New Insights into the LANCL2-ABA Binding Mode towards the Evaluation of New LANCL Agonists
Abstract
:1. Introduction
2. Materials and Methods
2.1. Computational Studies
2.1.1. Modelling of the Missing LANCL2 Residues
2.1.2. Molecular Docking Studies
2.1.3. MD Simulations
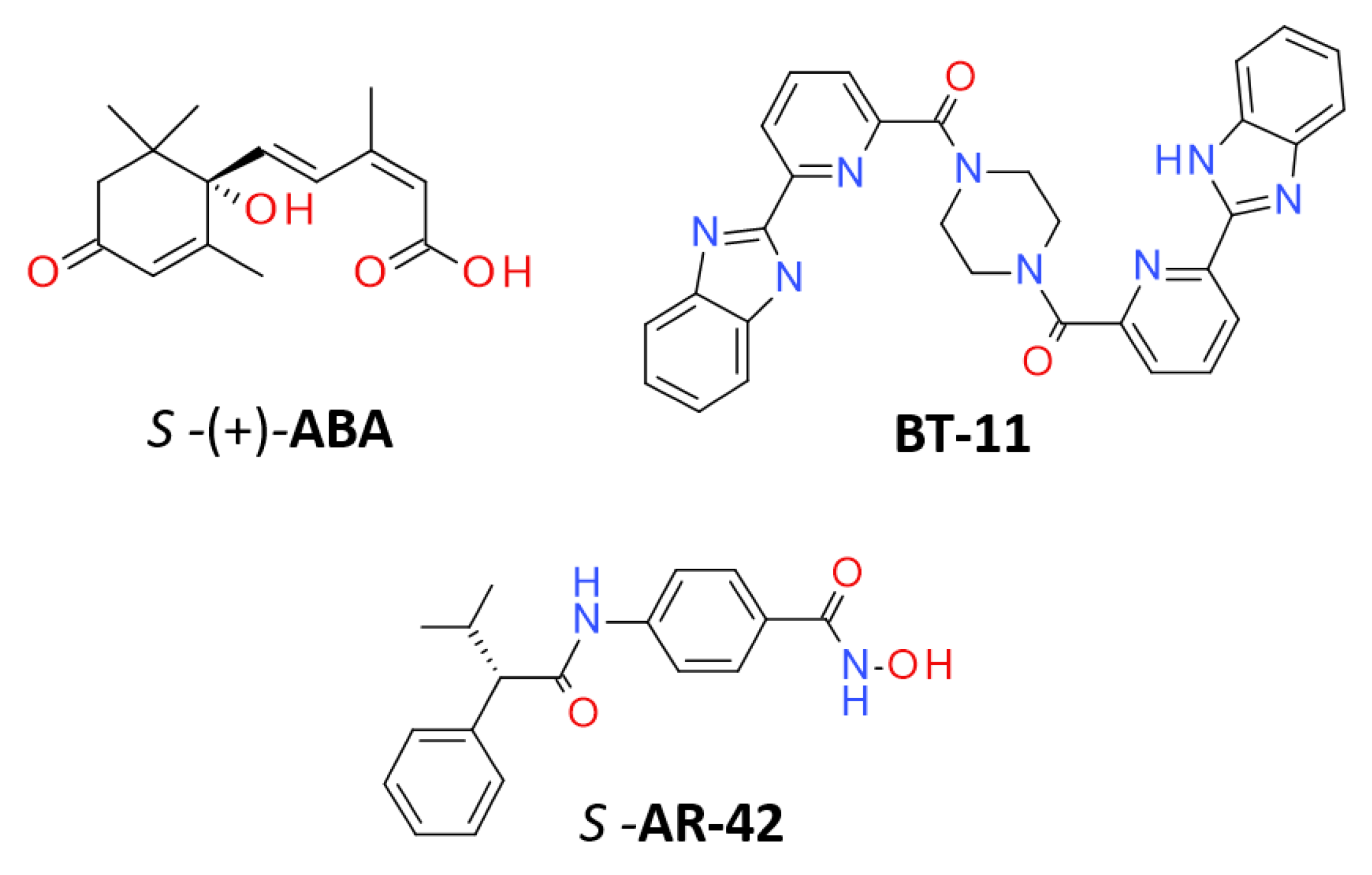
2.2. Chemical Structures
2.3. Biochemical Assays
2.3.1. Cell Culture
2.3.2. Cell Vitality Assay
2.3.3. JC-1 Analysis
2.3.4. qPCR Analysis
3. Results
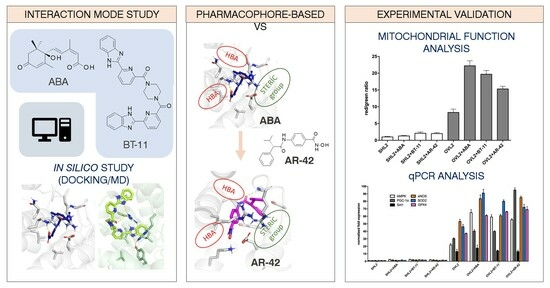
3.1. Computational Studies
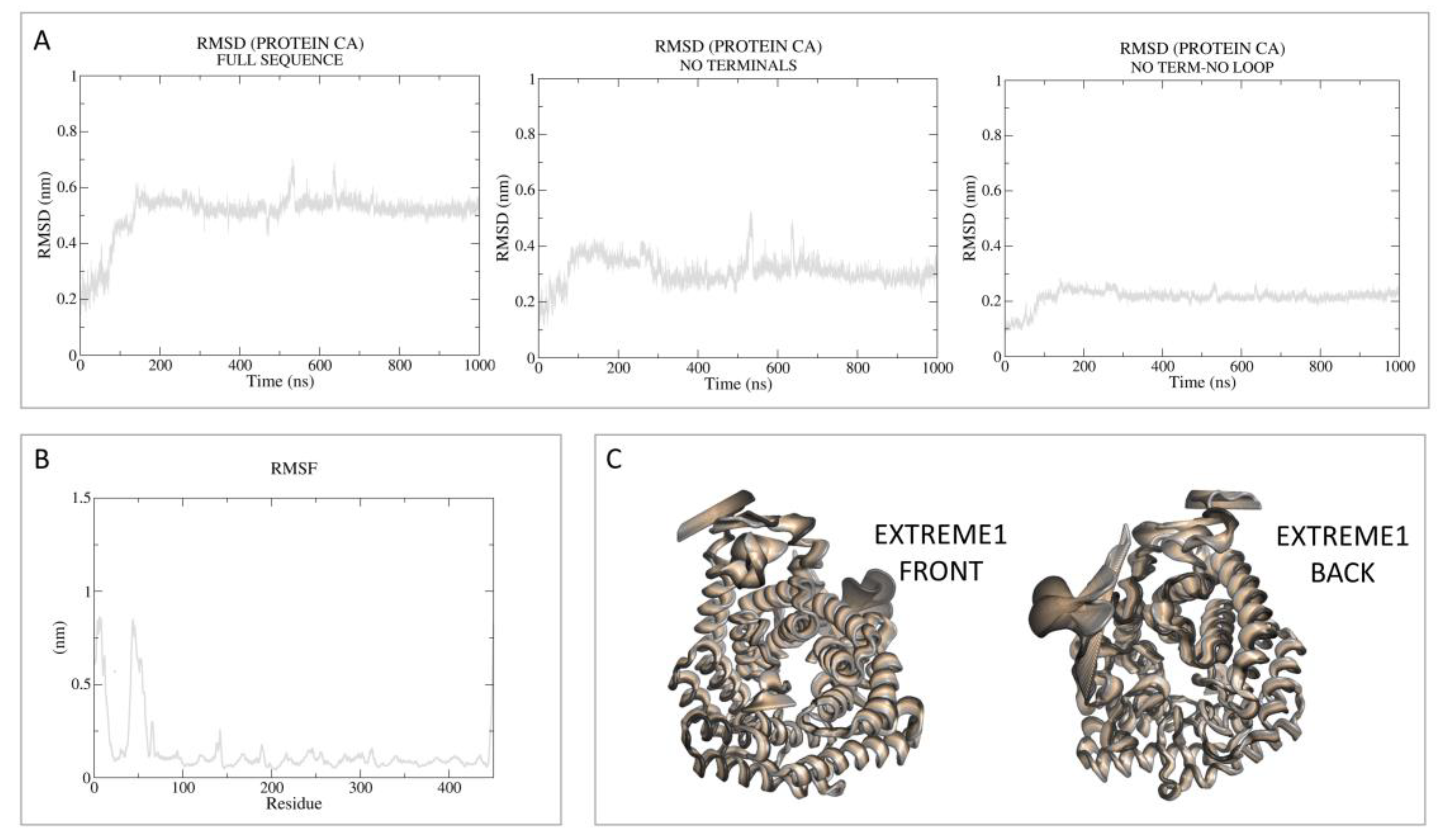
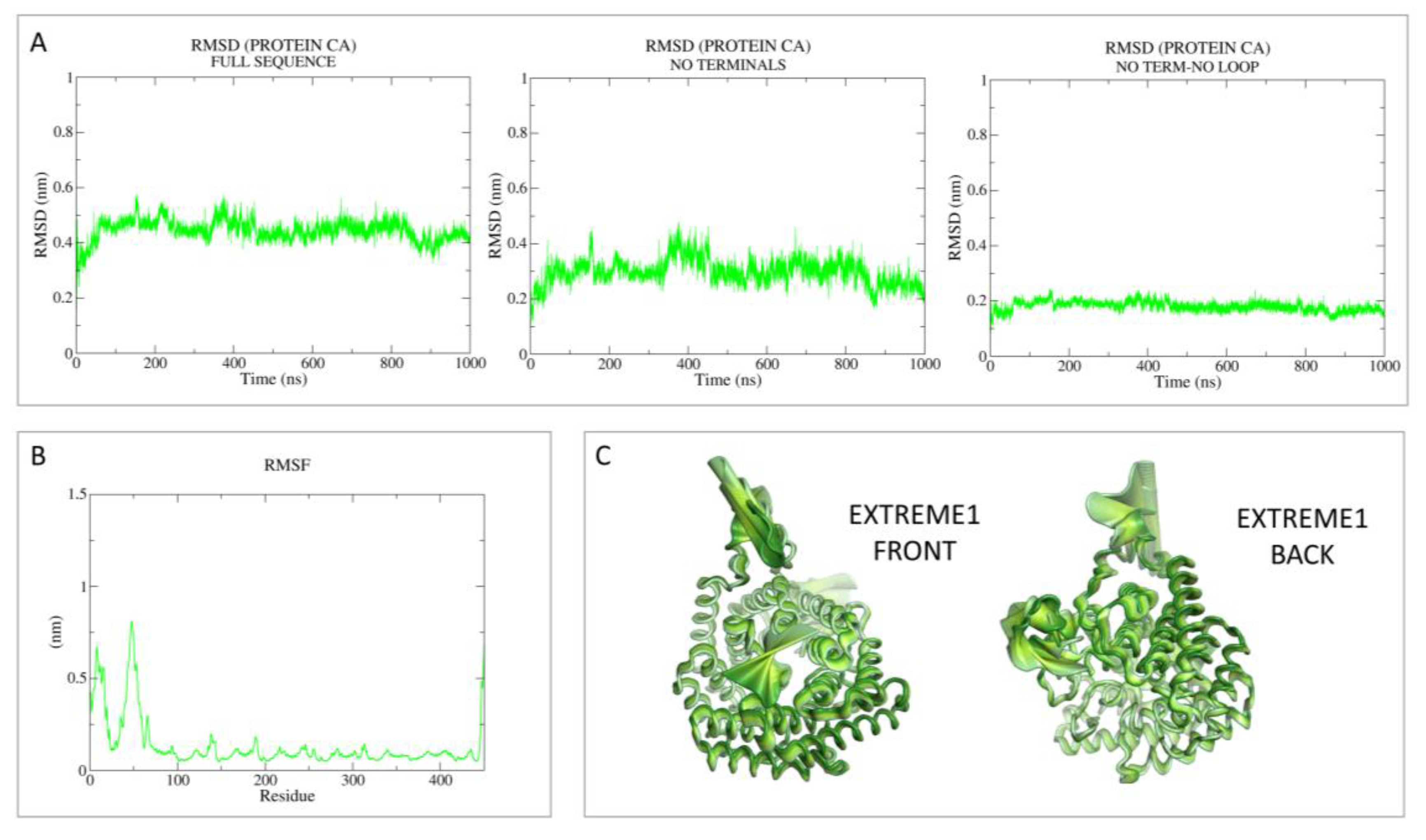
3.1.1. Apo-LANCL2 MD Simulations
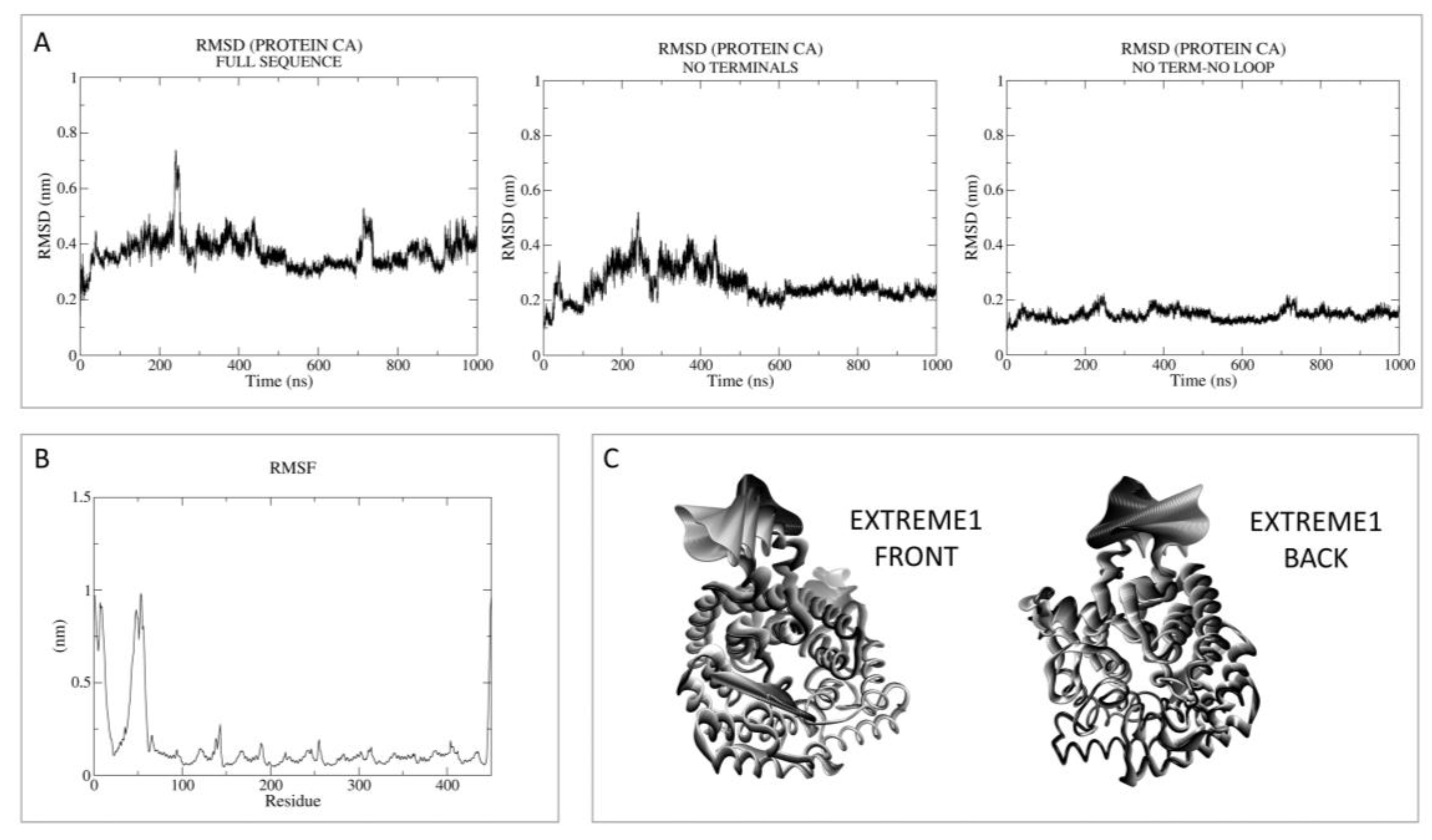
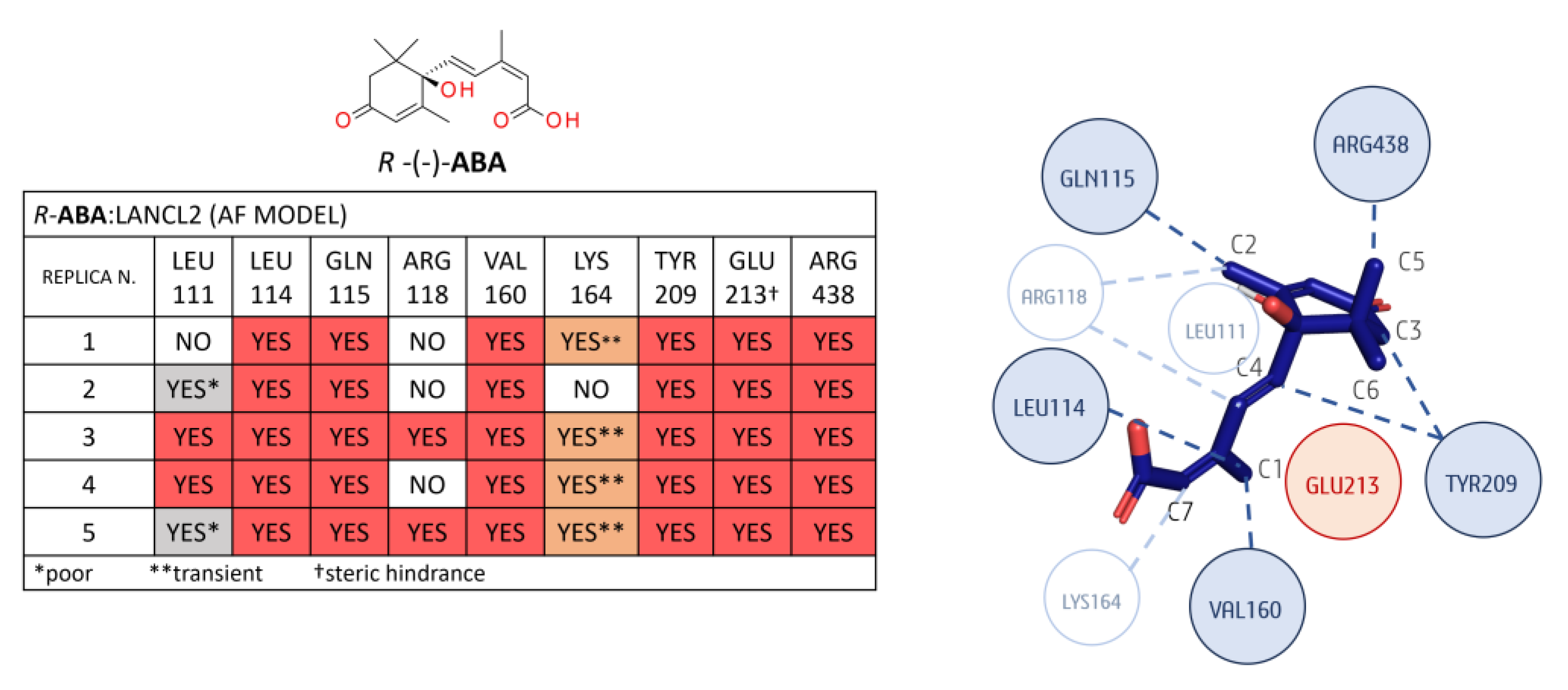
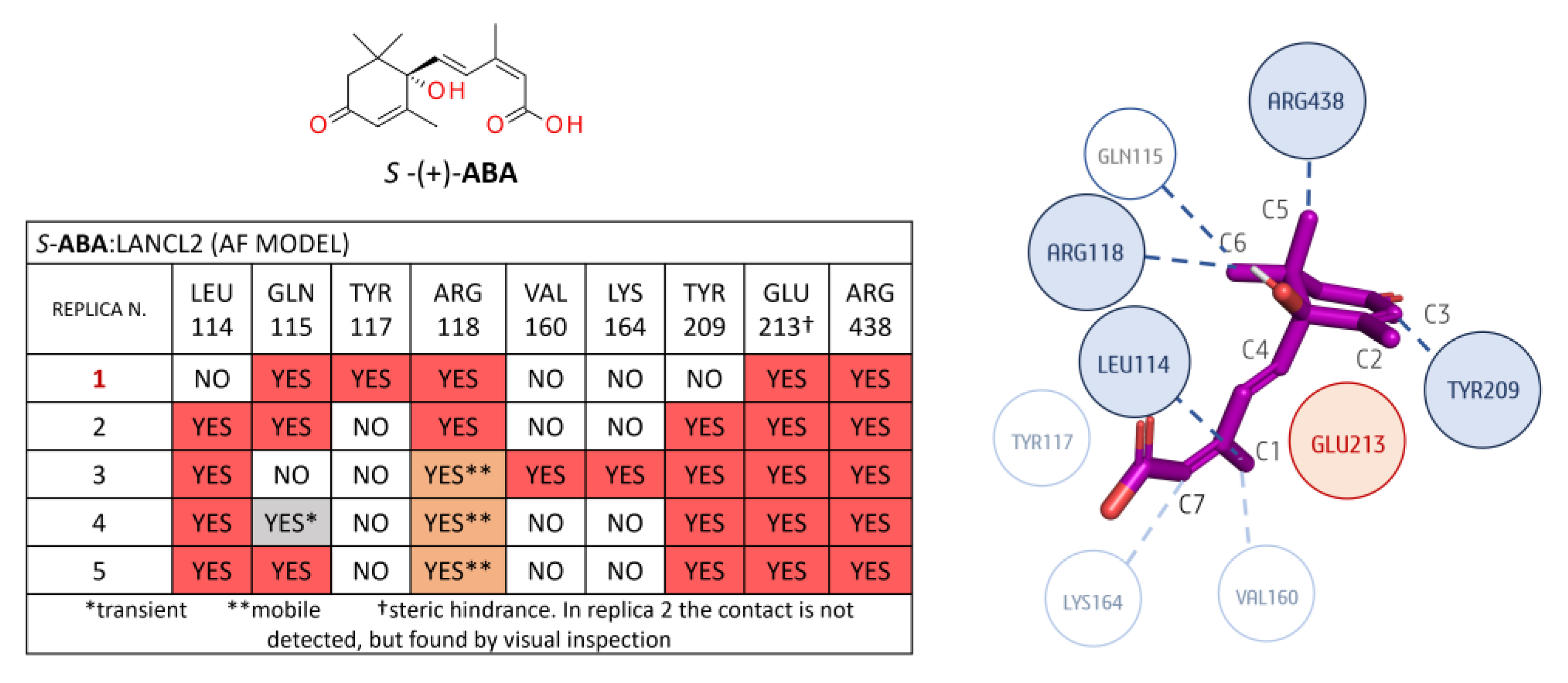
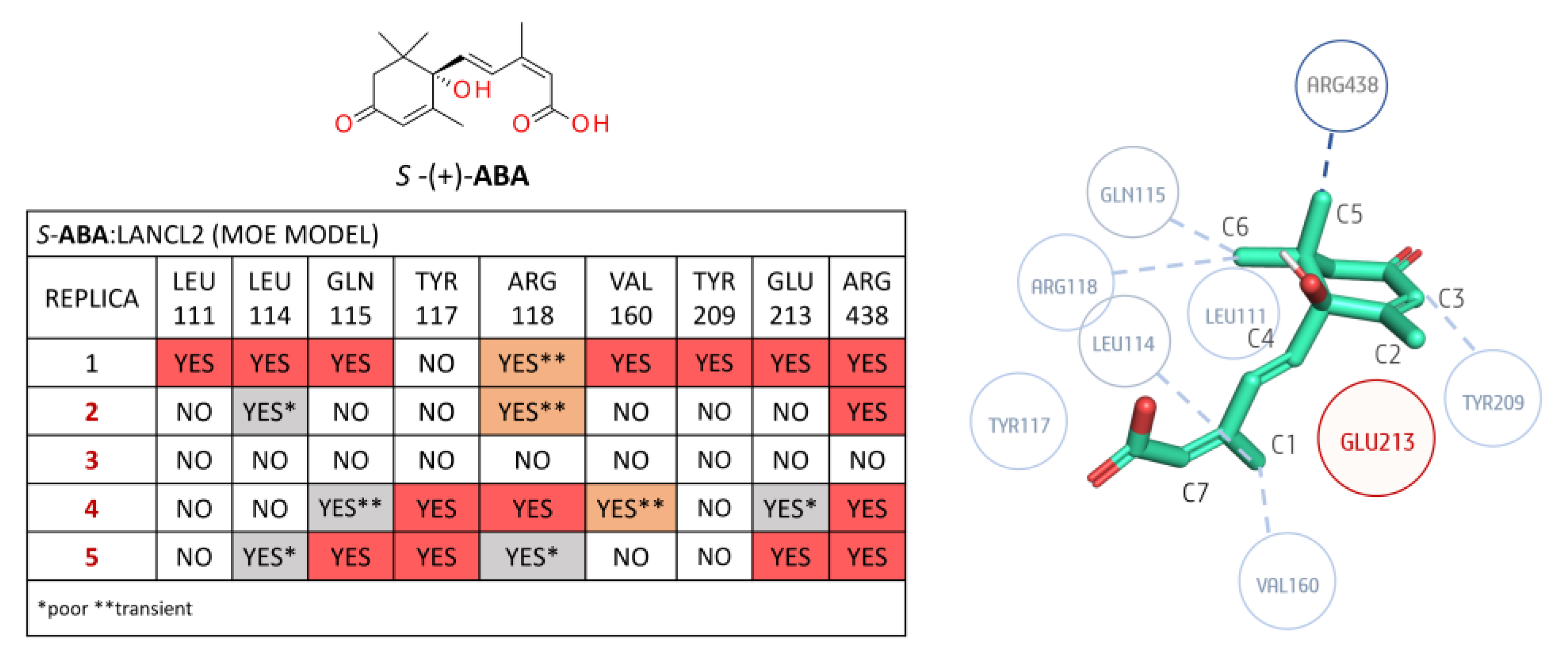
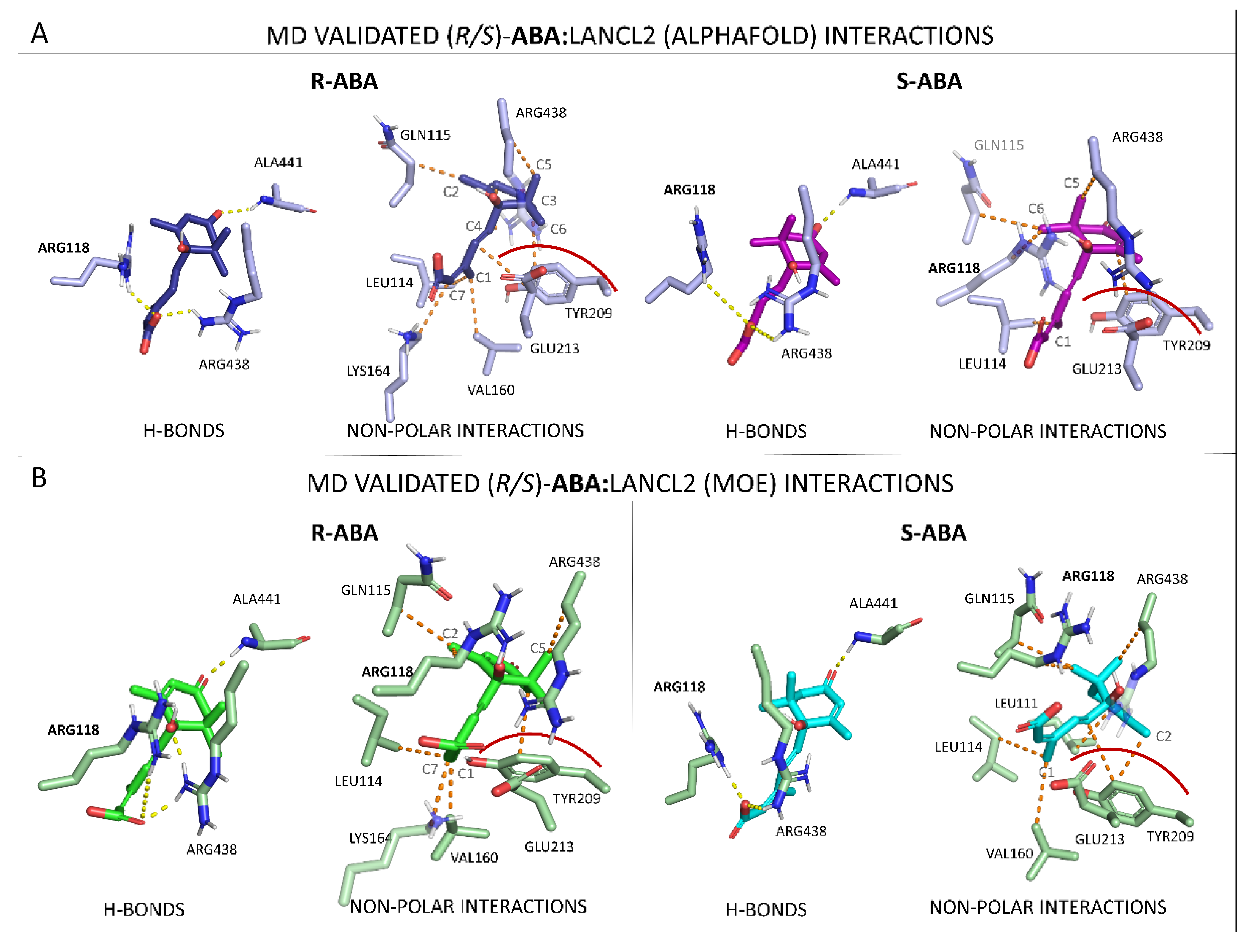
3.1.2. LANCL2-ABA Molecular Docking and MD Simulations
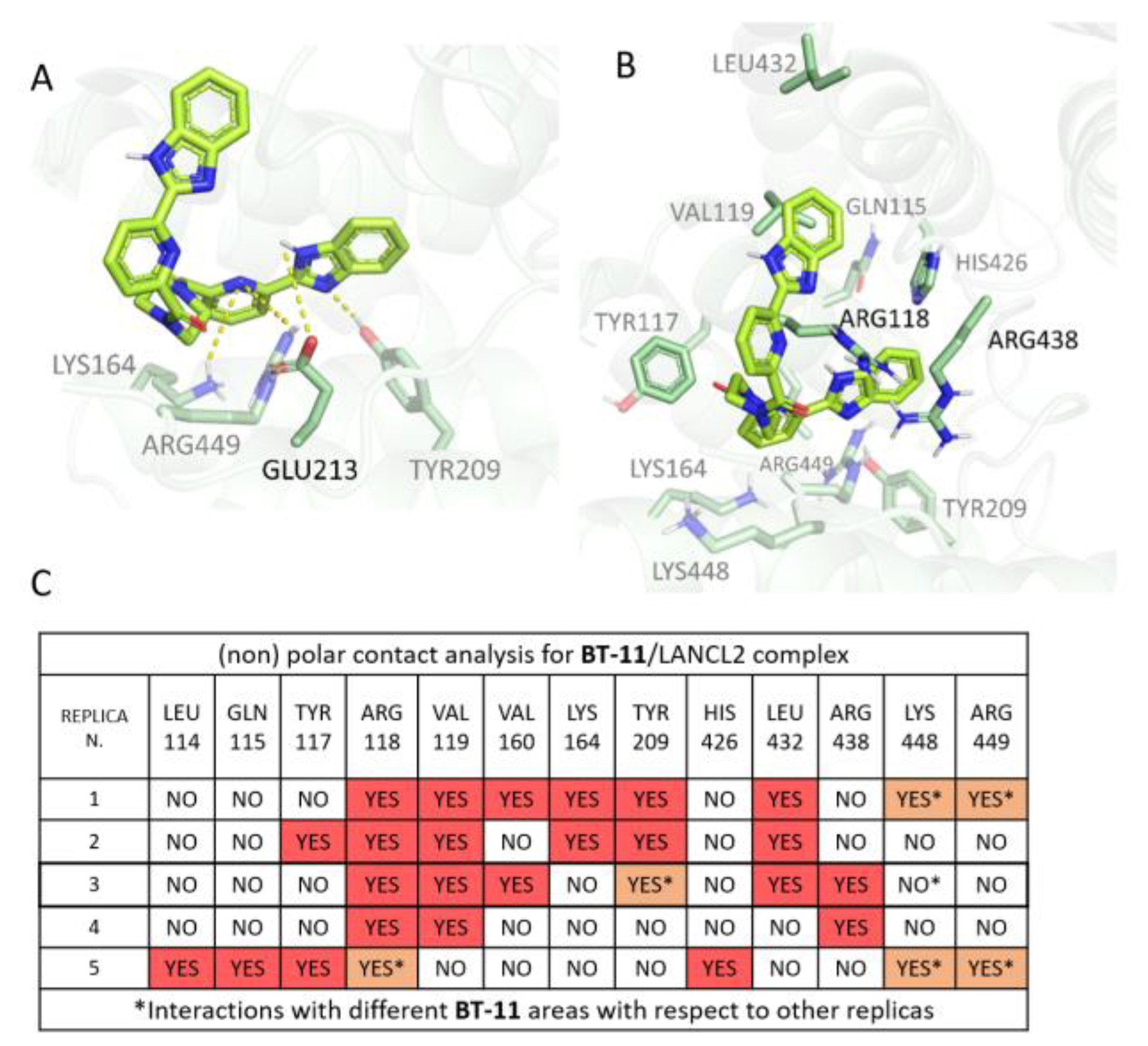
3.1.3. BT-11:LANCL2 MD Simulations
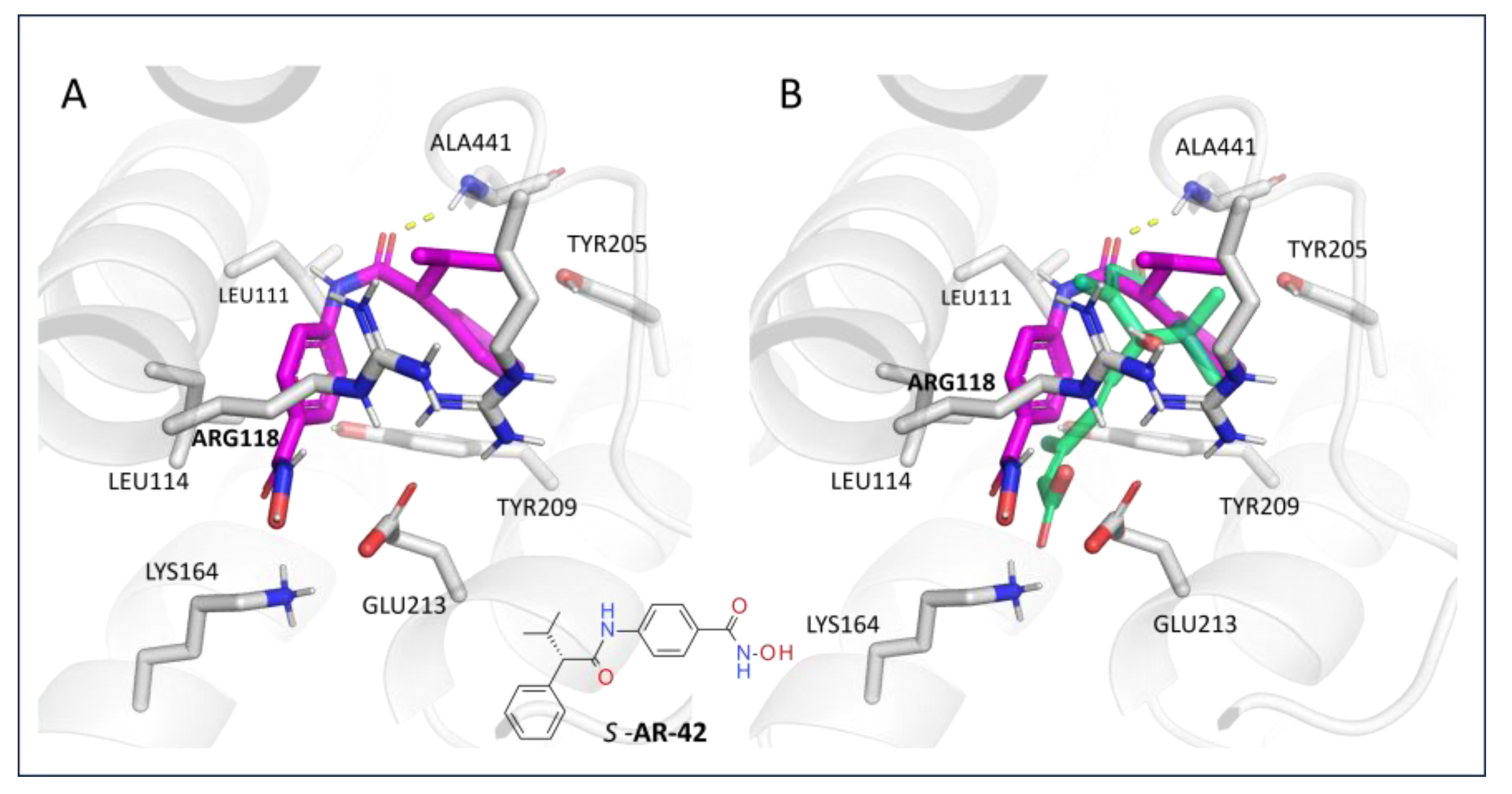
3.1.4. Virtual Screening
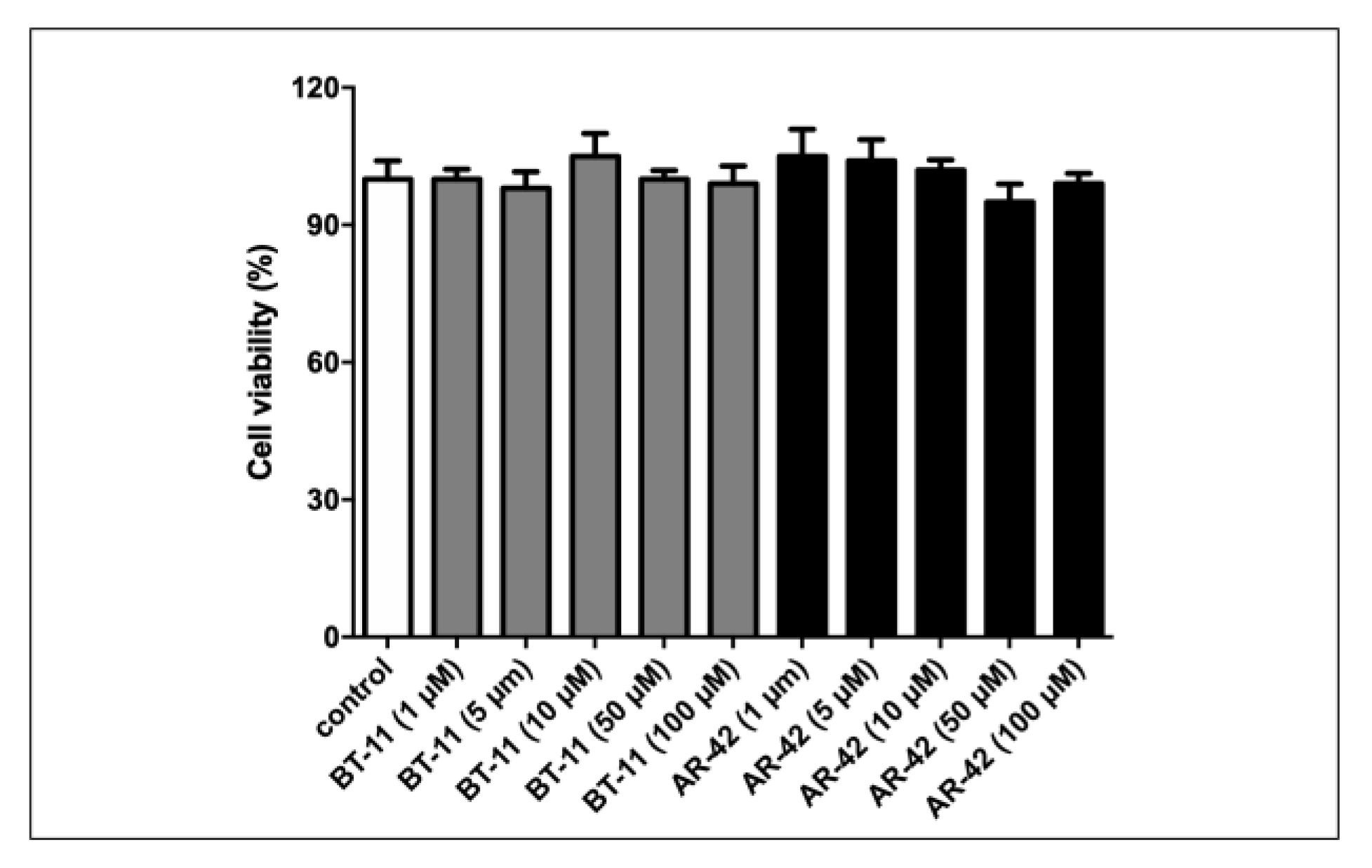
3.2. Biochemical Assays
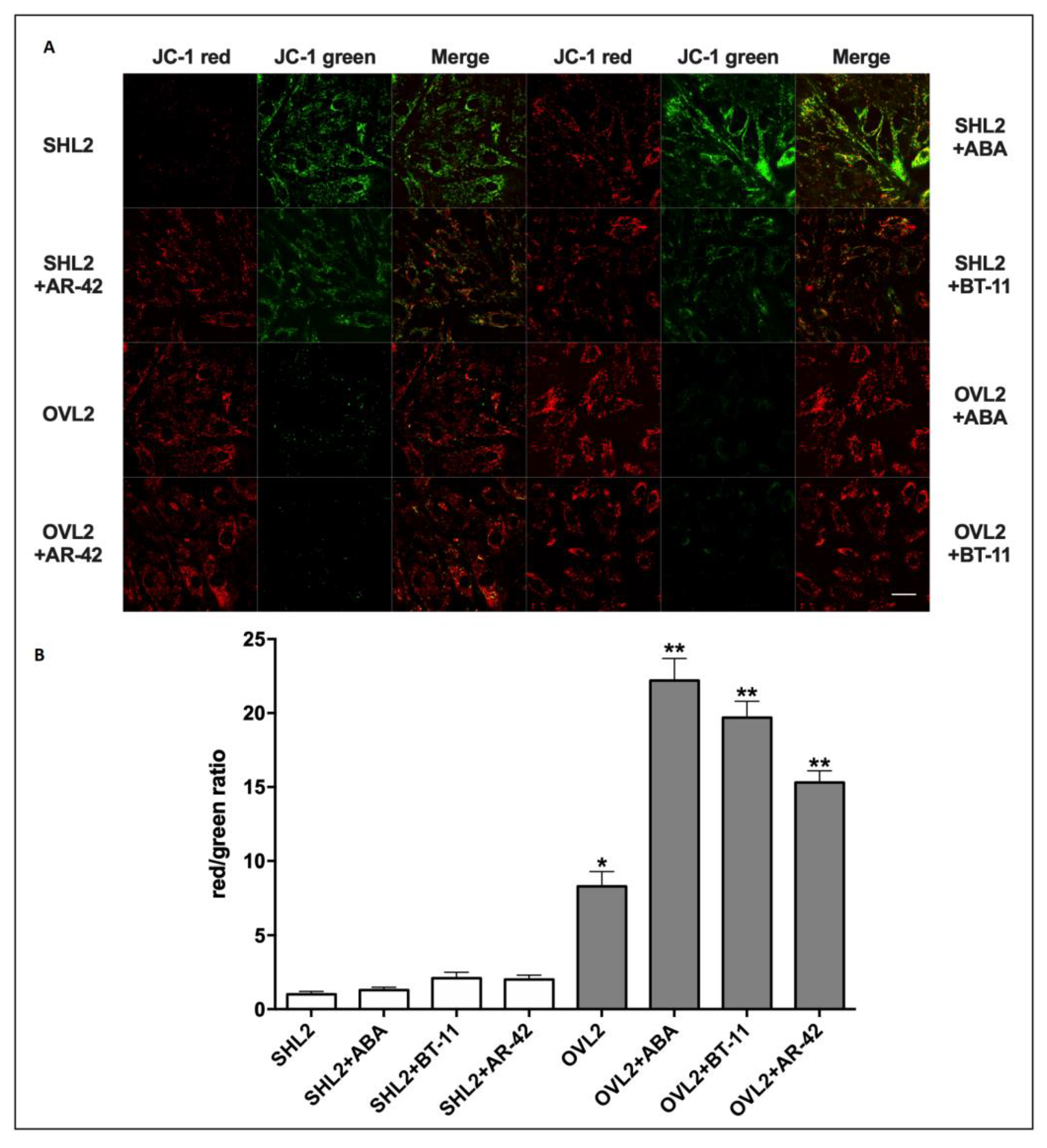
3.2.1. Mitochondrial Function Analysis in LANCL2-Overexpressing and LANCL2-Silenced Cells
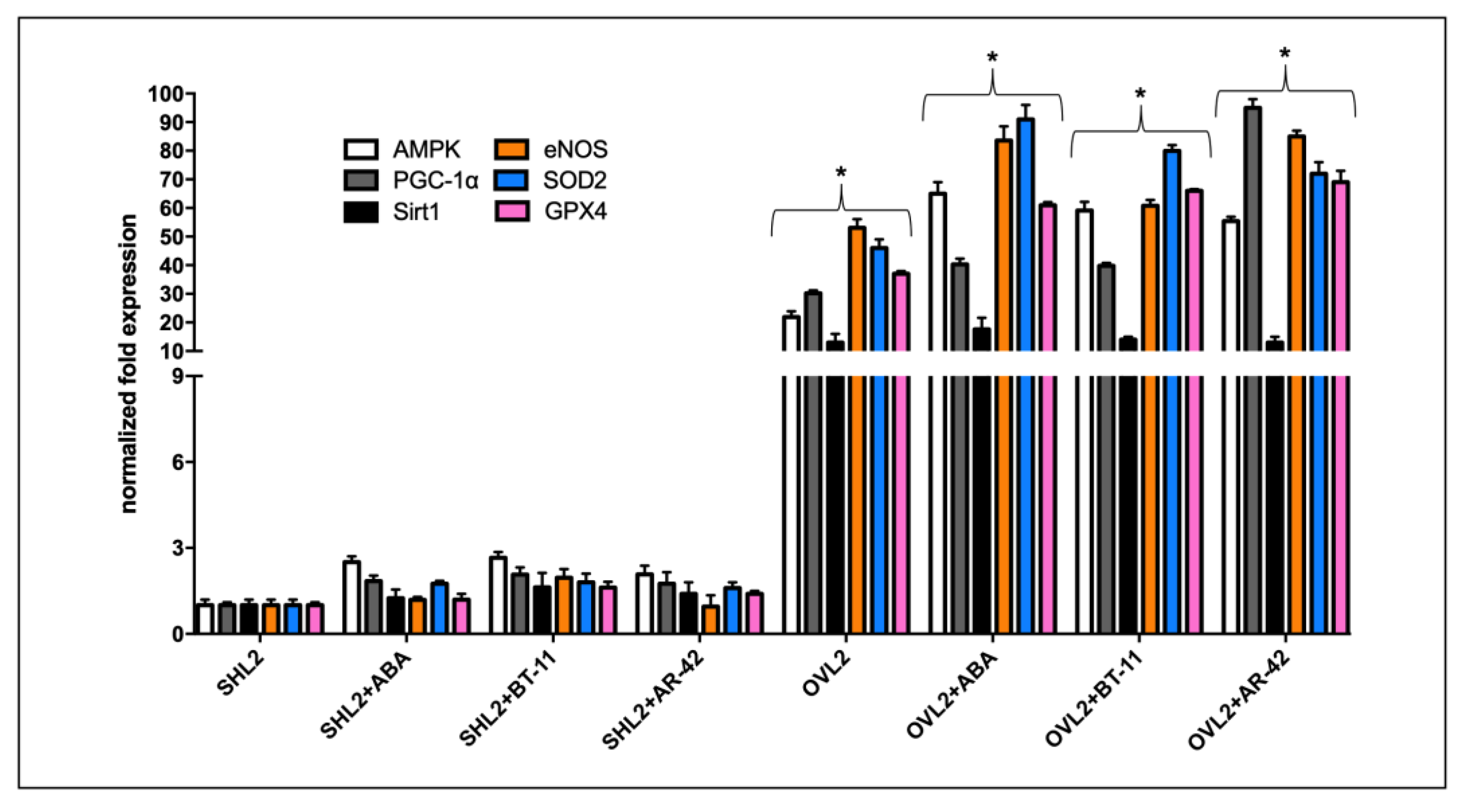
3.2.2. qPCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bauer, H.; Mayer, H.; Marchler-Bauer, A.; Salzer, U.; Prohaska, R. Characterization of P40/GPR69A as a Peripheral Membrane Protein Related to the Lantibiotic Synthetase Component C. Biochem. Biophys. Res. Commun. 2000, 275, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.