. Therapeutic drug monitoring of mycophenolate in adult solid organ transplant patients: An update. Expert Opin. Drug Metab. Toxicol. 2016, 12, 545–553. [Google Scholar] [CrossRef]
- Benjanuwattra, J.; Pruksakorn, D.; Koonrungsesomboon, N. Mycophenolic Acid and Its Pharmacokinetic Drug-Drug Interactions in Humans: Review of the Evidence and Clinical Implications. J. Clin. Pharmacol. 2020, 60, 295–311. [Google Scholar] [CrossRef] [PubMed]
- Shaw, L.M.; Figurski, M.; Milone, M.C.; Trofe, J.; Bloom, R.D. Therapeutic drug monitoring of mycophenolic acid. Clin. J. Am. Soc. Nephrol. 2007, 2, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Sherwin, C.M.T.; Fukuda, T.; Brunner, H.I.; Goebel, J.; Vinks, A.A. The evolution of population pharmacokinetic models to describe the enterohepatic recycling of mycophenolic acid in solid organ transplantation and autoimmune disease. Clin. Pharmacokinet. 2011, 50, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Bajinka, O.; Jarju, P.O.; Tan, Y.; Taal, A.M.; Ozdemir, G. The varying effects of antibiotics on gut microbiota. AMB Express 2021, 11, 116. [Google Scholar] [CrossRef]
- Vich Vila, A.; Collij, V.; Sanna, S.; Sinha, T.; Imhann, F.; Bourgonje, A.R.; Mujagic, Z.; Jonkers, D.M.A.E.; Masclee, A.A.M.; Fu, J.; et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 2020, 11, 362. [Google Scholar] [CrossRef]
- Naderer, O.J.; Dupuis, R.E.; Heinzen, E.L.; Wiwattanawongsa, K.; Johnson, M.W.; Smith, P.C. The Influence of Norfloxacin and Metronidazole on the Disposition of Mycophenolate Mofetil. J. Clin. Pharmacol. 2005, 45, 219–226. [Google Scholar] [CrossRef]
- Jha, V. Post-transplant infections: An ounce of prevention. Indian J. Nephrol. 2010, 20, 171–178. [Google Scholar] [CrossRef]
- Martin, S.I.; Fishman, J.A. Pneumocystis Pneumonia in Solid Organ Transplantation. Am. J. Transplant. 2013, 13, 272–279. [Google Scholar] [CrossRef]
- Bullingham, R.E.S.; Nicholls, A.J.; Kamm, B.R. Clinical Pharmacokinetics of Mycophenolate Mofetil. Clin. Pharmacokinet. 1998, 34, 429–455. [Google Scholar] [CrossRef] [PubMed]
- Willmann, M.; Vehreschild, M.J.G.T.; Biehl, L.M.; Vogel, W.; Dörfel, D.; Hamprecht, A.; Seifert, H.; Autenrieth, I.B.; Peter, S. Distinct impact of antibiotics on the gut microbiome and resistome: A longitudinal multicenter cohort study. BMC Biol. 2019, 17, 76. [Google Scholar] [CrossRef] [PubMed]
- Mavromanolakis, E.; Maraki, S.; Samonis, G.; Tselentis, Y.; Cranidis, A. Effect of Norfloxacin, Trimethoprim-Sulfamethoxazole and Nitrofurantoin on Fecal Flora of Women with Recurrent Urinary Tract Infections. J. Chemother. 1997, 9, 203–207. [Google Scholar] [CrossRef]
- Fazza, O.; Favard Ennachachibi, M.; Ennassiri, H.; Hmyene, A. Antibiotic Susceptibility of β-Glucuronidase-Positive Escherichia coli Isolated from Poultry Products in Morocco. Int. J. Food Sci. 2023, 2023, 7862168. [Google Scholar] [CrossRef]
- Hoffmann-La Roche Ltd. CellCept®mycophenolate Mofetil [Product Monograph]. Available online: https://www.rochecanada.com/PMs/CellCept/CellCept_PM_E.pdf (accessed on 20 May 2022).
