Macrophage Reprogramming via the Modulation of Unfolded Protein Response with siRNA-Loaded Magnetic Nanoparticles in a TAM-like Experimental Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation and Characterization of PDA-MNPs Nanoparticles
2.2. Obtention of PEMs
2.3. Cytocompatibility Tests
2.3.1. Cell Viability Assay
2.3.2. Detection of Reactive Oxygen Species (ROS) Production
2.3.3. Detection of ER Stress and the Activation of Unfolded Protein Response (UPR)
2.4. Internalization of PDA-MNPs in Cells in the Absence/Presence of a GMF
2.4.1. Prussian Blue Staining
2.4.2. Iron Quantification by Potassium Thiocyanate
2.4.3. Uptake of PDA-MNPs in PEMs
2.5. Functionalization of PDA-MNPs with siRNA Molecules
2.6. Cellular Uptake of the PDA-MNPs/siRNA
2.7. Gene Silencing Efficiency of PERK Protein via PDA-MNPs/siPERK
2.8. In Vitro Polarization of PEMs
2.9. Quantitative Real-Time PCR
2.10. Immunoblotting
2.11. Immunofluorescence Microscopy
2.12. Flow Cytometry
2.13. Statistical Analysis
3. Results and Discussion
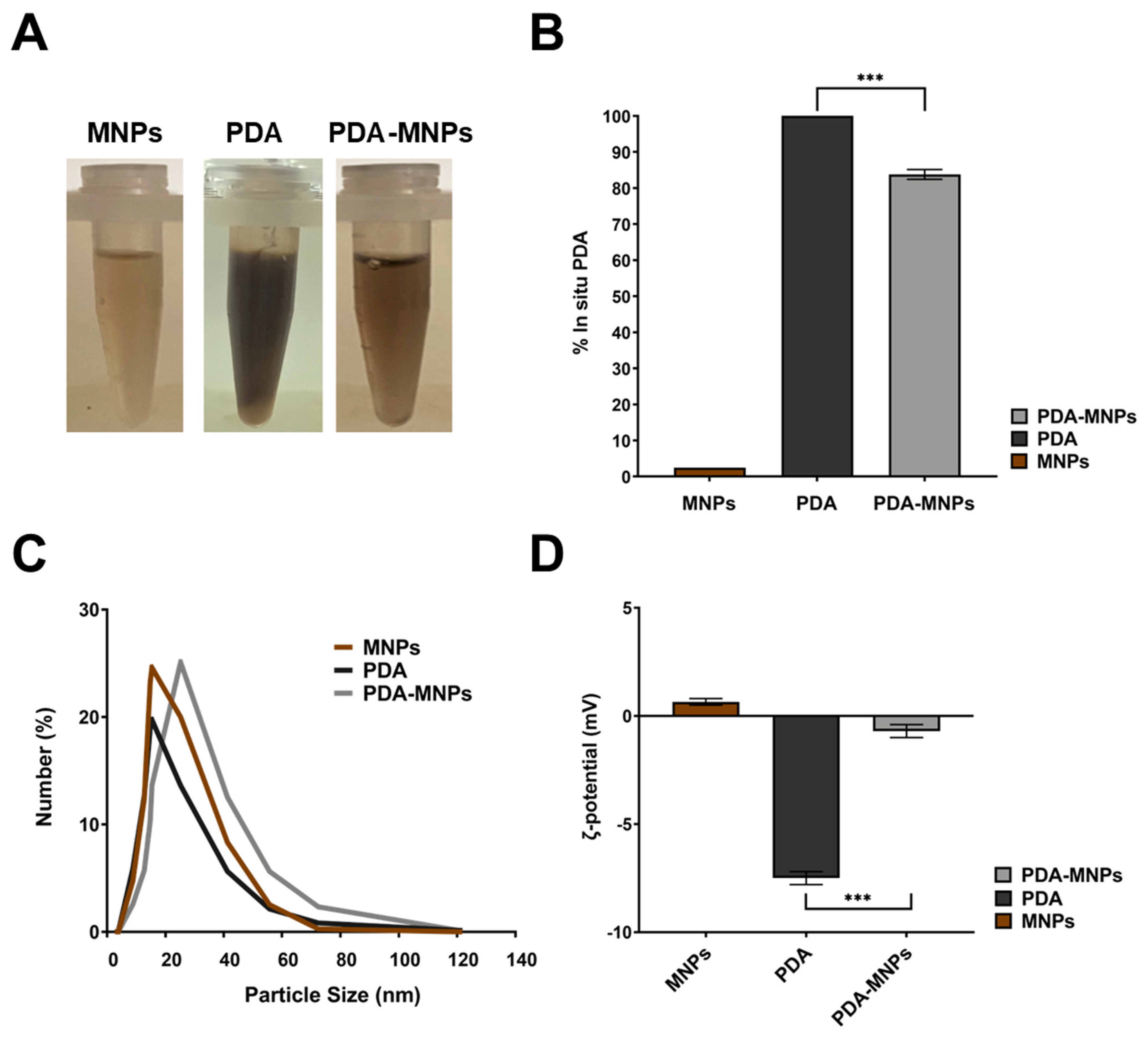
3.1. Preparation and Characterization of PDA-MNPs
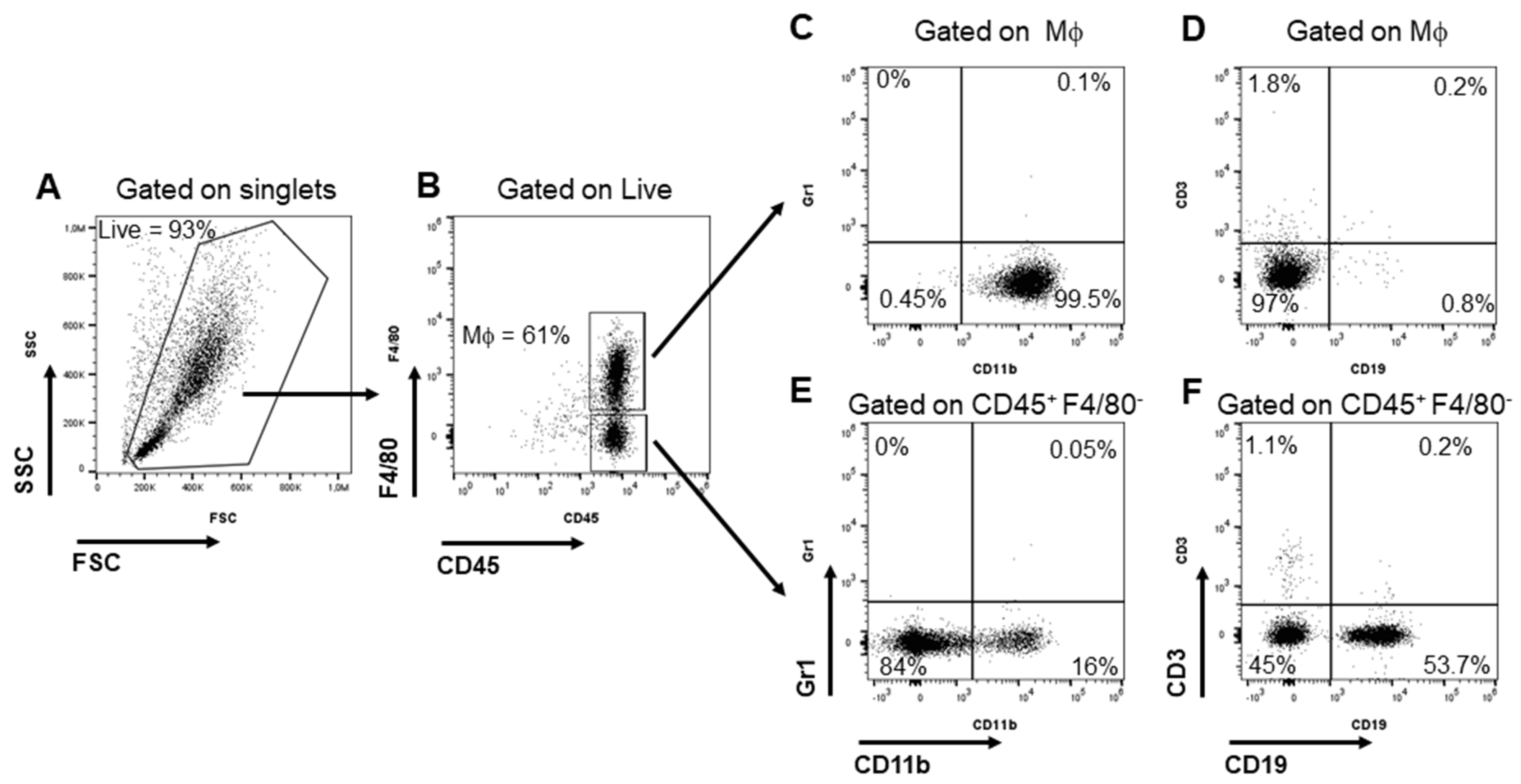
3.2. Isolation and Characterization of Peritoneal Macrophages (PEMs)
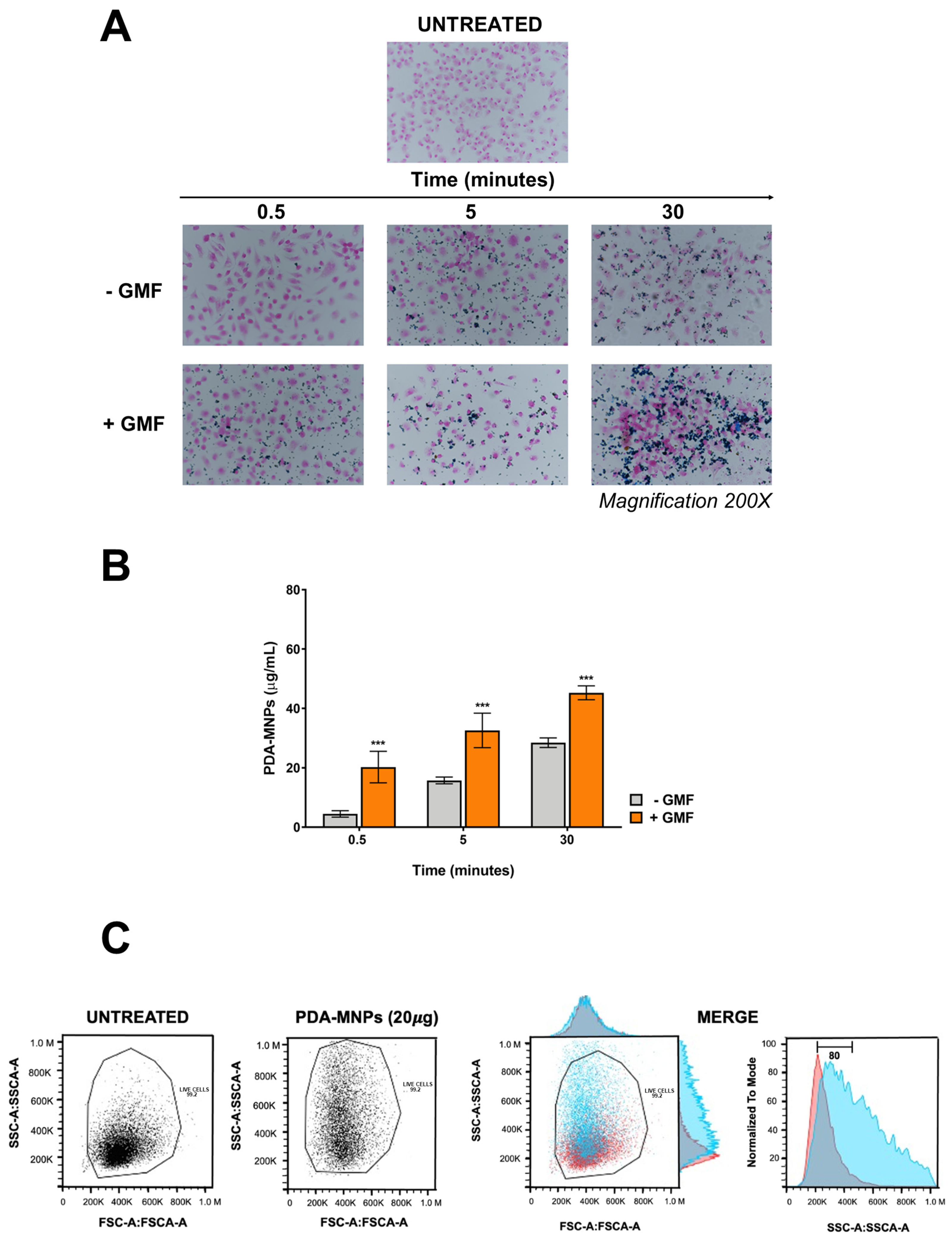
3.3. Cytocompatibility and Cellular Internalization of PDA-MNPs
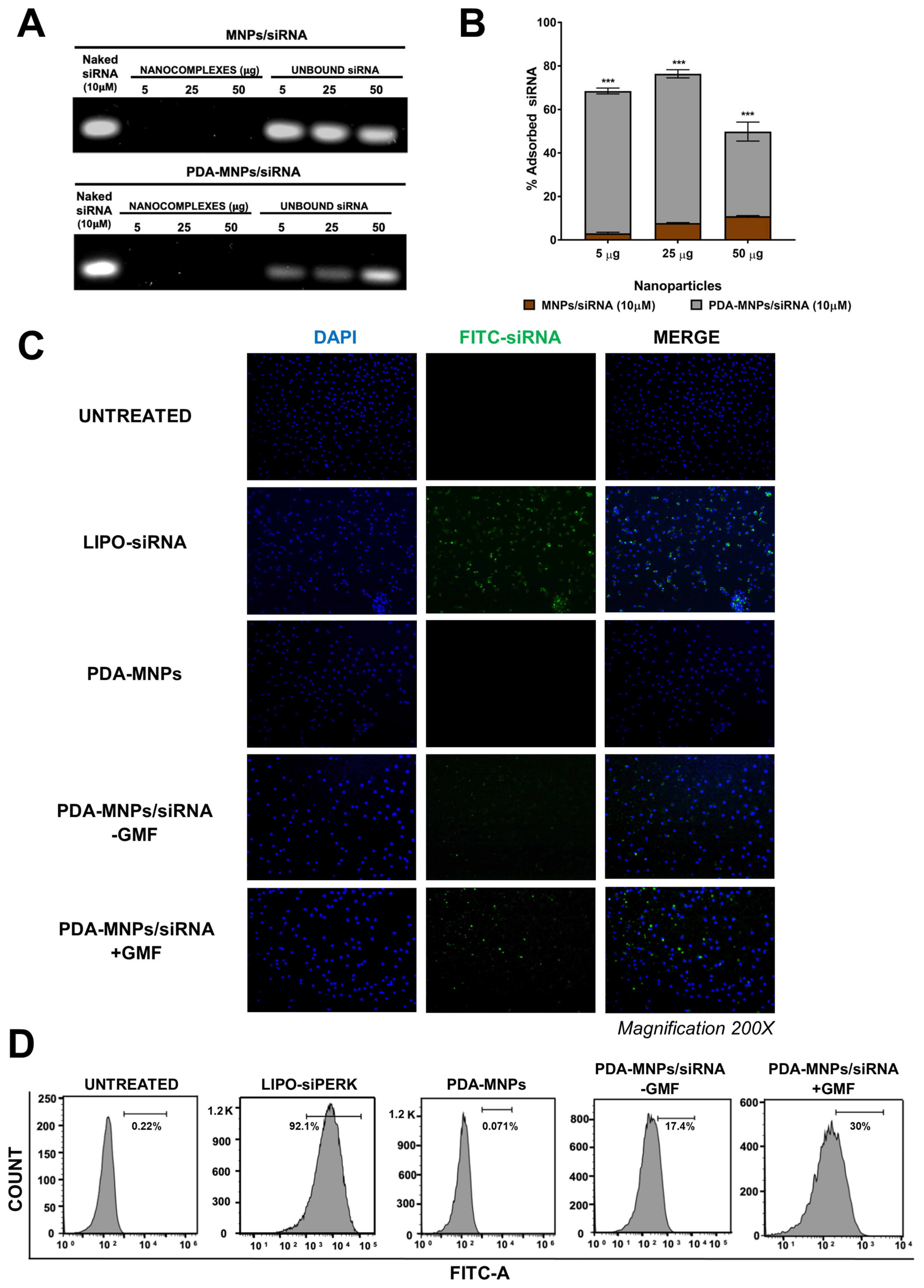
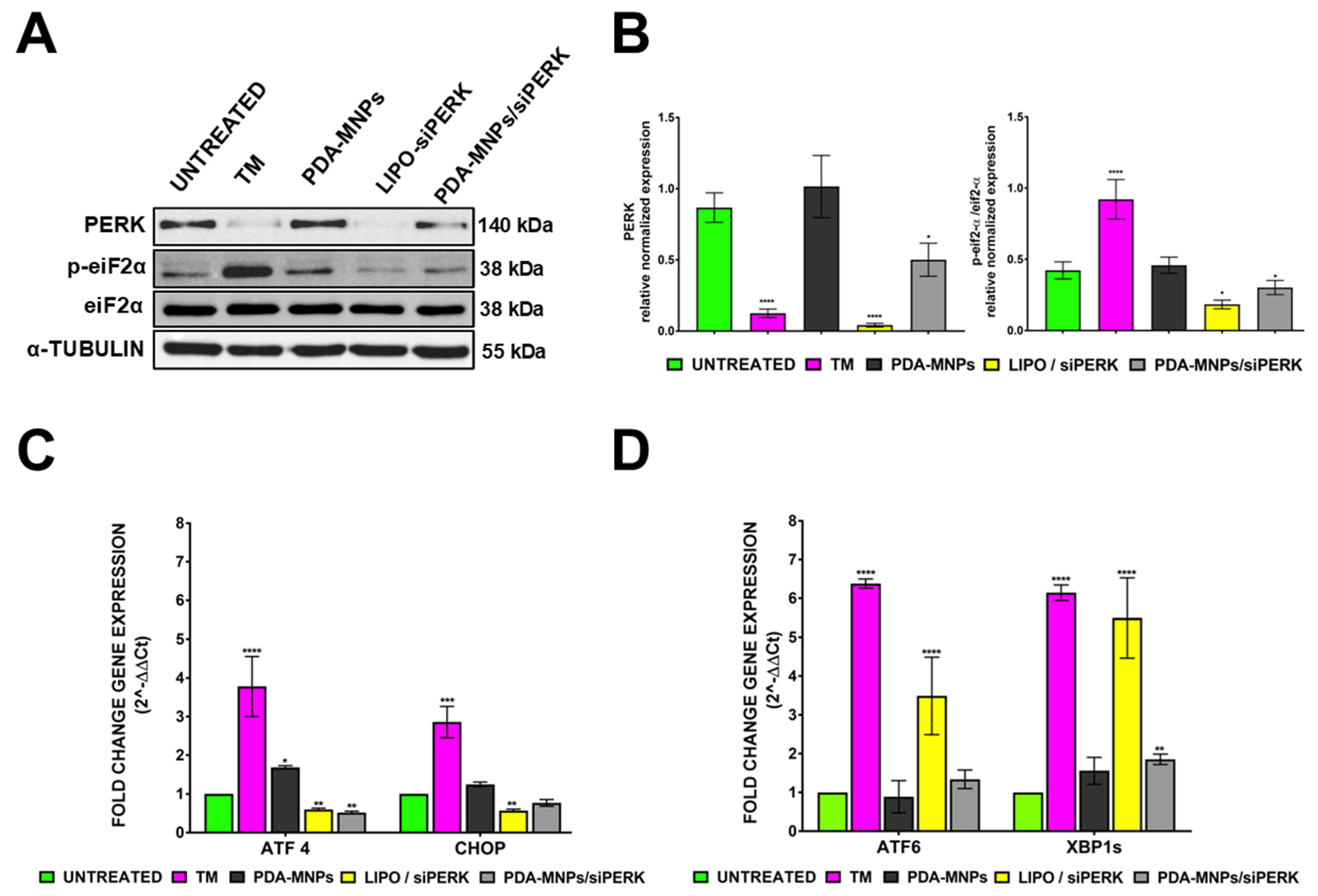
3.4. Gene Silencing Efficiency of siPERK via PDA-MNPs in PEMs
3.5. In Vitro Differentiation of PEM Macrophages into M1 or M2 Phenotypes
3.6. Reprogramming TAM-Like Macrophages toward the M1 Phenotype
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments

Conflicts of Interest
References
- Debela, D.T.; Muzazu, S.G.; Heraro, K.D.; Ndalama, M.T.; Mesele, B.W.; Haile, D.C.; Kitui, S.K.; Manyazewal, T. New approaches and procedures for cancer treatment: Current perspectives. SAGE Open Med. 2021, 9, 20503121211034366. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Tawfik, E.A.; Aldrak, N.A.; Albrahim, S.H.; Alzahrani, D.A.; Alfassam, H.A.; Alkoblan, S.M.; Almalik, A.M.; Chen, K.S.; Abou-Khalil, R.; Shah, K.; et al. Immunotherapy in hematological malignancies: Recent advances and open questions. Immunotherapy 2021, 13, 1215–1229. [Google Scholar] [CrossRef]