-Y.; Galan, S.R.G.; Zeng, Y.; Zhou, T.H.; He, C.; Raj, R.; Riedl, J.; Liu, S.; Chooi, K.P.; Garg, N.; et al. LanCLs Add Glutathione to Dehydroamino Acids Generated at Phosphorylated Sites in the Proteome. Cell 2021, 184, 2680–2695.e26. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Min, Z.; Debapriya, D.; Tong, H.K.; Jie, C.; Van Der Donk, W.A. LanCL proteins are not involved in Lanthionine Synthesis in Mammals. Sci. Rep. 2017, 7, 40980. [Google Scholar] [CrossRef]
- Mayer, H.; Breuss, J.; Ziegler, S.; Prohaska, R. Molecular Characterization and Tissue-Specific Expression of a Murine Putative G-Protein-Coupled Receptor. Biochim. Et Biophys. Acta BBA Gene Struct. Expr. 1998, 1399, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Magnone, M.; Guida, L.; Sturla, L.; Zocchi, E. The ABA/LANCL Hormone/Receptor System in the Control of Glycemia, of Cardiomyocyte Energy Metabolism, and in Neuroprotection: A New Ally in the Treatment of Diabetes Mellitus? Int. J. Mol. Sci. 2023, 24, 1199. [Google Scholar] [CrossRef] [PubMed]
- Mayer, H.; Pongratz, M.; Prohaska, R. Molecular Cloning, Characterization, and Tissue-Specific Expression of Human LANCL2, a Novel Member of the LanC-Like Protein Family. DNA Seq. 2001, 12, 161–166. [Google Scholar] [CrossRef]
- Landlinger, C.; Salzer, U.; Prohaska, R. Myristoylation of Human LanC-like Protein 2 (LANCL2) Is Essential for the Interaction with the Plasma Membrane and the Increase in Cellular Sensitivity to Adriamycin. Biochim. Et Biophys. Acta BBA Biomembr. 2006, 1758, 1759–1767. [Google Scholar] [CrossRef]
