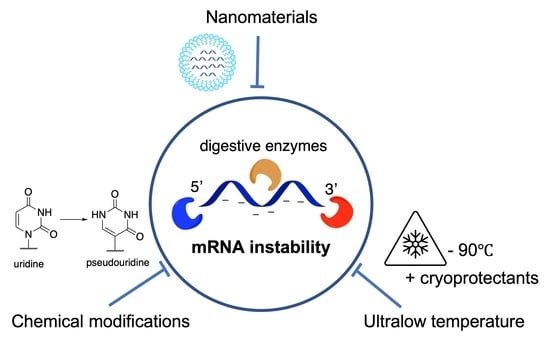
Nanobiotechnology-Enabled mRNA Stabilization
Abstract
1. Introduction
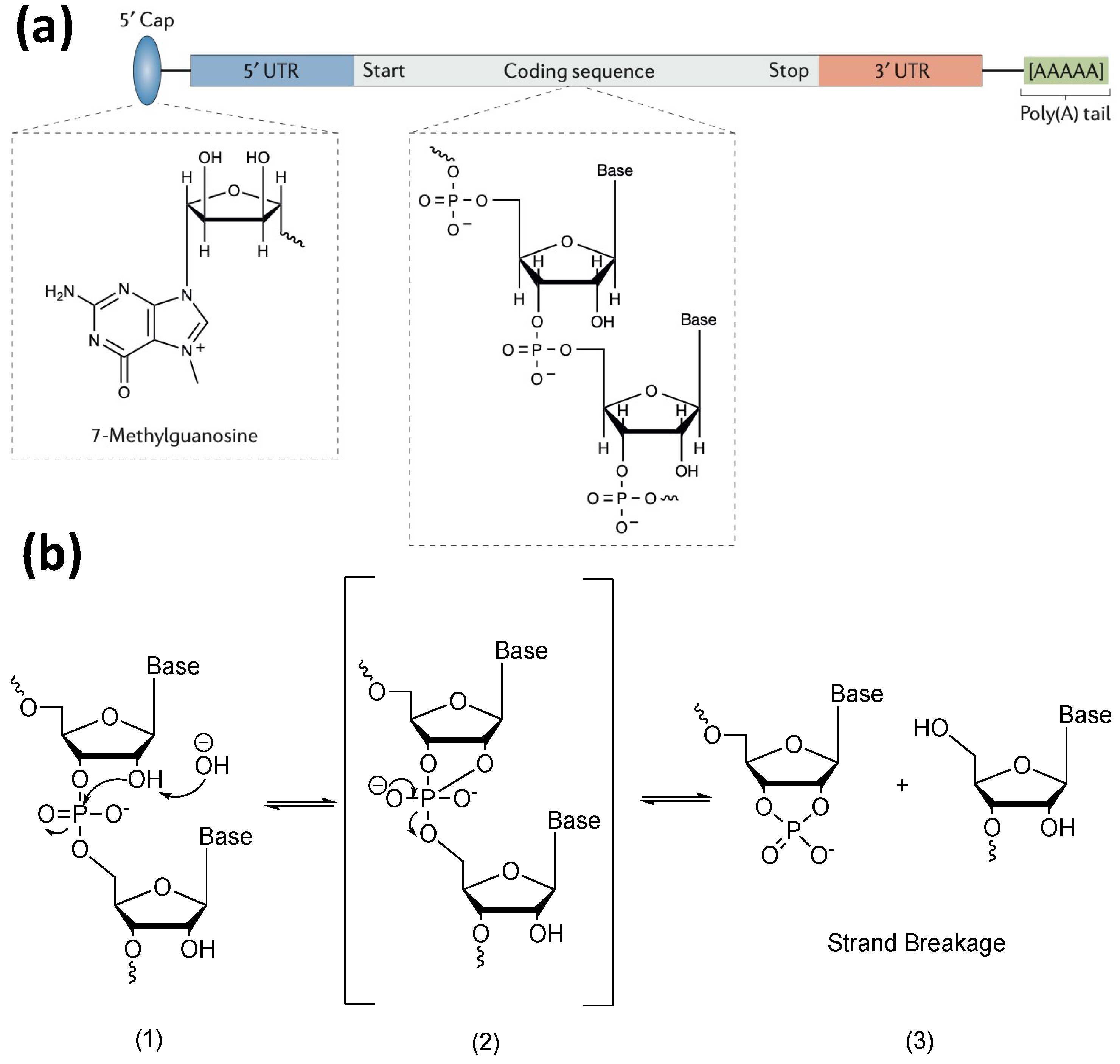
2. Factors Contributing to the Degradation of mRNA
3. Current Strategies for Improving mRNA Stability
3.1. Molecular Design of mRNA
3.1.1. Modification of mRNA Molecular Structure