- Lanier, O.L.; Pérez-Herrero, E.; Andrea, A.P.D.; Bahrami, K.; Lee, E.; Ward, D.M.; Ayala-Suárez, N.; Rodríguez-Méndez, S.M.; Peppas, N.A. Immunotherapy approaches for hematological cancers. iScience 2022, 25, 105326. [Google Scholar] [CrossRef]
- Paulson, K.G.; Lahman, M.C.; Chapuis, A.G.; Brownell, I. Immunotherapy for skin cancer. Int. Immunol. 2019, 31, 465–475. [Google Scholar] [CrossRef]
- Le Saux, O.; Lounici, Y.; Wajda, P.; Barrin, S.; Caux, C.; Dubois, B.; Ray-Coquard, I. Neoadjuvant immune checkpoint inhibitors in cancer, current state of the art. Crit. Rev. Oncol. 2021, 157, 103172. [Google Scholar] [CrossRef] [PubMed]
- Koustas, E.; Sarantis, P.; Papavassiliou, A.G.; Karamouzis, M.V. The Resistance Mechanisms of Checkpoint Inhibitors in Solid Tumors. Biomolecules 2020, 10, 666. [Google Scholar] [CrossRef]
- Murciano-Goroff, Y.R.; Warner, A.B.; Wolchok, J.D. The future of cancer immunotherapy: Microenvironment-targeting combinations. Cell Res. 2020, 30, 507–519. [Google Scholar] [CrossRef]
- Jin, M.-Z.; Jin, W.-L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 2020, 5, 166. [Google Scholar] [CrossRef]
- Cassim, S.; Pouyssegur, J. Tumor Microenvironment: A Metabolic Player that Shapes the Immune Response. Int. J. Mol. Sci. 2019, 21, 157. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Fang, Y.; Jin, X.; Zhang, M.; Shi, K. Modulating Repolarization of Tumor-Associated Macrophages with Targeted Therapeutic Nanoparticles as a Potential Strategy for Cancer Therapy. ACS Appl. Bio Mater. 2021, 4, 5871–5896. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-Y.; Song, X.-Y.; Li, Y.; Ye, L.-L.; Zhou, Q.; Yang, W.-B. Tumor-associated macrophages: A promising target for a cancer immunotherapeutic strategy. Pharmacol. Res. 2020, 161, 105111. [Google Scholar] [CrossRef]