- Spinelli, S.; Begani, G.; Guida, L.; Magnone, M.; Galante, D.; D’Arrigo, C.; Scotti, C.; Iamele, L.; De Jonge, H.; Zocchi, E.; et al. LANCL1 Binds Abscisic Acid and Stimulates Glucose Transport and Mitochondrial Respiration in Muscle Cells via the AMPK/PGC-1α/Sirt1 Pathway. Mol. Metab. 2021, 53, 101263. [Google Scholar] [CrossRef]
- Bassaganya-Riera, J.; Skoneczka, J.; Kingston, D.; Krishnan, A.; Misyak, S.; Guri, A.; Pereira, A.; Carter, A.; Minorsky, P.; Tumarkin, R.; et al. Mechanisms of Action and Medicinal Applications of Abscisic Acid. Curr. Med. Chem. 2010, 17, 467–478. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscis acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef]
- Bruzzone, S.; Bodrato, N.; Usai, C.; Guida, L.; Moreschi, I.; Nano, R.; Antonioli, B.; Fruscione, F.; Magnone, M.; Scarfì, S.; et al. Abscisic Acid Is an Endogenous Stimulator of Insulin Release from Human Pancreatic Islets with Cyclic ADP Ribose as Second Messenger. J. Biol. Chem. 2008, 283, 32188–32197. [Google Scholar] [CrossRef] [PubMed]
- Magnone, M.; Bruzzone, S.; Guida, L.; Damonte, G.; Millo, E.; Scarfì, S.; Usai, C.; Sturla, L.; Palombo, D.; De Flora, A.; et al. Abscisic Acid Released by Human Monocytes Activates Monocytes and Vascular Smooth Muscle Cell Responses Involved in Atherogenesis. J. Biol. Chem. 2009, 284, 17808–17818. [Google Scholar] [CrossRef] [PubMed]
- Bruzzone, S.; Moreschi, I.; Usai, C.; Guida, L.; Damonte, G.; Salis, A.; Scarfì, S.; Millo, E.; De Flora, A.; Zocchi, E. Abscisic Acid Is an Endogenous Cytokine in Human Granulocytes with Cyclic ADP-Ribose as Second Messenger. Proc. Natl. Acad. Sci. USA 2007, 104, 5759–5764. [Google Scholar] [CrossRef] [PubMed]