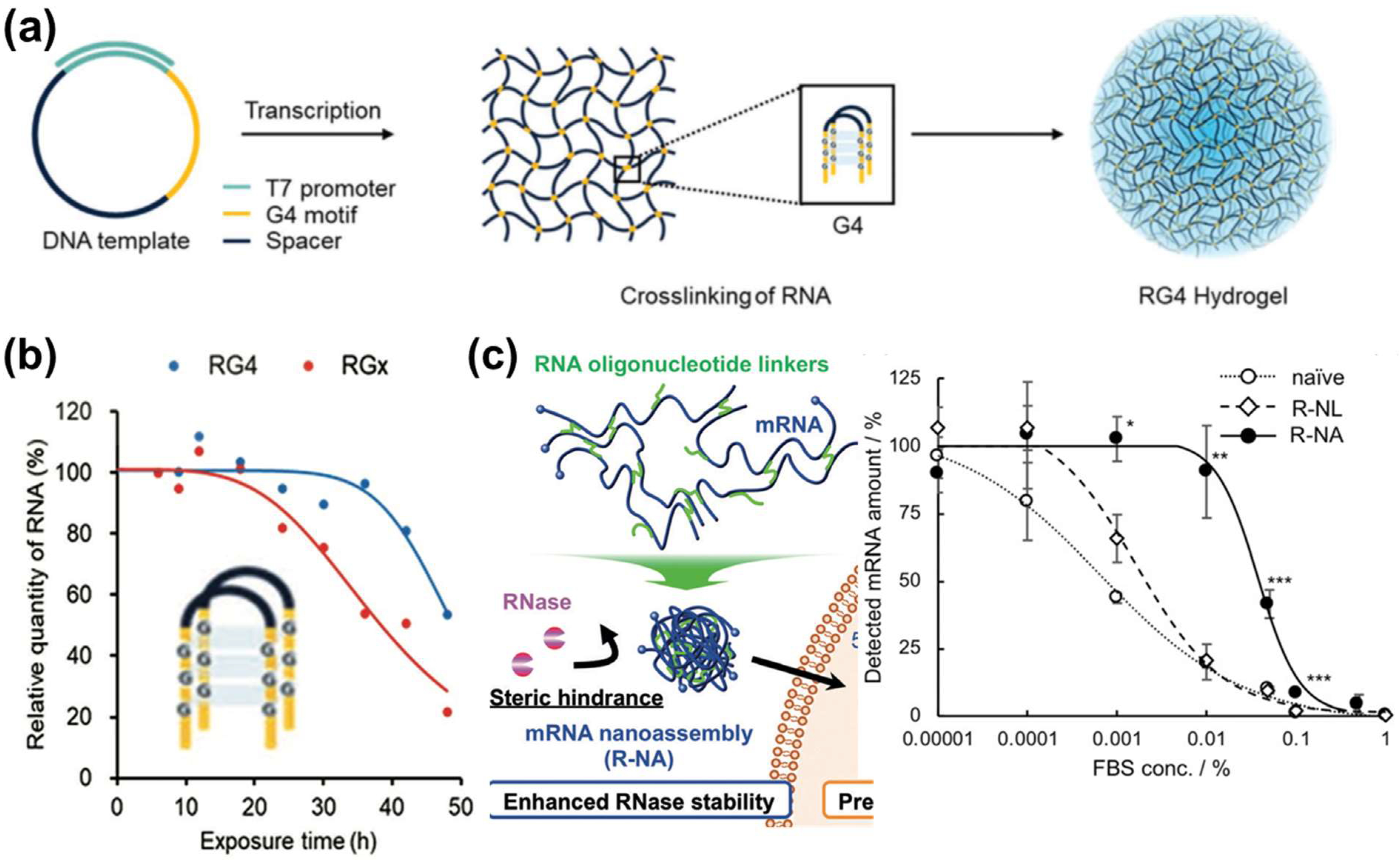
3.1.2. Construction of mRNA Nanoassemblies
3.2. Encapsulation of mRNA via Nanomaterials
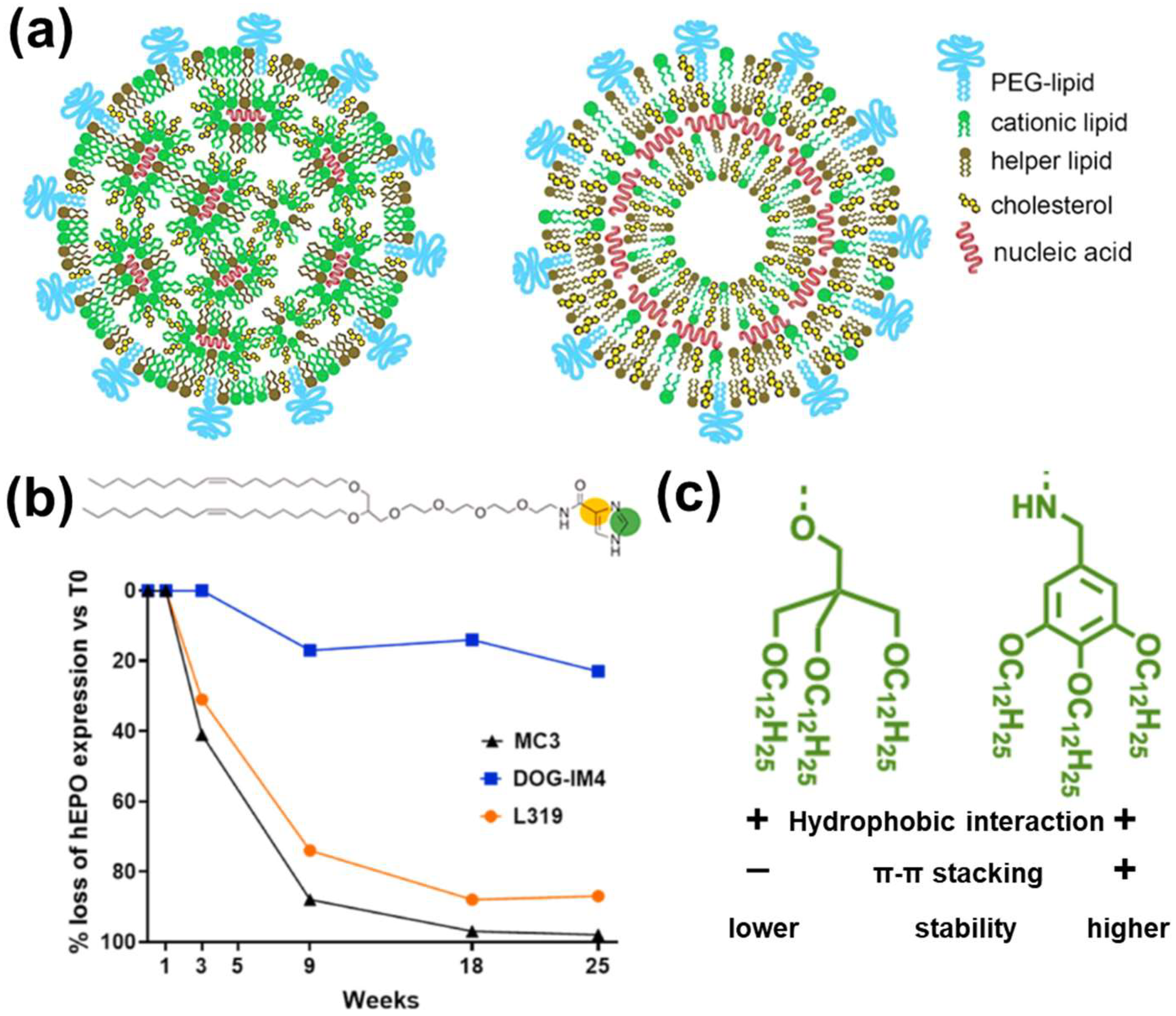
3.2.1. LNPs
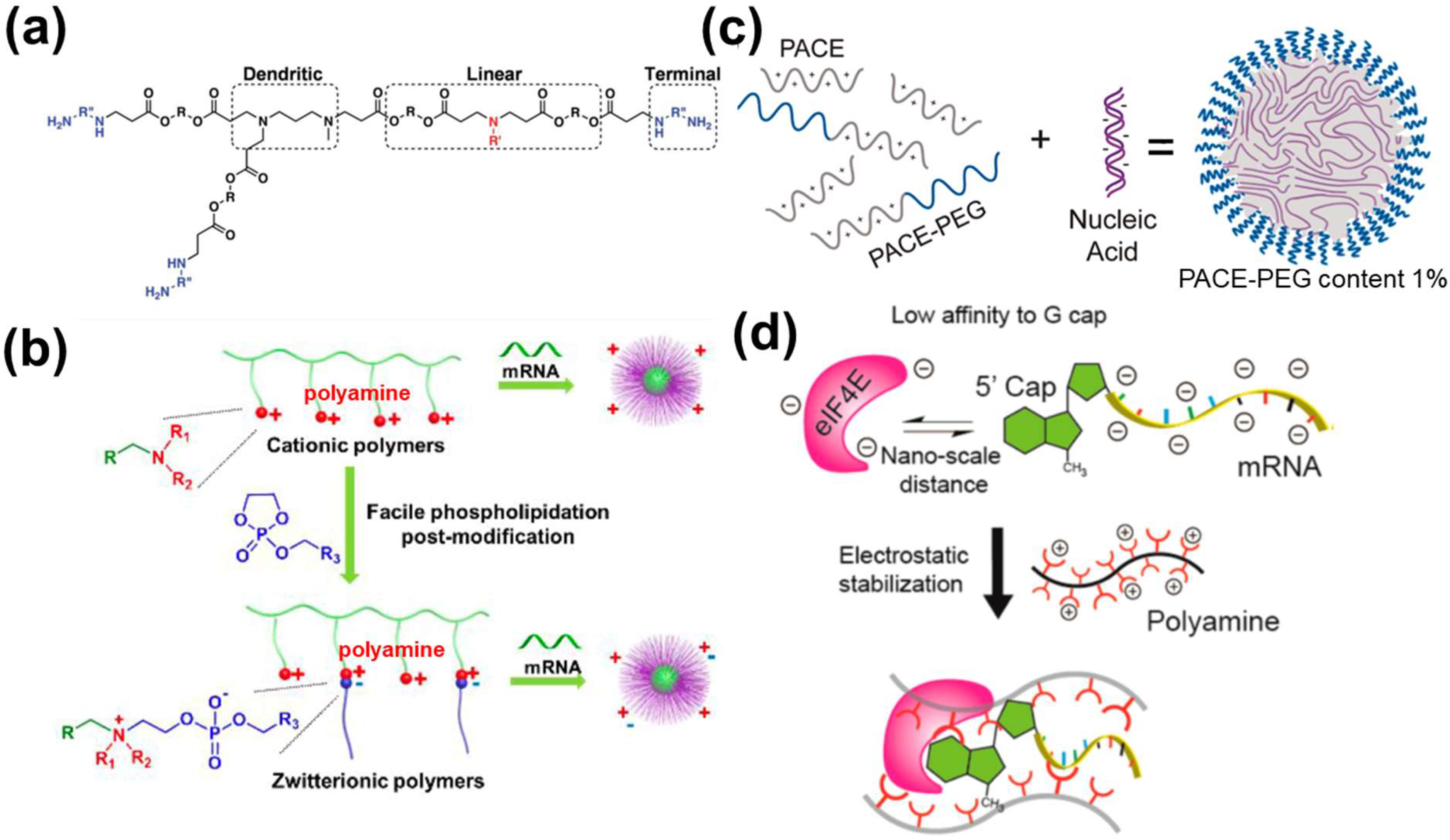
3.2.2. Polymer-Based NPs
3.2.3. Hybrid and Other Strategies
3.3. Other Considerations before Manufacturing
4. Conclusions and Outlook
- (1)
- The biological impact of mRNA with molecular designs is to be evaluated. Although mRNA sequence can be sophisticatedly designed with enhanced stability, comprehensive studies of the bio-distribution and bio-safety evaluations of such exogenous mRNA in vivo or in clinical studies are needed.