- Kowal, J.; Kornete, M.; Joyce, J.A.; Escobar, G.; Escobar, A.; Ascui, G.; Tempio, F.I.; Ortiz, M.C.; Pérez, C.A.; López, M.N.; et al. Re-education of macrophages as a therapeutic strategy in cancer. Immunotherapy 2019, 11, 677–689. [Google Scholar] [CrossRef]
- Anfray, C.; Ummarino, A.; Andón, F.T.; Allavena, P. Current Strategies to Target Tumor-Associated-Macrophages to Improve Anti-Tumor Immune Responses. Cells 2019, 9, 46. [Google Scholar] [CrossRef]
- Poltavets, A.S.; Vishnyakova, P.A.; Elchaninov, A.V.; Sukhikh, G.T.; Fatkhudinov, T.K. Macrophage Modification Strategies for Efficient Cell Therapy. Cells 2020, 9, 1535. [Google Scholar] [CrossRef] [PubMed]
- Van Dalen, F.J.; Van Stevendaal, M.H.M.E.; Fennemann, F.L.; Verdoes, M.; Ilina, O. Molecular Repolarisation of Tumour-Associated Macrophages. Molecules 2018, 24, 9. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Choi, S.H. Tumor-associated macrophages in cancer: Recent advancements in cancer nanoimmunotherapies. J. Exp. Clin. Cancer Res. 2022, 41, 68. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; He, L.; Zhu, J.; Zhang, P.; Liang, S. Targeting tumor-associated macrophages for cancer treatment. Cell Biosci. 2022, 12, 85. [Google Scholar] [CrossRef]
- Di Conza, G.; Tsai, C.-H.; Gallart-Ayala, H.; Yu, Y.-R.; Franco, F.; Zaffalon, L.; Xie, X.; Li, X.; Xiao, Z.; Raines, L.N.; et al. Tumor-induced reshuffling of lipid composition on the endoplasmic reticulum membrane sustains macrophage survival and pro-tumorigenic activity. Nat. Immunol. 2021, 22, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Raines, L.N.; Zhao, H.; Wang, Y.; Chen, H.-Y.; Gallart-Ayala, H.; Hsueh, P.-C.; Cao, W.; Koh, Y.; Alamonte-Loya, A.; Liu, P.-S.; et al. PERK is a critical metabolic hub for immunosuppressive function in macrophages. Nat. Immunol. 2022, 23, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Batista, A.; Rodvold, J.J.; Xian, S.; Searles, S.C.; Lew, A.; Iwawaki, T.; Almanza, G.; Waller, T.C.; Lin, J.; Jepsen, K.; et al. IRE1α regulates macrophage polarization, PD-L1 expression, and tumor survival. PLoS Biol. 2020, 18, e3000687. [Google Scholar] [CrossRef] [PubMed]