- Bodrato, N.; Franco, L.; Fresia, C.; Guida, L.; Usai, C.; Salis, A.; Moreschi, I.; Ferraris, C.; Verderio, C.; Basile, G.; et al. Abscisic Acid Activates the Murine Microglial Cell Line N9 through the Second Messenger Cyclic ADP-Ribose. J. Biol. Chem. 2009, 284, 14777–14787. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Guida, L.; Vigliarolo, T.; Passalacqua, M.; Begani, G.; Magnone, M.; Sturla, L.; Benzi, A.; Ameri, P.; Lazzarini, E.; et al. The ABA-LANCL1/2 Hormone-Receptors System Protects H9c2 Cardiomyocytes from Hypoxia-Induced Mitochondrial Injury via an AMPK- and NO-Mediated Mechanism. Cells 2022, 11, 2888. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Guida, L.; Passalacqua, M.; Magnone, M.; Cossu, V.; Sambuceti, G.; Marini, C.; Sturla, L.; Zocchi, E. Abscisic Acid and Its Receptors LANCL1 and LANCL2 Control Cardiomyocyte Mitochondrial Function, Expression of Contractile, Cytoskeletal and Ion Channel Proteins and Cell Proliferation via ERRα. Antioxidants 2023, 12, 1692. [Google Scholar] [CrossRef] [PubMed]
- Guri, A.J.; Hontecillas, R.; Si, H.; Liu, D.; Bassaganya-Riera, J. Dietary Abscisic Acid Ameliorates Glucose Tolerance and Obesity-Related Inflammation in Db/Db Mice Fed High-Fat Diets. Clin. Nutr. 2007, 26, 107–116. [Google Scholar] [CrossRef]
- Guri, A.J.; Hontecillas, R.; Ferrer, G.; Casagran, O.; Wankhade, U.; Noble, A.M.; Eizirik, D.L.; Ortis, F.; Cnop, M.; Liu, D.; et al. Loss of PPARγ in Immune Cells Impairs the Ability of Abscisic Acid to Improve Insulin Sensitivity by Suppressing Monocyte Chemoattractant Protein-1 Expression and Macrophage Infiltration into White Adipose Tissue. J. Nutr. Biochem. 2008, 19, 216–228. [Google Scholar] [CrossRef]