- Gao, J.-W.; Peng, Z.-H.; Li, X.-Y.; Sun, B.; Guo, Y.-K.; Liu, G.-L. simultaneous determination of mycophenolic acid and its metabolites by HPLC and pharmacokinetic studies in rat plasma and bile. Arch. Pharmacal Res. 2011, 34, 59. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 20 May 2022).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Dodge, Y. Spearman Rank Correlation Coefficient. In The Concise Encyclopedia of Statistics; Springer: New York, NY, USA, 2008; pp. 502–505. [Google Scholar]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef]
- Kodawara, T.; Masuda, S.; Yano, Y.; Matsubara, K.; Nakamura, T.; Masada, M. Inhibitory effect of ciprofloxacin on β-glucuronidase-mediated deconjugation of mycophenolic acid glucuronide. Biopharm. Drug Dispos. 2014, 35, 275–283. [Google Scholar] [CrossRef]
- Borrows, R.; Chusney, G.; James, A.; Stichbury, J.; Tromp, J.V.; Cairns, T.; Griffith, M.; Hakim, N.; McLean, A.; Palmer, A.; et al. Determinants of Mycophenolic Acid Levels after Renal Transplantation. Ther. Drug Monit. 2005, 27, 442–450. [Google Scholar] [CrossRef]
- Borrows, R.; Chusney, G.; Loucaidou, M.; James, A.; Tromp, J.V.; Cairns, T.; Griffith, M.; Hakim, N.; McLean, A.; Palmer, A.; et al. The Magnitude and Time Course of Changes in Mycophenolic Acid 12-Hour Predose Levels during Antibiotic Therapy in Mycophenolate Mofetil-Based Renal Transplantation. Ther. Drug Monit. 2007, 29, 122–126. [Google Scholar] [CrossRef]
- Ratna, P.; Mathew, B.S.; Annapandian, V.M.; Saravanakumar, K.; Basu, G.; Tamilarasi, V.; Fleming, D.H. Pharmacokinetic Drug Interaction of Mycophenolate with Co-Amoxiclav in Renal Transplant Patients. Transplantation 2011, 91, e36–e38. [Google Scholar] [CrossRef]
- Schmidt, L.E.; Rasmussen, A.; Nørrelykke, M.R.; Poulsen, H.E.; Hansen, B.A. The effect of selective bowel decontamination on the pharmacokinetics of mycophenolate mofetil in liver transplant recipients. Liver Transplant. 2001, 7, 739–742. [Google Scholar] [CrossRef]
- Taylor, M.R.; Flannigan, K.L.; Rahim, H.; Mohamud, A.; Lewis, I.A.; Hirota, S.A.; Greenway, S.C. Vancomycin relieves mycophenolate mofetil-induced gastrointestinal toxicity by eliminating gut bacterial β-glucuronidase activity. Sci. Adv. 2019, 5, eaax2358. [Google Scholar] [CrossRef] [PubMed]
- Candeliere, F.; Raimondi, S.; Ranieri, R.; Musmeci, E.; Zambon, A.; Amaretti, A.; Rossi, M. β-Glucuronidase Pattern Predicted From Gut Metagenomes Indicates Potentially Diversified Pharmacomicrobiomics. Front. Microbiol. 2022, 13, 826994. [Google Scholar] [CrossRef]
- Pollet, R.M.; D’Agostino, E.H.; Walton, W.G.; Xu, Y.; Little, M.S.; Biernat, K.A.; Pellock, S.J.; Patterson, L.M.; Creekmore, B.C.; Isenberg, H.N.; et al. An Atlas of β-Glucuronidases in the Human Intestinal Microbiome. Structure 2017, 25, 967–977.e5. [Google Scholar] [CrossRef] [PubMed]
- Methé, B.A.; Nelson, K.E.; Pop, M.; Creasy, H.H.; Giglio, M.G.; Huttenhower, C.; Gevers, D.; Petrosino, J.F.; Abubucker, S.; Badger, J.H.; et al. A framework for human microbiome research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef]