- (2)
- The contribution of nanomaterial-enhanced mRNA delivery or stability should be investigated and elucidated. Considering the multiple steps in mRNA delivery, e.g., organ accumulation, cell targeting, endosomal escape, and protein translation, tuning the nanocomposition/structure could also impact those delivery steps in addition to interaction/protection with mRNA molecules. To date, current research generally evaluated the final protein expression level as the indicator to evaluate the so-called protection effect or stability, which may not be accurate. For example, an mRNA molecule may be stable in its structure, but has low efficacy in translation into proteins. Using biotechnology methods such as polymerase chain reaction (PCR) may provide mRNA sequence/structure information to better understand the stability change in various delivery steps and contribution to final enhancement in protein expression.
- (1)
- To tackle base mutation generated by oxidation. For example, in cancer microenvironment, high reactive oxygen species may be a factor leading to mRNA mutation and hindering mRNA function [94]. To date, there are rare reports integrating antioxidant molecules to alleviate the oxidative stress in those mRNA formulations for better protection, which is worth of future study.
- (2)
- To tackle RNases-mediated in-line hydrolysis. Allosteric regulators may be considered to inhibit RNase activity in a nanoformulation [95].
- (3)
- To tackle intracellular mRNA decapping. As discussed in Section 2, mRNA 5′ decapping can be counterbalanced by the binding of translation initiation factors. Despite mixing cationic polymers, translation initiation protein and mRNA have shown to enhance mRNA stability [23]; this strategy is limited to one type of translation initiation protein. It is expected that activating the cellular pathway (mechanistic target of rapamycin complex 1 (mTORC1)) responsible for recruiting multiple translation initiation factors, e.g., eIF4E and eIF4G [15] may provide better performance. Glutamine is a reported biomolecule to activate the mTORC1 [96]. It is expected that incorporating glutamine into nanomaterials may enhance mRNA stability.
- (4)
- For multi-stranded mRNA self-assembly, current building blocks are generally limited to mRNA expressing one type of protein. Future study on design and assembly of mRNA expressing different proteins may be of interest to applications such as multivalent vaccine application.
- (5)
- For lipid-based mRNA formulations, future effort may be devoted to the enhancement of mRNA-LNP structural stability as it prevents mRNA leakage before arriving at the targets. Inspired by current research on control over cationic lipids for tuned surface charge etc., other lipid components such as helper lipids and PEG may be tuned to control the shell structure of lipid for better colloidal and mRNA stability.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Sahin, U.; Kariko, K.; Tureci, O. mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Wu, K.; Choi, A.; Koch, M.; Elbashir, S.; Ma, L.; Lee, D.; Woods, A.; Henry, C.; Palandjian, C.; Hill, A.; et al. Variant SARS-CoV-2 mRNA vaccines confer broad neutralization as primary or booster series in mice. bioRxiv 2021. [Google Scholar] [CrossRef]
- Freyn, A.W.; Ramos da Silva, J.; Rosado, V.C.; Bliss, C.M.; Pine, M.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; de Souza Ferreira, L.C.; Weissman, D.; et al. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020, 28, 1569–1584. [Google Scholar] [CrossRef] [PubMed]
- Kis, Z.; Kontoravdi, C.; Dey, A.K.; Shattock, R.; Shah, N. Rapid development and deployment of high-volume vaccines for pandemic response. J. Adv. Manuf. Process. 2020, 2, e10060. [Google Scholar] [CrossRef]
- Midoux, P.; Pichon, C. Lipid-based mRNA vaccine delivery systems. Expert Rev. Vaccines 2015, 14, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Schlake, T.; Thess, A.; Fotin-Mleczek, M.; Kallen, K.J. Developing mRNA-vaccine technologies. RNA Biol. 2012, 9, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. mRNA vaccines for infectious diseases: Principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021, 20, 817–838. [Google Scholar] [CrossRef] [PubMed]
- Crommelin, D.J.A.; Anchordoquy, T.J.; Volkin, D.B.; Jiskoot, W.; Mastrobattista, E. Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci. 2021, 110, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef] [PubMed]
- Klauer, A.A.; van Hoof, A. Degradation of mRNAs that lack a stop codon: A decade of nonstop progress. Wiley Interdiscip. Rev. RNA 2012, 3, 649–660. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, C. Emerging Concepts of Nanobiotechnology in mRNA Delivery. Angew. Chem. Int. Ed. Engl. 2020, 59, 23374–23385. [Google Scholar] [CrossRef] [PubMed]