- Guri, A.J.; Hontecillas, R.; Bassaganya-Riera, J. Abscisic Acid Ameliorates Experimental IBD by Downregulating Cellular Adhesion Molecule Expression and Suppressing Immune Cell Infiltration. Clin. Nutr. 2010, 29, 824–831. [Google Scholar] [CrossRef]
- Guri, A.J.; Misyak, S.A.; Hontecillas, R.; Hasty, A.; Liu, D.; Si, H.; Bassaganya-Riera, J. Abscisic Acid Ameliorates Atherosclerosis by Suppressing Macrophage and CD4+ T Cell Recruitment into the Aortic Wall. J. Nutr. Biochem. 2010, 21, 1178–1185. [Google Scholar] [CrossRef]
- Leber, A.; Hontecillas, R.; Tubau-Juni, N.; Zoccoli-Rodriguez, V.; Goodpaster, B.; Bassaganya-Riera, J. Abscisic Acid Enriched Fig Extract Promotes Insulin Sensitivity by Decreasing Systemic Inflammation and Activating LANCL2 in Skeletal Muscle. Sci. Rep. 2020, 10, 10463. [Google Scholar] [CrossRef] [PubMed]
- Bissel, P.; Boes, K.; Hinckley, J.; Jortner, B.S.; Magnin-Bissel, G.; Werre, S.R.; Ehrich, M.; Carbo, A.; Philipson, C.; Hontecillas, R.; et al. Exploratory Studies With BT-11. Int. J. Toxicol. 2016, 35, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.H.; Kim, N.; Ju, Y.-J.; Gee, M.S.; Lee, D.; Lee, J.K. Phytohormone Abscisic Acid Improves Memory Impairment and Reduces Neuroinflammation in 5xFAD Mice by Upregulation of LanC-Like Protein 2. Int. J. Mol. Sci. 2020, 21, 8425. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Sarasúa, S.; Moustafa, S.; García-Avilés, Á.; López-Climent, M.F.; Gómez-Cadenas, A.; Olucha-Bordonau, F.E.; Sánchez-Pérez, A.M. The Effect of Abscisic Acid Chronic Treatment on Neuroinflammatory Markers and Memory in a Rat Model of High-Fat Diet Induced Neuroinflammation. Nutr. Metab. 2016, 13, 73. [Google Scholar] [CrossRef]