- Elmassry, M.M.; Kim, S.; Busby, B. Predicting drug-metagenome interactions: Variation in the microbial β-glucuronidase level in the human gut metagenomes. PLoS ONE 2021, 16, e0244876. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, J.; Wang, R. Gut microbiota modulates drug pharmacokinetics. Drug Metab. Rev. 2018, 50, 357–368. [Google Scholar] [CrossRef]
- Gloux, K.; Berteau, O.; El Oumami, H.; Béguet, F.; Leclerc, M.; Doré, J. A metagenomic β-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4539–4546. [Google Scholar] [CrossRef]
- Dabek, M.; McCrae, S.I.; Stevens, V.J.; Duncan, S.H.; Louis, P. Distribution of β-glucosidase and β-glucuronidase activity and of β-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol. Ecol. 2008, 66, 487–495. [Google Scholar] [CrossRef]
- Saqr, A.; Carlson, B.; Staley, C.; Rashidi, A.; Al-Kofahi, M.; Kaiser, T.; Holtan, S.; MacMillan, M.; Young, J.-A.; Jurdi, N.E.; et al. Reduced Enterohepatic Recirculation of Mycophenolate and Lower Blood Concentrations Are Associated with the Stool Bacterial Microbiome after Hematopoietic Cell Transplantation. Transplant. Cell. Ther. 2022, 28, 372.e1–372.e9. [Google Scholar] [CrossRef] [PubMed]
- Beaud, D.; Tailliez, P.; Anba-Mondoloni, J. Genetic characterization of the β-glucuronidase enzyme from a human intestinal bacterium, Ruminococcus gnavus. Microbiology 2005, 151, 2323–2330. [Google Scholar] [CrossRef] [PubMed]
- Biernat, K.A.; Pellock, S.J.; Bhatt, A.P.; Bivins, M.M.; Walton, W.G.; Tran, B.N.T.; Wei, L.; Snider, M.C.; Cesmat, A.P.; Tripathy, A.; et al. Structure, function, and inhibition of drug reactivating human gut microbial β-glucuronidases. Sci. Rep. 2019, 9, 825. [Google Scholar] [CrossRef] [PubMed]
- Christians, U.; Strom, T.; Zhang, Y.L.; Steudel, W.; Schmitz, V.; Trump, S.; Haschke, M. Active Drug Transport of Immunosuppressants: New Insights for Pharmacokinetics and Pharmacodynamics. Ther. Drug Monit. 2006, 28, 39–44. [Google Scholar] [CrossRef]
- Karlgren, M.; Bergström, C.A.S. CHAPTER 1 How Physicochemical Properties of Drugs Affect Their Metabolism and Clearance. In New Horizons in Predictive Drug Metabolism and Pharmacokinetics; The Royal Society of Chemistry: London, UK, 2016; pp. 1–26. [Google Scholar]
- Pedersen, J.M.; Matsson, P.; Bergström, C.A.S.; Hoogstraate, J.; Norén, A.; LeCluyse, E.L.; Artursson, P. Early identification of clinically relevant drug interactions with the human bile salt export pump (BSEP/ABCB11). Toxicol. Sci. 2013, 136, 328–343. [Google Scholar] [CrossRef]
- Guirong, Y.E.; Minjie, Z.; Lixin, Y.U.; Junsheng, Y.E.; Lin, Y.; Lisha, S. Gut microbiota in renal transplant recipients, patients with chronic kidney disease and healthy subjects. Nan Fang Yi Ke Da Xue Xue Bao 2018, 38, 1401–1408. [Google Scholar] [CrossRef]
- Blum, K. Gut Microbiome Altered in Kidney Transplant Recipients. Available online: https://www.kidneynews.org/view/post/education-12/gut-microbiome-altered-in-kidney-transplant-recipients-.xml (accessed on 15 May 2022).