- Pogocki, D.; Schoneich, C. Chemical stability of nucleic acid-derived drugs. J. Pharm. Sci. 2000, 89, 443–456. [Google Scholar] [CrossRef]
- Wu, Q.; Medina, S.G.; Kushawah, G.; DeVore, M.L.; Castellano, L.A.; Hand, J.M.; Wright, M.; Bazzini, A.A. Translation affects mRNA stability in a codon-dependent manner in human cells. eLife 2019, 8, e45396. [Google Scholar] [CrossRef]
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef]
- Kose, N.; Fox, J.M.; Sapparapu, G.; Bombardi, R.; Tennekoon, R.N.; de Silva, A.D.; Elbashir, S.M.; Theisen, M.A.; Humphris-Narayanan, E.; Ciaramella, G.; et al. A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against chikungunya infection. Sci. Immunol. 2019, 4, eaaw6647. [Google Scholar] [CrossRef]
- Benner, N.L.; McClellan, R.L.; Turlington, C.R.; Haabeth, O.A.W.; Waymouth, R.M.; Wender, P.A. Oligo (serine ester) Charge-Altering Releasable Transporters: Organocatalytic Ring-Opening Polymerization and their Use for In Vitro and In Vivo mRNA Delivery. J. Am. Chem. Soc. 2019, 141, 8416–8421. [Google Scholar] [CrossRef]
- Parhiz, H.; Shuvaev, V.V.; Pardi, N.; Khoshnejad, M.; Kiseleva, R.Y.; Brenner, J.S.; Uhler, T.; Tuyishime, S.; Mui, B.L.; Tam, Y.K.; et al. PECAM-1 directed re-targeting of exogenous mRNA providing two orders of magnitude enhancement of vascular delivery and expression in lungs independent of apolipoprotein E-mediated uptake. J. Control. Release 2018, 291, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Xu, Y.; Tao, W.; Ubellacker, J.M.; Lim, M.; Aum, D.; Lee, G.Y.; Zhou, K.; Zope, H.; Yu, M.; et al. Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat. Biomed. Eng. 2018, 2, 850–864. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, W.; He, Y.; Li, Y.; Yan, E.Z.; Zhang, K.; Irvine, D.J.; Hammond, P.T. Structurally Programmed Assembly of Translation Initiation Nanoplex for Superior mRNA Delivery. ACS Nano 2017, 11, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Tang, Z.; Huang, X.; Chen, W.; Zhou, J.; Liu, H.; Liu, C.; Kong, N.; Tao, W. Emerging mRNA technologies: Delivery strategies and biomedical applications. Chem. Soc. Rev. 2022, 51, 3828–3845. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.A.; Neshat, S.Y.; Green, J.J.; Santos, J.L.; Tuesca, A.D. Targeting strategies for mRNA delivery. Mater. Today Adv. 2022, 14, 100240. [Google Scholar] [CrossRef]
- Ho, W.; Gao, M.; Li, F.; Li, Z.; Zhang, X.Q.; Xu, X. Next-Generation Vaccines: Nanoparticle-Mediated DNA and mRNA Delivery. Adv. Healthc. Mater. 2021, 10, e2001812. [Google Scholar] [CrossRef]
- Uddin, M.N.; Roni, M.A. Challenges of Storage and Stability of mRNA-Based COVID-19 Vaccines. Vaccines 2021, 9, 1033. [Google Scholar] [CrossRef]
- Pei, H.; Deng, H.Z.; Zhou, Y.F.; Chen, X.Y. The roles of polymers in mRNA delivery. Matter 2022, 5, 1670–1699. [Google Scholar] [CrossRef]
- Houseley, J.; Tollervey, D. The many pathways of RNA degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef]
- Jaquet, V.; Wallerich, S.; Voegeli, S.; Turos, D.; Viloria, E.C.; Becskei, A. Determinants of the temperature adaptation of mRNA degradation. Nucleic Acids Res. 2022, 50, 1092–1110. [Google Scholar] [CrossRef]
- Zeng, W.; Yu, M.; Chen, T.; Liu, Y.; Yi, Y.; Huang, C.; Tang, J.; Li, H.; Ou, M.; Wang, T.; et al. Polypyrrole Nanoenzymes as Tumor Microenvironment Modulators to Reprogram Macrophage and Potentiate Immunotherapy. Adv. Sci. 2022, 9, e2201703. [Google Scholar] [CrossRef] [PubMed]
- Simms, C.L.; Yan, L.L.; Zaher, H.S. Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol. Cell 2017, 68, 361–373.e5. [Google Scholar] [CrossRef] [PubMed]
- Dirisala, A.; Uchida, S.; Tockary, T.A.; Yoshinaga, N.; Li, J.; Osawa, S.; Gorantla, L.; Fukushima, S.; Osada, K.; Kataoka, K. Precise tuning of disulphide crosslinking in mRNA polyplex micelles for optimising extracellular and intracellular nuclease tolerability. J. Drug Target 2019, 27, 670–680. [Google Scholar] [CrossRef] [PubMed]
- Yen, A.; Cheng, Y.; Sylvestre, M.; Gustafson, H.H.; Puri, S.; Pun, S.H. Serum Nuclease Susceptibility of mRNA Cargo in Condensed Polyplexes. Mol. Pharm. 2018, 15, 2268–2276. [Google Scholar] [CrossRef]
- Lin, Y.; Wagner, E.; Lachelt, U. Non-viral delivery of the CRISPR/Cas system: DNA versus RNA versus RNP. Biomater. Sci. 2022, 10, 1166–1192. [Google Scholar] [CrossRef]
- Leppek, K.; Byeon, G.W.; Kladwang, W.; Wayment-Steele, H.K.; Kerr, C.H.; Xu, A.F.; Kim, D.S.; Topkar, V.V.; Choe, C.; Rothschild, D.; et al. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Nat. Commun. 2022, 13, 1536. [Google Scholar] [CrossRef]
- Poole, T.L.; Stevens, A. Structural modifications of RNA influence the 5′ exoribonucleolytic hydrolysis by XRN1 and HKE1 of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1997, 235, 799–805. [Google Scholar] [CrossRef]