- Lu, P.; Hontecillas, R.; Horne, W.T.; Carbo, A.; Viladomiu, M.; Pedragosa, M.; Bevan, D.R.; Lewis, S.N.; Bassaganya-Riera, J. Computational Modeling-Based Discovery of Novel Classes of Anti-Inflammatory Drugs That Target Lanthionine Synthetase C-Like Protein 2. PLoS ONE 2012, 7, e34643. [Google Scholar] [CrossRef]
- Diddi, N.; Lai, L.; Nguyen, C.H.; Yan, D.; Nambara, E.; Abrams, S. An Efficient and Scalable Synthesis of a Persistent Abscisic Acid Analog (+)-Tetralone ABA. Org. Biomol. Chem. 2023, 21, 3014–3019. [Google Scholar] [CrossRef]
- Carbo, A.; Gandour, R.D.; Hontecillas, R.; Philipson, N.; Uren, A.; Bassaganya-Riera, J. An N,N-Bis(Benzimidazolylpicolinoyl)Piperazine (BT-11): A Novel Lanthionine Synthetase C-Like 2-Based Therapeutic for Inflammatory Bowel Disease. J. Med. Chem. 2016, 59, 10113–10126. [Google Scholar] [CrossRef]
- Leber, A.; Hontecillas, R.; Zoccoli-Rodriguez, V.; Bassaganya-Riera, J. Activation of LANCL2 by BT-11 Ameliorates IBD by Supporting Regulatory T Cell Stability Through Immunometabolic Mechanisms. Inflamm. Bowel Dis. 2018, 24, 1978–1991. [Google Scholar] [CrossRef]
- Leber, A.; Hontecillas, R.; Zoccoli-Rodriguez, V.; Ehrich, M.; Davis, J.; Chauhan, J.; Bassaganya-Riera, J. Nonclinical Toxicology and Toxicokinetic Profile of an Oral Lanthionine Synthetase C-Like 2 (LANCL2) Agonist, BT-11. Int. J. Toxicol. 2019, 38, 96–109. [Google Scholar] [CrossRef]