- Junjappa, R.P.; Patil, P.; Bhattarai, K.R.; Kim, H.-R.; Chae, H.-J. IRE1α Implications in Endoplasmic Reticulum Stress-Mediated Development and Pathogenesis of Autoimmune Diseases. Front. Immunol. 2018, 9, 1289. [Google Scholar] [CrossRef] [PubMed]
- Sanna, V.; Sechi, M. Therapeutic Potential of Targeted Nanoparticles and Perspective on Nanotherapies. ACS Med. Chem. Lett. 2020, 11, 1069–1073. [Google Scholar] [CrossRef]
- Ni, Q.; Xu, F.; Wang, Y.; Li, Y.; Qing, G.; Zhang, Y.; Zhong, J.; Li, J.; Liang, X.-J. Nanomaterials with changeable physicochemical property for boosting cancer immunotherapy. J. Control. Release 2022, 342, 210–227. [Google Scholar] [CrossRef]
- Timon-David, E.; Perez, C.; Rodallec, A. Nanotherapeutics Plus Immunotherapy in Oncology: Who Brings What to the Table? Pharmaceutics 2022, 14, 2326. [Google Scholar] [CrossRef]
- Dias, A.M.M.; Courteau, A.; Bellaye, P.-S.; Kohli, E.; Oudot, A.; Doulain, P.-E.; Petitot, C.; Walker, P.-M.; Decréau, R.; Collin, B. Superparamagnetic Iron Oxide Nanoparticles for Immunotherapy of Cancers through Macrophages and Magnetic Hyperthermia. Pharmaceutics 2022, 14, 2388. [Google Scholar] [CrossRef]
- Peigneux, A.; Oltolina, F.; Colangelo, D.; Iglesias, G.R.; Delgado, A.V.; Prat, M.; Jimenez-Lopez, C. Functionalized Biomimetic Magnetic Nanoparticles as Effective Nanocarriers for Targeted Chemotherapy. Part. Part. Syst. Charact. 2019, 36, 1900057. [Google Scholar] [CrossRef]
- Garcia-Pinel, B.; Jabalera, Y.; Ortiz, R.; Cabeza, L.; Jimenez-Lopez, C.; Melguizo, C.; Prados, J. Biomimetic Magnetoliposomes as Oxaliplatin Nanocarriers: In Vitro Study for Potential Application in Colon Cancer. Pharmaceutics 2020, 12, 589. [Google Scholar] [CrossRef]
- Oltolina, F.; Colangelo, D.; Miletto, I.; Clemente, N.; Miola, M.; Verné, E.; Prat, M.; Follenzi, A. Tumor Targeting by Monoclonal Antibody Functionalized Magnetic Nanoparticles. Nanomaterials 2019, 9, 1575. [Google Scholar] [CrossRef]
- Mu, X.; Li, J.; Yan, S.; Zhang, H.; Zhang, W.; Zhang, F.; Jiang, J. siRNA Delivery with Stem Cell Membrane-Coated Magnetic Nanoparticles for Imaging-Guided Photothermal Therapy and Gene Therapy. ACS Biomater. Sci. Eng. 2018, 4, 3895–3905. [Google Scholar] [CrossRef] [PubMed]