- Grudzien-Nogalska, E.; Kiledjian, M. New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscip. Rev. RNA 2017, 8, e1379. [Google Scholar] [CrossRef]
- Meyer, S.; Temme, C.; Wahle, E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 197–216. [Google Scholar] [CrossRef]
- Edri, S.; Tuller, T. Quantifying the effect of ribosomal density on mRNA stability. PLoS ONE 2014, 9, e102308. [Google Scholar] [CrossRef]
- Juszkiewicz, S.; Slodkowicz, G.; Lin, Z.; Freire-Pritchett, P.; Peak-Chew, S.Y.; Hegde, R.S. Ribosome collisions trigger cis-acting feedback inhibition of translation initiation. eLife 2020, 9, e60038. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 2020, 182, 404–416.e14. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, Y.; Doi, Y.; Hosoda, N.; Uchida, N.; Osawa, M.; Shimada, I.; Tsujimoto, M.; Suzuki, T.; Katada, T.; Hoshino, S. Mechanism of mRNA deadenylation: Evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007, 21, 3135–3148. [Google Scholar] [CrossRef] [PubMed]
- Hajj, K.A.; Whitehead, K.A. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017, 2, 17056. [Google Scholar] [CrossRef]
- Wojtczak, B.A.; Sikorski, P.J.; Fac-Dabrowska, K.; Nowicka, A.; Warminski, M.; Kubacka, D.; Nowak, E.; Nowotny, M.; Kowalska, J.; Jemielity, J. 5′-Phosphorothiolate Dinucleotide Cap Analogues: Reagents for Messenger RNA Modification and Potent Small-Molecular Inhibitors of Decapping Enzymes. J. Am. Chem. Soc. 2018, 140, 5987–5999. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Passmore, L.A.; Coller, J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell. Biol. 2022, 23, 93–106. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Zhang, J.; Kuo, C.C.; Chen, L. GC content around splice sites affects splicing through pre-mRNA secondary structures. BMC Genom. 2011, 12, 90. [Google Scholar] [CrossRef]
- Wayment-Steele, H.K.; Kim, D.S.; Choe, C.A.; Nicol, J.J.; Wellington-Oguri, R.; Watkins, A.M.; Parra Sperberg, R.A.; Huang, P.S.; Participants, E.; Das, R. Theoretical basis for stabilizing messenger RNA through secondary structure design. Nucleic Acids Res. 2021, 49, 10604–10617. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.Y.; Kim, J.; Vellampatti, S.; Oh, S.; Lim, Y.T.; Park, S.H.; Luo, D.; Chung, J.; Um, S.H. Protein-Encoding Free-Standing RNA Hydrogel for Sub-Compartmentalized Translation. Adv. Mater. 2022, 34, e2110424. [Google Scholar] [CrossRef] [PubMed]
- Poliskey, J.A.; Crowley, S.T.; Ramanathan, R.; White, C.W.; Mathew, B.; Rice, K.G. Metabolically stabilized double-stranded mRNA polyplexes. Gene Ther. 2018, 25, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, N.; Cho, E.; Koji, K.; Mochida, Y.; Naito, M.; Osada, K.; Kataoka, K.; Cabral, H.; Uchida, S. Bundling mRNA Strands to Prepare Nano-Assemblies with Enhanced Stability Towards RNase for In Vivo Delivery. Angew. Chem. Int. Ed. Engl. 2019, 58, 11360–11363. [Google Scholar] [CrossRef] [PubMed]
- Uzgun, S.; Nica, G.; Pfeifer, C.; Bosinco, M.; Michaelis, K.; Lutz, J.F.; Schneider, M.; Rosenecker, J.; Rudolph, C. PEGylation improves nanoparticle formation and transfection efficiency of messenger RNA. Pharm. Res. 2011, 28, 2223–2232. [Google Scholar] [CrossRef]
- Patel, A.K.; Kaczmarek, J.C.; Bose, S.; Kauffman, K.J.; Mir, F.; Heartlein, M.W.; DeRosa, F.; Langer, R.; Anderson, D.G. Inhaled Nanoformulated mRNA Polyplexes for Protein Production in Lung Epithelium. Adv. Mater. 2019, 31, e1805116. [Google Scholar] [CrossRef]
- Liu, S.; Wang, X.; Yu, X.; Cheng, Q.; Johnson, L.T.; Chatterjee, S.; Zhang, D.; Lee, S.M.; Sun, Y.; Lin, T.C.; et al. Zwitterionic Phospholipidation of Cationic Polymers Facilitates Systemic mRNA Delivery to Spleen and Lymph Nodes. J. Am. Chem. Soc. 2021, 143, 21321–21330. [Google Scholar] [CrossRef]
- Grun, M.K.; Suberi, A.; Shin, K.; Lee, T.; Gomerdinger, V.; Moscato, Z.M.; Piotrowski-Daspit, A.S.; Saltzman, W.M. PEGylation of poly(amine-co-ester) polyplexes for tunable gene delivery. Biomaterials 2021, 272, 120780. [Google Scholar] [CrossRef]
- Dirisala, A.; Uchida, S.; Li, J.; Van Guyse, J.F.R.; Hayashi, K.; Vummaleti, S.V.C.; Kaur, S.; Mochida, Y.; Fukushima, S.; Kataoka, K. Effective mRNA Protection by Poly(l-ornithine) Synergizes with Endosomal Escape Functionality of a Charge-Conversion Polymer toward Maximizing mRNA Introduction Efficiency. Macromol. Rapid Commun. 2022, 43, e2100754. [Google Scholar] [CrossRef]
- Yoshinaga, N.; Uchida, S.; Dirisala, A.; Naito, M.; Osada, K.; Cabral, H.; Kataoka, K. mRNA loading into ATP-responsive polyplex micelles with optimal density of phenylboronate ester crosslinking to balance robustness in the biological milieu and intracellular translational efficiency. J. Control. Release 2021, 330, 317–328. [Google Scholar] [CrossRef]