- Mohammadpour, A. Evaluation of a modified salt-out method for DNA extraction from whole blood lymphocytes: A simple and economical method for gene polymorphism. Pharm. Biomed. Res. 2018, 4, 28–32. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Hall, A.B.; Tolonen, A.C.; Xavier, R.J. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 2017, 18, 690–699. [Google Scholar] [CrossRef]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Blekhman, R.; Goodrich, J.K.; Huang, K.; Sun, Q.; Bukowski, R.; Bell, J.T.; Spector, T.D.; Keinan, A.; Ley, R.E.; Gevers, D.; et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015, 16, 191. [Google Scholar] [CrossRef] [PubMed]
- Lopera-Maya, E.A.; Kurilshikov, A.; van der Graaf, A.; Hu, S.; Andreu-Sánchez, S.; Chen, L.; Vila, A.V.; Gacesa, R.; Sinha, T.; Collij, V.; et al. Effect of host genetics on the gut microbiome in 7738 participants of the Dutch Microbiome Project. Nat. Genet. 2022, 54, 143–151. [Google Scholar] [CrossRef]
- Cook, D.E.; Andersen, E.C. VCF-kit: Assorted utilities for the variant call format. Bioinformatics 2017, 33, 1581–1582. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]



| Characteristics | Value (n = 16) |
|---|---|
| Sex, n (%) | |
| Male | 10 (62.5%) |
| Female | 6 (37.5%) |
| Age (year) | 28.88 ± 2.66 |
| Weight (kg) | 59.19 ± 9.57 |
| BMI (kg/m2) | 21.52 ± 2.62 |
| Heart rate (beats/min) | 76.94 ± 11.35 |
| Blood pressure; Systolic/Diastolic (mmHg) | 118.63 ± 11.98/70.31 ± 8.61 |
| Laboratory parameters | |
| Complete blood count | |
| Hemoglobin (g/dL) | 13.10 ± 1.15 |
| Hematocrit (%) | 40.99 ± 3.88 |
| White blood cell count (cells/μL) | 6346.25 ± 1028.87 |
| Platelets (cells/μL) | 282,937.50 ± 55,789.45 |
| Serum electrolytes | |
| Na+ (mmol/L) | 138.06 ± 1.69 |
| K+ (mmol/L) | 3.94 ± 0.22 |
| Cl− (mmol/L) | 100.44 ± 1.90 |
| HCO3− (mmol/L) | 23.50 ± 1.46 |
| Liver function test | |
| Albumin (g/dL) | 4.55 ± 0.16 |
| Total bilirubin (mg/dL) | 1.03 ± 1.35 |
| Direct bilirubin (mg/dL) | 0.40 ± 0.28 |
| ALT (U/L) | 17.25 ± 9.06 |
| AST (U/L) | 20.56 ± 6.41 |
| ALP (U/L) | 59.50 ± 17.68 |
| Renal function test | |
| BUN (mg/dL) | 12.31 ± 2.85 |
| Creatinine (mg/dL) | 0.85 ± 0.19 |
| eGFR (mL/min/1.73 m2) | 109.78 ± 13.14 |
| Parameters | Mean ± Standard Deviation | p Value b | |
|---|---|---|---|
| MMF | MMF + TMP-SMX | ||
| MPA | |||
| Cmax (µg/mL) | 25.89 ± 8.65 | 23.41 ± 9.99 | 0.446 |
| Tmax (h) a | 0.67 (0.33–1.5) | 0.67 (0.33–2) | 0.942 |
| AUC0–6 (µg·h/mL) | 33.03 ± 13.17 | 28.98 ± 12.38 | 0.227 |