- Borroni, E.; Miola, M.; Ferraris, S.; Ricci, G.; Rožman, K.Ž.; Kostevšek, N.; Catizone, A.; Rimondini, L.; Prat, M.; Verné, E.; et al. Tumor targeting by lentiviral vectors combined with magnetic nanoparticles in mice. Acta Biomater. 2017, 59, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Oltolina, F.; Peigneux, A.; Colangelo, D.; Clemente, N.; D’urso, A.; Valente, G.; Iglesias, G.R.; Jiménez-Lopez, C.; Prat, M. Biomimetic Magnetite Nanoparticles as Targeted Drug Nanocarriers and Mediators of Hyperthermia in an Experimental Cancer Model. Cancers 2020, 12, 2564. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, C.S.; Alves, A.R.; de Melo, C.P.; Corrêa-Oliveira, R.; Calzavara-Silva, C.E. Immunotherapy for cancer: Effects of iron oxide nanoparticles on polarization of tumor-associated macrophages. Nanomedicine 2021, 16, 2633–2650. [Google Scholar] [CrossRef]
- Reichel, D.; Tripathi, M.; Perez, J.M. Biological Effects of Nanoparticles on Macrophage Polarization in the Tumor Microenvironment. Nanotheranostics 2019, 3, 66–88. [Google Scholar] [CrossRef]
- Jin, A.; Wang, Y.; Lin, K.; Jiang, L. Nanoparticles Modified by Polydopamine: Working as “Drug” Carriers. Bioact. Mater. 2020, 5, 522–541. [Google Scholar] [CrossRef]
- Gu, X.; Zhang, Y.; Sun, H.; Song, X.; Fu, C.; Dong, P. Mussel-Inspired Polydopamine Coated Iron Oxide Nanoparticles for Biomedical Application. J. Nanomater. 2015, 2015, 3. [Google Scholar] [CrossRef]
- Lin, L.-S.; Cong, Z.-X.; Cao, J.-B.; Ke, K.-M.; Peng, Q.-L.; Gao, J.; Yang, H.-H.; Liu, G.; Chen, X. Multifunctional Fe3O4@Polydopamine Core–Shell Nanocomposites for Intracellular mRNA Detection and Imaging-Guided Photothermal Therapy. ACS Nano 2014, 8, 3876–3883. [Google Scholar] [CrossRef]
- Tatiparti, K.; Sau, S.; Kashaw, S.K.; Iyer, A.K. siRNA Delivery Strategies: A Comprehensive Review of Recent Developments. Nanomaterials 2017, 7, 77. [Google Scholar] [CrossRef]
- Nieto, C.; Vega, M.A.; Marcelo, G.; Del Valle, E.M.M. Polydopamine nanoparticles kill cancer cells. RSC Adv. 2018, 8, 36201–36208. [Google Scholar] [CrossRef]