- Jackson, M.A.; Werfel, T.A.; Curvino, E.J.; Yu, F.; Kavanaugh, T.E.; Sarett, S.M.; Dockery, M.D.; Kilchrist, K.V.; Jackson, A.N.; Giorgio, T.D.; et al. Zwitterionic Nanocarrier Surface Chemistry Improves siRNA Tumor Delivery and Silencing Activity Relative to Polyethylene Glycol. ACS Nano 2017, 11, 5680–5696. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, Q.; Jiang, Q.; Huang, Y.; Liu, H.; Zhao, Y.; Cao, W.; Ma, G.; Dai, F.; Liang, X.; et al. Enhanced endosomal/lysosomal escape by distearoyl phosphoethanolamine-polycarboxybetaine lipid for systemic delivery of siRNA. J. Control. Release 2014, 176, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Siegwart, D.J.; Whitehead, K.A.; Nuhn, L.; Sahay, G.; Cheng, H.; Jiang, S.; Ma, M.; Lytton-Jean, A.; Vegas, A.; Fenton, P.; et al. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proc. Natl. Acad. Sci. USA 2011, 108, 12996–13001. [Google Scholar] [CrossRef] [PubMed]
- Moghimi, S.M.; Symonds, P.; Murray, J.C.; Hunter, A.C.; Debska, G.; Szewczyk, A. A two-stage poly(ethylenimine)-mediated cytotoxicity: Implications for gene transfer/therapy. Mol. Ther. 2005, 11, 990–995. [Google Scholar] [CrossRef]
- Jiang, Y.; Gaudin, A.; Zhang, J.; Agarwal, T.; Song, E.; Kauffman, A.C.; Tietjen, G.T.; Wang, Y.; Jiang, Z.; Cheng, C.J.; et al. A “top-down” approach to actuate poly(amine-co-ester) terpolymers for potent and safe mRNA delivery. Biomaterials 2018, 176, 122–130. [Google Scholar] [CrossRef]
- Uchida, H.; Itaka, K.; Nomoto, T.; Ishii, T.; Suma, T.; Ikegami, M.; Miyata, K.; Oba, M.; Nishiyama, N.; Kataoka, K. Modulated protonation of side chain aminoethylene repeats in N-substituted polyaspartamides promotes mRNA transfection. J. Am. Chem. Soc. 2014, 136, 12396–12405. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, J.C.; Patel, A.K.; Kauffman, K.J.; Fenton, O.S.; Webber, M.J.; Heartlein, M.W.; DeRosa, F.; Anderson, D.G. Polymer-Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew. Chem. Int. Ed. Engl. 2016, 55, 13808–13812. [Google Scholar] [CrossRef]
- Jia, F.; Huang, W.; Yin, Y.; Jiang, Y.; Yang, Q.; Huang, H.; Nie, G.; Wang, H. Stabilizing RNA Nanovaccines with Transformable Hyaluronan Dynamic Hydrogel for Durable Cancer Immunotherapy. Adv. Funct. Mater. 2022, 33, 2204636. [Google Scholar] [CrossRef]
- Gao, C.; Cheng, K.; Li, Y.; Gong, R.; Zhao, X.; Nie, G.; Ren, H. Injectable Immunotherapeutic Hydrogel Containing RNA-Loaded Lipid Nanoparticles Reshapes Tumor Microenvironment for Pancreatic Cancer Therapy. Nano Lett. 2022, 22, 8801–8809. [Google Scholar] [CrossRef]
- Yin, Y.; Li, X.; Ma, H.; Zhang, J.; Yu, D.; Zhao, R.; Yu, S.; Nie, G.; Wang, H. In Situ Transforming RNA Nanovaccines from Polyethylenimine Functionalized Graphene Oxide Hydrogel for Durable Cancer Immunotherapy. Nano Lett. 2021, 21, 2224–2231. [Google Scholar] [CrossRef]
- Zhao, P.; Hou, X.; Yan, J.; Du, S.; Xue, Y.; Li, W.; Xiang, G.; Dong, Y. Long-term storage of lipid-like nanoparticles for mRNA delivery. Bioact. Mater. 2020, 5, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, H.; Lam, K.; Bajusz, C.; Laczko, D.; Kariko, K.; Schreiner, P.; Martin, A.; Lutwyche, P.; Heyes, J.; Pardi, N. Lyophilization provides long-term stability for a lipid nanoparticle-formulated, nucleoside-modified mRNA vaccine. Mol. Ther. 2022, 30, 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Anchordoquy, T.J.; Allison, S.D.; Molina, M.C.; Girouard, L.G.; Carson, T.K. Physical stabilization of DNA-based therapeutics. Drug Discov. Today 2001, 6, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Boafo, G.F.; Magar, K.T.; Ekpo, M.D.; Qian, W.; Tan, S.W.; Chen, C.P. The Role of Cryoprotective Agents in Liposome Stabilization and Preservation. Int. J. Mol. Sci. 2022, 23, 12487. [Google Scholar] [CrossRef]
- Ripoll, M.; Bernard, M.C.; Vaure, C.; Bazin, E.; Commandeur, S.; Perkov, V.; Lemdani, K.; Nicolai, M.C.; Bonifassi, P.; Kichler, A.; et al. An imidazole modified lipid confers enhanced mRNA-LNP stability and strong immunization properties in mice and non-human primates. Biomaterials 2022, 286, 121570. [Google Scholar] [CrossRef]
- Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014, 24, 374–387. [Google Scholar] [CrossRef]
- Iwamoto, N.; Butler, D.C.D.; Svrzikapa, N.; Mohapatra, S.; Zlatev, I.; Sah, D.W.Y.; Meena; Standley, S.M.; Lu, G.; Apponi, L.H.; et al. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat. Biotechnol. 2017, 35, 845–851. [Google Scholar] [CrossRef]
- Anderson, B.R.; Muramatsu, H.; Jha, B.K.; Silverman, R.H.; Weissman, D.; Kariko, K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011, 39, 9329–9338. [Google Scholar] [CrossRef]
- Morais, P.; Adachi, H.; Yu, Y.T. The Critical Contribution of Pseudouridine to mRNA COVID-19 Vaccines. Front. Cell Dev. Biol. 2021, 9, 789427. [Google Scholar] [CrossRef]
- Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Dewitte, H. Three decades of messenger RNA vaccine development. Nano Today 2019, 28, 100766. [Google Scholar] [CrossRef]