| AUC6–12 (µg·h/mL) | 5.45 ± 3.22 | 3.79 ± 3.18 | 0.082 |
| AUC0–12 (µg·h/mL) | 38.48 ± 14.71 | 32.76 ± 13.31 | 0.119 |
| AUC0-∞ (µg·h/mL) | 54.00 ± 27.33 | 39.45 ± 17.81 | 0.041 * |
| Vz/F (L) | 166.29 ± 69.83 | 161.58 ± 61.40 | 0.853 |
| t1/2 (h) | 6.25 ± 3.91 | 4.47 ± 2.98 | 0.122 |
| CL/F (L/h) | 22.51 ± 9.69 | 36.63 ± 34.27 | 0.104 |
| MPAG | |||
| Cmax (µg/mL) | 54.87 ± 30.36 | 52.67 ± 18.61 | 0.704 |
| Tmax (h) a | 1.5 (1–2) | 1.5 (1–2) | 0.963 |
| AUC0–6 (µg·h/mL) | 204.36 ± 110.23 | 201.57 ± 74.45 | 0.890 |
| AUC6–12 (µg·h/mL) | 96.04 ± 49.30 | 103.50 ± 37.46 | 0.582 |
| AUC0–12 (µg·h/mL) | 300.39 ± 157.32 | 305.07 ± 101.75 | 0.887 |
| AUC0-∞ (µg·h/mL) | 542.71 ± 225.14 | 719.23 ± 137.91 | 0.016 * |
| Vz/F (L) | 36.32 ± 17.51 | 37.48 ± 16.77 | 0.797 |
| t1/2 (h) | 11.49 ± 4.39 | 18.35 ± 8.32 | 0.020 * |
| CL/F (L/h) | 2.31 ± 1.29 | 1.44 ± 0.28 | 0.015 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dukaew, N.; Thongkumkoon, P.; Sirikaew, N.; Dissook, S.; Sakuludomkan, W.; Tongjai, S.; Thiennimitr, P.; Na Takuathung, M.; Benjanuwattra, J.; Kongthaweelert, P.; et al. Gut Microbiota-Mediated Pharmacokinetic Drug–Drug Interactions between Mycophenolic Acid and Trimethoprim-Sulfamethoxazole in Humans. Pharmaceutics 2023, 15, 1734. https://doi.org/10.3390/pharmaceutics15061734
Dukaew N, Thongkumkoon P, Sirikaew N, Dissook S, Sakuludomkan W, Tongjai S, Thiennimitr P, Na Takuathung M, Benjanuwattra J, Kongthaweelert P, et al. Gut Microbiota-Mediated Pharmacokinetic Drug–Drug Interactions between Mycophenolic Acid and Trimethoprim-Sulfamethoxazole in Humans. Pharmaceutics. 2023; 15(6):1734. https://doi.org/10.3390/pharmaceutics15061734
Chicago/Turabian StyleDukaew, Nahathai, Patcharawadee Thongkumkoon, Nutnicha Sirikaew, Sivamoke Dissook, Wannachai Sakuludomkan, Siripong Tongjai, Parameth Thiennimitr, Mingkwan Na Takuathung, Juthipong Benjanuwattra, Prachya Kongthaweelert, and et al. 2023. "Gut Microbiota-Mediated Pharmacokinetic Drug–Drug Interactions between Mycophenolic Acid and Trimethoprim-Sulfamethoxazole in Humans" Pharmaceutics 15, no. 6: 1734. https://doi.org/10.3390/pharmaceutics15061734
APA StyleDukaew, N., Thongkumkoon, P., Sirikaew, N., Dissook, S., Sakuludomkan, W., Tongjai, S., Thiennimitr, P., Na Takuathung, M., Benjanuwattra, J., Kongthaweelert, P., & Koonrungsesomboon, N. (2023). Gut Microbiota-Mediated Pharmacokinetic Drug–Drug Interactions between Mycophenolic Acid and Trimethoprim-Sulfamethoxazole in Humans. Pharmaceutics, 15(6), 1734. https://doi.org/10.3390/pharmaceutics15061734