- Rios, F.J.; Touyz, R.M.; Montezano, A.C. Isolation and Differentiation of Murine Macrophages. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2017; Volume 1527, pp. 297–309. [Google Scholar]
- Hamidzadeh, K.; Belew, A.T.; El-Sayed, N.M.; Mosser, D.M. The transition of M-CSF–derived human macrophages to a growth-promoting phenotype. Blood Adv. 2020, 4, 5460–5472. [Google Scholar] [CrossRef] [PubMed]
- Feito, M.J.; Diez-Orejas, R.; Cicuéndez, M.; Casarrubios, L.; Rojo, J.M.; Portolés, M.T. Characterization of M1 and M2 polarization phenotypes in peritoneal macrophages after treatment with graphene oxide nanosheets. Colloids Surf. B Biointerfaces 2018, 176, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-L.; Tian, P.-X.; Han, F.; Zheng, J.; Xia, X.-X.; Xue, W.-J.; Ding, X.-M.; Ding, C.-G. Comparison of the characteristics of macrophages derived from murine spleen, peritoneal cavity, and bone marrow. J. Zhejiang Univ. B 2017, 18, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Cassado, A.A.; D’Império Lima, M.R.; Bortoluci, K.R. Revisiting Mouse Peritoneal Macrophages: Heterogeneity, Development, and Function. Front. Immunol. 2015, 6, 225. [Google Scholar] [CrossRef]
- Altin, J.G.; Sloan, E. The role of CD45 and CD45-associated molecules in T cell activation. Immunol. Cell Biol. 1997, 75, 430–445. [Google Scholar] [CrossRef]
- Ge, R.; Li, X.; Lin, M.; Wang, D.; Li, S.; Liu, S.; Tang, Q.; Liu, Y.; Jiang, J.; Liu, L.; et al. Fe3O4@polydopamine Composite Theranostic Superparticles Employing Preassembled Fe3O4 Nanoparticles as the Core. ACS Appl. Mater. Interfaces 2016, 8, 22942–22952. [Google Scholar] [CrossRef]
- Siciliano, G.; Monteduro, A.G.; Turco, A.; Primiceri, E.; Rizzato, S.; Depalo, N.; Curri, M.L.; Maruccio, G. Polydopamine-Coated Magnetic Iron Oxide Nanoparticles: From Design to Applications. Nanomaterials 2022, 12, 1145. [Google Scholar] [CrossRef]
- ISO 10993-5:2009; Biological Evaluation of Medical Devices—Part 5: Tests for Cytotoxicity: In Vitro Methods. International Organization for Standardization: Geneva, Switzerland, 2009.