- Tenchov, R.; Bird, R.; Curtze, A.E.; Zhou, Q. Lipid Nanoparticles horizontal line from Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. [Google Scholar] [CrossRef] [PubMed]
- Kranz, L.M.; Diken, M.; Haas, H.; Kreiter, S.; Loquai, C.; Reuter, K.C.; Meng, M.; Fritz, D.; Vascotto, F.; Hefesha, H.; et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Atochina-Vasserman, E.N.; Maurya, D.S.; Huang, N.; Xiao, Q.; Ona, N.; Liu, M.; Shahnawaz, H.; Ni, H.; Kim, K.; et al. One-Component Multifunctional Sequence-Defined Ionizable Amphiphilic Janus Dendrimer Delivery Systems for mRNA. J. Am. Chem. Soc. 2021, 143, 12315–12327. [Google Scholar] [CrossRef]
- Alsaiari, S.K.; Patil, S.; Alyami, M.; Alamoudi, K.O.; Aleisa, F.A.; Merzaban, J.S.; Li, M.; Khashab, N.M. Endosomal Escape and Delivery of CRISPR/Cas9 Genome Editing Machinery Enabled by Nanoscale Zeolitic Imidazolate Framework. J. Am. Chem. Soc. 2018, 140, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Lokugamage, M.P.; Vanover, D.; Beyersdorf, J.; Hatit, M.Z.C.; Rotolo, L.; Echeverri, E.S.; Peck, H.E.; Ni, H.; Yoon, J.K.; Kim, Y.; et al. Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat. Biomed. Eng. 2021, 5, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Paunovska, K.; Da Silva Sanchez, A.J.; Sago, C.D.; Gan, Z.; Lokugamage, M.P.; Islam, F.Z.; Kalathoor, S.; Krupczak, B.R.; Dahlman, J.E. Nanoparticles Containing Oxidized Cholesterol Deliver mRNA to the Liver Microenvironment at Clinically Relevant Doses. Adv. Mater. 2019, 31, e1807748. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Lokugamage, M.P.; Hatit, M.Z.C.; Loughrey, D.; Paunovska, K.; Sato, M.; Cristian, A.; Dahlman, J.E. Nanoparticles containing constrained phospholipids deliver mRNA to liver immune cells in vivo without targeting ligands. Bioeng. Transl. Med. 2020, 5, e10161. [Google Scholar] [CrossRef]
- Ball, R.L.; Hajj, K.A.; Vizelman, J.; Bajaj, P.; Whitehead, K.A. Lipid Nanoparticle Formulations for Enhanced Co-delivery of siRNA and mRNA. Nano Lett. 2018, 18, 3814–3822. [Google Scholar] [CrossRef]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef]
- Yoshinaga, N.; Uchida, S.; Dirisala, A.; Naito, M.; Koji, K.; Osada, K.; Cabral, H.; Kataoka, K. Bridging mRNA and Polycation Using RNA Oligonucleotide Derivatives Improves the Robustness of Polyplex Micelles for Efficient mRNA Delivery. Adv. Healthc. Mater. 2022, 11, e2102016. [Google Scholar] [CrossRef]
- Rezaee, M.; Oskuee, R.K.; Nassirli, H.; Malaekeh-Nikouei, B. Progress in the development of lipopolyplexes as efficient non-viral gene delivery systems. J. Control. Release 2016, 236, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ball, R.L.; Bajaj, P.; Whitehead, K.A. Achieving long-term stability of lipid nanoparticles: Examining the effect of pH, temperature, and lyophilization. Int. J. Nanomed. 2017, 12, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Han, D.; Cai, C.; Tang, X. An overview of liposome lyophilization and its future potential. J. Control. Release 2010, 142, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, F.; Ramnath, N.; Nagrath, D. Reactive Oxygen Species in the Tumor Microenvironment: An Overview. Cancers 2019, 11, 1191. [Google Scholar] [CrossRef]
- Ghosh, R.; Wang, L.; Wang, E.S.; Perera, B.G.; Igbaria, A.; Morita, S.; Prado, K.; Thamsen, M.; Caswell, D.; Macias, H.; et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 2014, 158, 534–548. [Google Scholar] [CrossRef]
- Choi, Y.K.; Park, K.G. Targeting Glutamine Metabolism for Cancer Treatment. Biomol. Ther. 2018, 26, 19–28. [Google Scholar] [CrossRef]


| Factors Destabilizing mRNA | Possible Counteracting Strategies | Reference |
|---|---|---|
| RNase-mediated in-line hydrolysis |
| [4,5,11,13,18,27,45,46,47,48,49] [50,51,52,53,54] [22,23,28,33,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70] |
| High temperature | Freezing or lyophilization of nanoformualtions with the addition of cryoprotectants | [16,29,30,71,72,73,74] |
| Oxidative species | Introducing chemical groups (e.g., imidazole) with antioxidant properties | [75] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xian, H.; Zhang, Y.; Yu, C.; Wang, Y. Nanobiotechnology-Enabled mRNA Stabilization. Pharmaceutics 2023, 15, 620. https://doi.org/10.3390/pharmaceutics15020620
Xian H, Zhang Y, Yu C, Wang Y. Nanobiotechnology-Enabled mRNA Stabilization. Pharmaceutics. 2023; 15(2):620. https://doi.org/10.3390/pharmaceutics15020620
Chicago/Turabian StyleXian, He, Yue Zhang, Chengzhong Yu, and Yue Wang. 2023. "Nanobiotechnology-Enabled mRNA Stabilization" Pharmaceutics 15, no. 2: 620. https://doi.org/10.3390/pharmaceutics15020620
APA StyleXian, H., Zhang, Y., Yu, C., & Wang, Y. (2023). Nanobiotechnology-Enabled mRNA Stabilization. Pharmaceutics, 15(2), 620. https://doi.org/10.3390/pharmaceutics15020620