- Cichero, E.; Fresia, C.; Guida, L.; Booz, V.; Millo, E.; Scotti, C.; Iamele, L.; de Jonge, H.; Galante, D.; De Flora, A.; et al. Identification of a High Affinity Binding Site for Abscisic Acid on Human Lanthionine Synthetase Component C-like Protein 2. Int. J. Biochem. Cell Biol. 2018, 97, 52–61. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Fenger, J.; Murahari, S.; Bear, M.D.; Kulp, S.K.; Wang, D.; Chen, C.-S.; Kisseberth, W.C.; London, C.A. AR-42, a Novel HDAC Inhibitor, Exhibits Biologic Activity against Malignant Mast Cell Lines via down-Regulation of Constitutively Activated Kit. Blood 2010, 115, 4217–4225. [Google Scholar] [CrossRef]
- Chen, G.; Yang, Y.; Xu, C.; Gao, S. A Flow Cytometry-Based Assay for Measuring Mitochondrial Membrane Potential in Cardiac Myocytes After Hypoxia/Reoxygenation. J. Vis. Exp. 2018, 137, e57725. [Google Scholar] [CrossRef]
- Ashleigh, T.; Swerdlow, R.H.; Beal, M.F. The Role of Mitochondrial Dysfunction in Alzheimer’s Disease Pathogenesis. Alzheimer’s Dement. 2023, 19, 333–342. [Google Scholar] [CrossRef]
- Dewanjee, S.; Chakraborty, P.; Bhattacharya, H.; Chacko, L.; Singh, B.; Chaudhary, A.; Javvaji, K.; Pradhan, S.R.; Vallamkondu, J.; Dey, A.; et al. Altered Glucose Metabolism in Alzheimer’s Disease: Role of Mitochondrial Dysfunction and Oxidative Stress. Free. Radic. Biol. Med. 2022, 193, 134–157. [Google Scholar] [CrossRef]
- Chen, M.; Huang, N.; Liu, J.; Huang, J.; Shi, J.; Jin, F. AMPK: A Bridge between Diabetes Mellitus and Alzheimer’s Disease. Behav. Brain Res. 2021, 400, 113043. [Google Scholar] [CrossRef]
- Dubey, H.; Gulati, K.; Ray, A. Alzheimer’s Disease: A Contextual Link with Nitric Oxide Synthase. Curr. Mol. Med. 2020, 20, 505–515. [Google Scholar] [CrossRef]
- Chemical Computing Group ULC Molecular Operating Environment (MOE2019.01) 2021. Available online: http://www.Chemcomp.Com/ (accessed on 1 September 2022).