- Jia, P.-P.; Sun, T.; Junaid, M.; Yang, L.; Ma, Y.-B.; Cui, Z.-S.; Wei, D.-P.; Shi, H.-F.; Pei, D.-S. Nanotoxicity of different sizes of graphene (G) and graphene oxide (GO) in vitro and in vivo. Environ. Pollut. 2019, 247, 595–606. [Google Scholar] [CrossRef]
- Khan, A.A.; Allemailem, K.S.; Almatroudi, A.; Almatroodi, S.A.; Mahzari, A.; Alsahli, M.A.; Rahmani, A.H. Endoplasmic Reticulum Stress Provocation by Different Nanoparticles: An Innovative Approach to Manage the Cancer and Other Common Diseases. Molecules 2020, 25, 5336. [Google Scholar] [CrossRef]
- Sicari, D.; Delaunay-Moisan, A.; Combettes, L.; Chevet, E.; Igbaria, A. A guide to assessing endoplasmic reticulum homeostasis and stress in mammalian systems. FEBS J. 2019, 287, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.-T.; Yu, J.; Wang, H.-J.; Shi, Y.; Zhao, T.-S.; He, B.-X.; Qiao, B.; Feng, Z.-W. Effects of endoplasmic reticulum stress on the autophagy, apoptosis, and chemotherapy resistance of human breast cancer cells by regulating the PI3K/AKT/mTOR signaling pathway. Tumor Biol. 2017, 39, 1010428317697562. [Google Scholar] [CrossRef] [PubMed]
- Di Conza, G.; Ho, P.-C. ER Stress Responses: An Emerging Modulator for Innate Immunity. Cells 2020, 9, 695. [Google Scholar] [CrossRef]
- Chang, T.-K.; Lawrence, D.A.; Lu, M.; Tan, J.; Harnoss, J.M.; Marsters, S.A.; Liu, P.; Sandoval, W.; Martin, S.E.; Ashkenazi, A. Coordination between Two Branches of the Unfolded Protein Response Determines Apoptotic Cell Fate. Mol. Cell 2018, 71, 629–636.e5. [Google Scholar] [CrossRef]
- Lara-Reyna, S.; Scambler, T.; Holbrook, J.; Wong, C.; Jarosz-Griffiths, H.H.; Martinon, F.; Savic, S.; Peckham, D.; McDermott, M.F. Metabolic Reprograming of Cystic Fibrosis Macrophages via the IRE1α Arm of the Unfolded Protein Response Results in Exacerbated Inflammation. Front. Immunol. 2019, 10, 1789. [Google Scholar] [CrossRef]
- Kashfi, K.; Kannikal, J.; Nath, N. Macrophage Reprogramming and Cancer Therapeutics: Role of INOS-Derived NO. Cells 2021, 10, 3194. [Google Scholar] [CrossRef]
- Hörhold, F.; Eisel, D.; Oswald, M.; Kolte, A.; Röll, D.; Osen, W.; Eichmüller, S.B.; König, R. Reprogramming of macrophages employing gene regulatory and metabolic network models. PLoS Comput. Biol. 2020, 16, e1007657. [Google Scholar] [CrossRef]
- Otero, K.; Turnbull, I.R.; Poliani, P.L.; Vermi, W.; Cerutti, E.; Aoshi, T.; Tassi, I.; Takai, T.; Stanley, S.L.; Miller, M.; et al. Macrophage colony-stimulating factor induces the proliferation and survival of macrophages via a pathway involving DAP12 and β-catenin. Nat. Immunol. 2009, 10, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, K.A.; Amici, S.A.; Webb, L.M.; Ruiz-Rosado, J.D.D.; Popovich, P.G.; Partida-Sanchez, S.; Guerau-De-Arellano, M. Novel Markers to Delineate Murine M1 and M2 Macrophages. PLoS ONE 2015, 10, e0145342. [Google Scholar] [CrossRef]
- Mulens-Arias, V.; Rojas, J.M.; Barber, D.F. The Use of Iron Oxide Nanoparticles to Reprogram Macrophage Responses and the Immunological Tumor Microenvironment. Front. Immunol. 2021, 12, 693709. [Google Scholar] [CrossRef]
- Mulens-Arias, V.; Rojas, J.M.; Barber, D.F. The Intrinsic Biological Identities of Iron Oxide Nanoparticles and Their Coatings: Unexplored Territory for Combinatorial Therapies. Nanomaterials 2020, 10, 837. [Google Scholar] [CrossRef] [PubMed]
- Pratap, U.P.; Vadlamudi, R.K. PERK promotes immunosuppressive M2 macrophage phenotype by metabolic reprogramming and epigenetic modifications through the PERK-ATF4-PSAT1 axis. Immunometabolism 2022, 4, e00007. [Google Scholar] [CrossRef] [PubMed]
- Sheshadri, N.; Poria, D.K.; Sharan, S.; Hu, Y.; Yan, C.; Koparde, V.N.; Balamurugan, K.; Sterneck, E. PERK signaling through C/EBPδ contributes to ER stress-induced expression of immunomodulatory and tumor promoting chemokines by cancer cells. Cell Death Dis. 2021, 12, 1038. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Sun, J.; Song, B.; Zhang, L.; Shao, Q.; Liu, Y.; Yuan, D.; Zhang, Y.; Qu, X. Fucoidan inhibits CCL22 production through NF-κB pathway in M2 macrophages: A potential therapeutic strategy for cancer. Sci. Rep. 2016, 6, 35855. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Li, X.; Zhang, J.; Lu, Y.; Shi, Y.; Zhu, C.; Liu, Y.; Qin, B.; Luo, Z.; Du, Y.; et al. Dual Inhibition of Endoplasmic Reticulum Stress and Oxidation Stress Manipulates the Polarization of Macrophages under Hypoxia to Sensitize Immunotherapy. ACS Nano 2021, 15, 14522–14534. [Google Scholar] [CrossRef]
- Yin, Z.; Ma, T.; Lin, Y.; Lu, X.; Zhang, C.; Chen, S.; Jian, Z. Retracted: IL-6/STAT3 pathway intermediates M1/M2 macrophage polarization during the development of hepatocellular carcinoma. J. Cell. Biochem. 2018, 119, 9419–9432. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhang, W.; Zhong, W.-Q.; Liu, Z.-J.; Li, H.-M.; Yu, Z.-L.; Zhao, Y.-F. Tumor associated macrophages induce epithelial to mesenchymal transition via the EGFR/ERK1/2 pathway in head and neck squamous cell carcinoma. Oncol. Rep. 2018, 40, 2558–2572. [Google Scholar] [CrossRef] [PubMed]



| Target Gene | Forward Sequence | Reverse Sequence | Product Length (bp) |
|---|---|---|---|
| PERK | CCAGGCATTGTGAGGTATTT | TCTGTGCTTTCGTCTTTGAG | 98 |
| ATF4 | GTTTAGAGCTAGGCAGTGAAG | CCTTTACACATGGAGGGATTAG | 93 |
| Chop | ACACGCACATCCCAAAG | ACCACTCTGTTTCCGTTTC | 108 |
| Bip | GAGAGAGGGAGAGAAGAACA | GCCACCACTTCAAAGACA | 99 |
| XBP1s | AGTCCGCAGCAGGTG | GGTCCAACTTGTCCAGAATG | 93 |
| ATF6 | GGTCCAACTTGTCCAGAATG | TGGAGGTGGAGGCATATAA | 109 |
| Arg–I | ATCCCACCTAGGAGACAAAG | GGGACCTGGAATCTGTCTAT | 116 |
| TGF–β | CTCCCGTGGCTTCTAGTGC | GCCTTAGTTTGGACAGGATCTG | 133 |
| PPARγ | GTGACTCTGCTCAAGTATGG | GAACTCCCTGGTCATGAATC | 114 |
| iNOS | GTTCTCAGCCCAACAATACAAG | GTGGACGGGTCGATGTCAC | 127 |
| TNF–α | AGCCCCCAGTCTGTATCCTT | CTCCCTTTGCAGAACTCAGG | 212 |
| Cox–2 | CTCCCTTTGCAGAACTCAGG | AGTGCTGGGCAAAGAATG | 125 |
| β-actina | GATGACCCAGATCATGTTTGA | GGAGAGCATAGCCCTCGTAG | 161 |
| Antigen | Species | Dilutions | Expected Band | Source | Cat. Number |
|---|---|---|---|---|---|
| PERK | Rabbit polyclonal | 1/500 | 140 kDa | Cell Signaling Technology | C33E10 |
| eif2–𝛼 | Mouse monoclonal | 1/500 | 38 kDa | Cell Signaling Technology | 2103 |
| Phospho eif2–𝛼 | Rabbit polyclonal | 1/500 | 38 kDa | Cell Signaling Technology | 3398 |
| STAT3 | Mouse monoclonal | 1/500 | 80 kDa | Cell Signaling Technology | 9139 |
| Phospho STAT3 | Rabbit polyclonal | 1/500 | 80 kDa | Cell Signaling Technology | 9145 |
| NF-κB p65 | Rabbit polyclonal | 1/500 | 65 kDa | Santa Cruz Technology | sc-7151 |
| Phospho NF-κB p65 | Mouse monoclonal | 1/500 | 65 kDa | Santa Cruz Technology | sc-166748 |
| MAPKK1/2 | Rabbit polyclonal | 1/500 | 42–44 kDa | Millipore | ABS44 |
| Phospho MAPKK1/2 | Rabbit polyclonal | 1/500 | 42–44 kDa | Millipore | 04-797 |
| 𝛼–Tubulin | Mouse monoclonal | 1/500 | 55–60 kDa | Millipore | 05-829 |
| Vinculin | Mouse monoclonal | 1/500 | 120 kDa | Santa Cruz Technology | sc-73614 |
| Antigen | Species | Dilutions | Source | Cat. Number |
|---|---|---|---|---|
| F4/80-BIOTIN | Mouse monoclonal | 1/400 | Miltenyi Biotec | 130-116-514 |
| CD206 | Rabbit polyclonal | 1/50 | Abcam | ab64693 |
| CD86 | Rat | 1/50 | eBioscience | 14-0862-82 |
| CD80 | Rat | 1/50 | eBioscience | 553368 |
| Antigen | Species | Dilutions | Source | Cat. Number |
|---|---|---|---|---|
| F4/80-FITC | Mouse monoclonal | 1/100 | eBioscience | 11–4801–82 |
| CD206–PE–Cy7 | Mouse monoclonal | 1/3600 | eBioscience | 25–2061–82 |
| CD86–PE | Mouse monoclonal | 1/100 | eBioscience | 12–0862–82 |
| CD80–PE | Mouse monoclonal | 1/100 | eBioscience | 16–10A1 |
| CD11b–FITC | Mouse monoclonal | 1/100 | eBioscience | 11–0112–82 |
| CD11b–PE | Mouse monoclonal | 1/100 | eBioscience | 12–0112–83 |
| CD11b-PE-Cy7 | Mouse monoclonal | 1/100 | eBioscience | 25–0112–82 |
| CD45–APC–eFluor 780 | Mouse monoclonal | 1/100 | eBioscience | 47–0451–82 |
| CD3 FITC | Mouse monoclonal | 1/50 | eBioscience | 11–0032–82 |
| CD19 APC | Mouse monoclonal | 1/50 | eBioscience | 17–0193–82 |
| Ly–6G/Ly–6C (Gr1)PerCP-Cyanine 5.5 | Mouse monoclonal | 1/300 | eBioscience | 45-5931–80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Urso, A.; Oltolina, F.; Borsotti, C.; Prat, M.; Colangelo, D.; Follenzi, A. Macrophage Reprogramming via the Modulation of Unfolded Protein Response with siRNA-Loaded Magnetic Nanoparticles in a TAM-like Experimental Model. Pharmaceutics 2023, 15, 1711. https://doi.org/10.3390/pharmaceutics15061711
D’Urso A, Oltolina F, Borsotti C, Prat M, Colangelo D, Follenzi A. Macrophage Reprogramming via the Modulation of Unfolded Protein Response with siRNA-Loaded Magnetic Nanoparticles in a TAM-like Experimental Model. Pharmaceutics. 2023; 15(6):1711. https://doi.org/10.3390/pharmaceutics15061711
Chicago/Turabian StyleD’Urso, Annarita, Francesca Oltolina, Chiara Borsotti, Maria Prat, Donato Colangelo, and Antonia Follenzi. 2023. "Macrophage Reprogramming via the Modulation of Unfolded Protein Response with siRNA-Loaded Magnetic Nanoparticles in a TAM-like Experimental Model" Pharmaceutics 15, no. 6: 1711. https://doi.org/10.3390/pharmaceutics15061711
APA StyleD’Urso, A., Oltolina, F., Borsotti, C., Prat, M., Colangelo, D., & Follenzi, A. (2023). Macrophage Reprogramming via the Modulation of Unfolded Protein Response with siRNA-Loaded Magnetic Nanoparticles in a TAM-like Experimental Model. Pharmaceutics, 15(6), 1711. https://doi.org/10.3390/pharmaceutics15061711