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Alphafold Protein Structure Database. Available online: https://Alphafold.Ebi.Ac.Uk/July2022Release (accessed on 1 September 2022).
- Decherchi, S.; Bottegoni, G.; Spitaleri, A.; Rocchia, W.; Cavalli, A. BiKi Life Sciences: A New Suite for Molecular Dynamics and Related Methods in Drug Discovery. J. Chem. Inf. Model. 2018, 58, 219–224. [Google Scholar] [CrossRef]
- Schrödinger Release 2023-2: Maestro; Schrödinger LLC: New York, NY, USA, 2023.
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Available online: https://www.Schrodinger.Com/Science-Articles/Docking-and-Scoring (accessed on 3 November 2022).
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef] [PubMed]
- Sherman, W.; Beard, H.S.; Farid, R. Use of an Induced Fit Receptor Structure in Virtual Screening. Chem. Biol. Drug. Des. 2006, 67, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, E.; Abraham, M.J.; Hess, B.; van der Spoel, D. GROMACS 2020 Manual; Version 2020; Zenodo: Meyrin, Switzerland, 2020. [Google Scholar] [CrossRef]
- Macchiagodena, M.; Pagliai, M.; Andreini, C.; Rosato, A.; Procacci, P. Upgraded AMBER Force Field for Zinc-Binding Residues and Ligands for Predicting Structural Properties and Binding Affinities in Zinc-Proteins. ACS Omega 2020, 5, 15301–15310. [Google Scholar] [CrossRef] [PubMed]
- Macchiagodena, M.; Pagliai, M.; Andreini, C.; Rosato, A.; Procacci, P. Upgrading and Validation of the AMBER Force Field for Histidine and Cysteine Zinc(II)-Binding Residues in Sites with Four Protein Ligands. J. Chem. Inf. Model. 2019, 59, 3803–3816. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Berryman, J.T.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cisneros, G.A.; Cruzeiro, V.W.D.; et al. Amber Manuals. Available online: https://ambermd.org/Manuals.php (accessed on 14 July 2023).
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Jakalian, A.; Bush, B.L.; Jack, D.B.; Bayly, C.I. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: I. Method. J. Comput. Chem. 2000, 21, 132–146. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: II. Parameterization and Validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, H.C.S.; Hu, Z. Using PyMOL as a Platform for Computational Drug Design. WIREs Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Grace User’s Guide (for Grace-5.1.22). Available online: https://plasma-gate.weizmann.ac.il/Grace/doc/UsersGuide.html (accessed on 15 July 2023).
- Soares, R.O.S. GitHub Repository of the ReadHBmap. Py Script. Available online: https://github.com/quytruong1808/vilas/blob/master/vilas/analyzer/readHBmap.py (accessed on 3 November 2022).
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for theassessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Zhu, Y.; Yuan, T.; Zhang, Y.; Shi, J.; Bai, L.; Duan, X.; Tong, R.; Zhong, L. AR-42: A Pan-HDAC Inhibitor with Antitumor and Antiangiogenic Activities in Esophageal Squamous Cell Carcinoma. Drug. Des. Devel. Ther. 2019, 13, 4321–4330. [Google Scholar] [CrossRef]
- Collier, K.A.; Valencia, H.; Newton, H.; Hade, E.M.; Sborov, D.W.; Cavaliere, R.; Poi, M.; Phelps, M.A.; Liva, S.G.; Coss, C.C.; et al. A Phase 1 Trial of the Histone Deacetylase Inhibitor AR-42 in Patients with Neurofibromatosis Type 2-Associated Tumors and Advanced Solid Malignancies. Cancer Chemother. Pharmacol. 2021, 87, 599–611. [Google Scholar] [CrossRef]
- Gu, M.; Wei, Z.; Wang, X.; Gao, Y.; Wang, D.; Liu, X.; Bai, C.; Su, G.; Yang, L.; Li, G. Myostatin Knockout Affects Mitochondrial Function by Inhibiting the AMPK/SIRT1/PGC1α Pathway in Skeletal Muscle. Int. J. Mol. Sci. 2022, 23, 13703. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, M.; Ying, X.; Yuan, J.; Wang, C.; Zhang, W.; Tian, S.; Yan, X. Caloric Restriction Increases the Resistance of Aged Heart to Myocardial Ischemia/Reperfusion Injury via Modulating AMPK–SIRT1–PGC1a Energy Metabolism Pathway. Sci. Rep. 2023, 13, 2045. [Google Scholar] [CrossRef] [PubMed]
- Hajam, Y.A.; Rani, R.; Ganie, S.Y.; Sheikh, T.A.; Javaid, D.; Qadri, S.S.; Pramodh, S.; Alsulimani, A.; Alkhanani, M.F.; Harakeh, S.; et al. Oxidative Stress in Human Pathology and Aging: Molecular Mechanisms and Perspectives. Cells 2022, 11, 552. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scarano, N.; Di Palma, F.; Origlia, N.; Musumeci, F.; Schenone, S.; Spinelli, S.; Passalacqua, M.; Zocchi, E.; Sturla, L.; Cichero, E.; et al. New Insights into the LANCL2-ABA Binding Mode towards the Evaluation of New LANCL Agonists. Pharmaceutics 2023, 15, 2754. https://doi.org/10.3390/pharmaceutics15122754
Scarano N, Di Palma F, Origlia N, Musumeci F, Schenone S, Spinelli S, Passalacqua M, Zocchi E, Sturla L, Cichero E, et al. New Insights into the LANCL2-ABA Binding Mode towards the Evaluation of New LANCL Agonists. Pharmaceutics. 2023; 15(12):2754. https://doi.org/10.3390/pharmaceutics15122754
Chicago/Turabian StyleScarano, Naomi, Francesco Di Palma, Nicola Origlia, Francesca Musumeci, Silvia Schenone, Sonia Spinelli, Mario Passalacqua, Elena Zocchi, Laura Sturla, Elena Cichero, and et al. 2023. "New Insights into the LANCL2-ABA Binding Mode towards the Evaluation of New LANCL Agonists" Pharmaceutics 15, no. 12: 2754. https://doi.org/10.3390/pharmaceutics15122754
APA StyleScarano, N., Di Palma, F., Origlia, N., Musumeci, F., Schenone, S., Spinelli, S., Passalacqua, M., Zocchi, E., Sturla, L., Cichero, E., & Cavalli, A. (2023). New Insights into the LANCL2-ABA Binding Mode towards the Evaluation of New LANCL Agonists. Pharmaceutics, 15(12), 2754. https://doi.org/10.3390/pharmaceutics15122754