Population Pharmacodynamic Modelling of the CD19+ Suppression Effects of Rituximab in Paediatric Patients with Neurological and Autoimmune Diseases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design, Patients, and Drug Administration
2.2. Rituximab Pharmacologic Response: Blood Sampling and Flow Cytometry
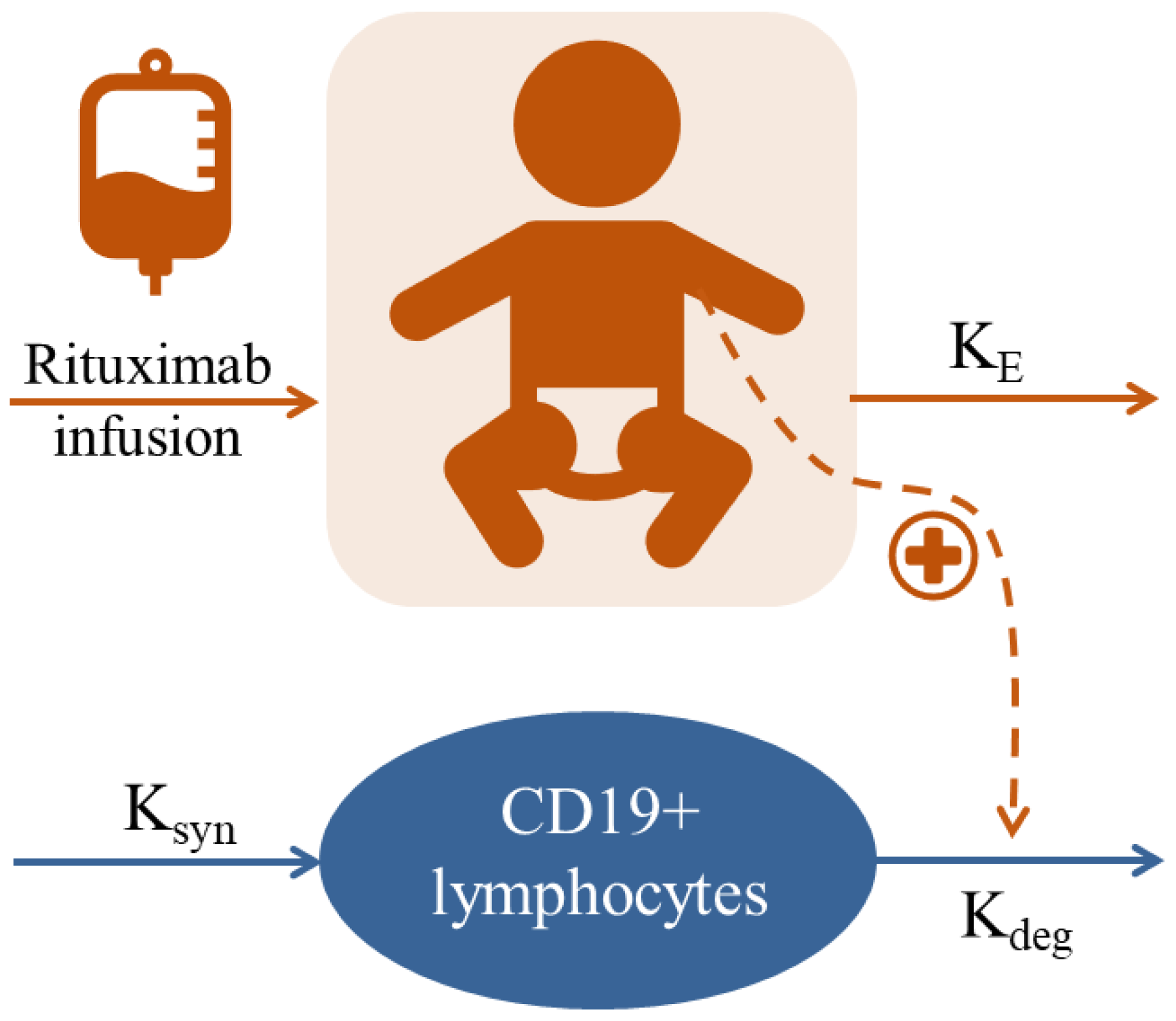
2.3. Population Kinetic-Pharmacodynamic Analysis
2.3.1. Base Population Model
2.3.2. Covariate Selection
2.3.3. Model Evaluation, External Predictive Performance Evaluation and Exploration
2.4. Clinical Response and Predictive Factors
3. Results
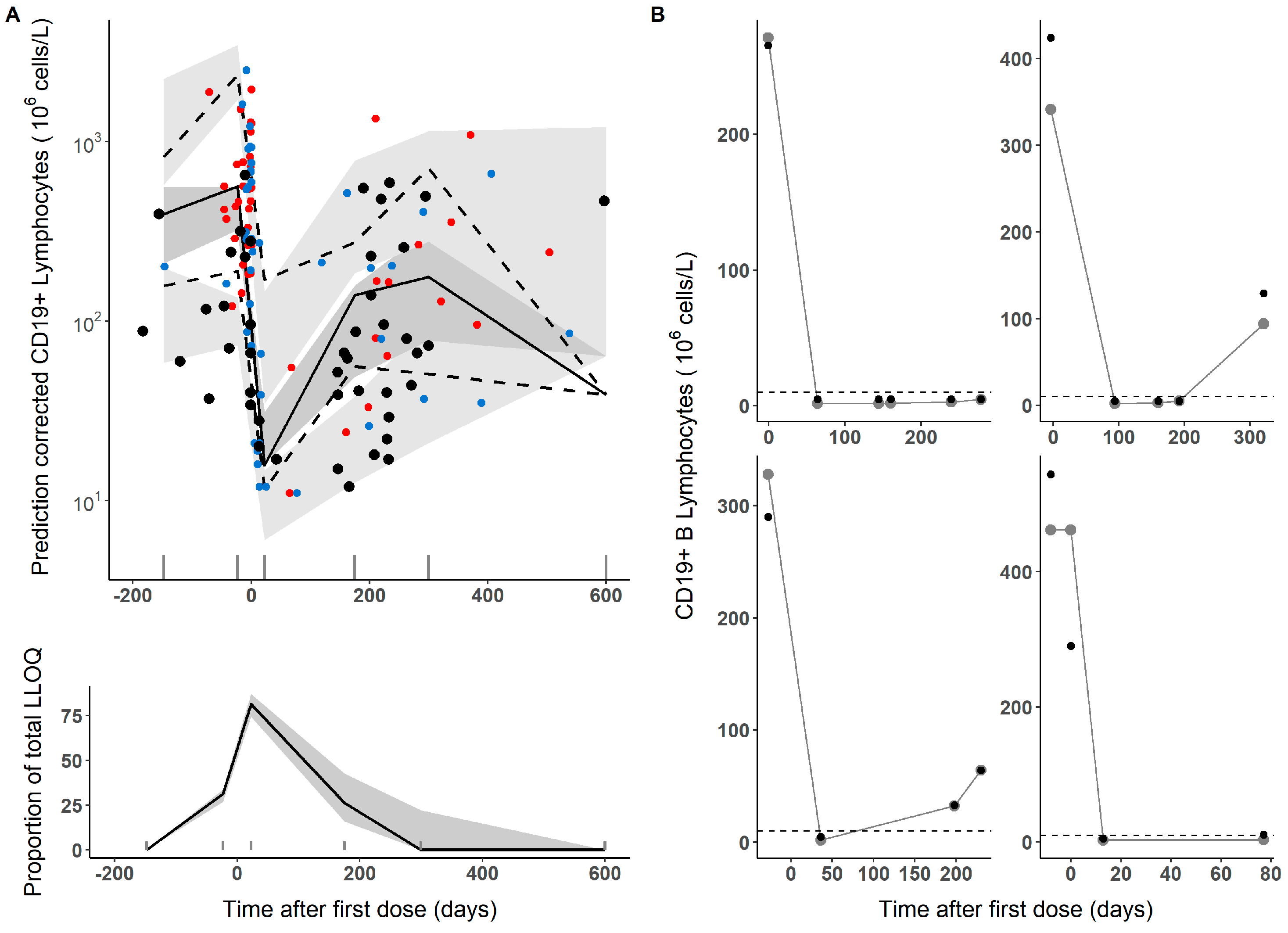
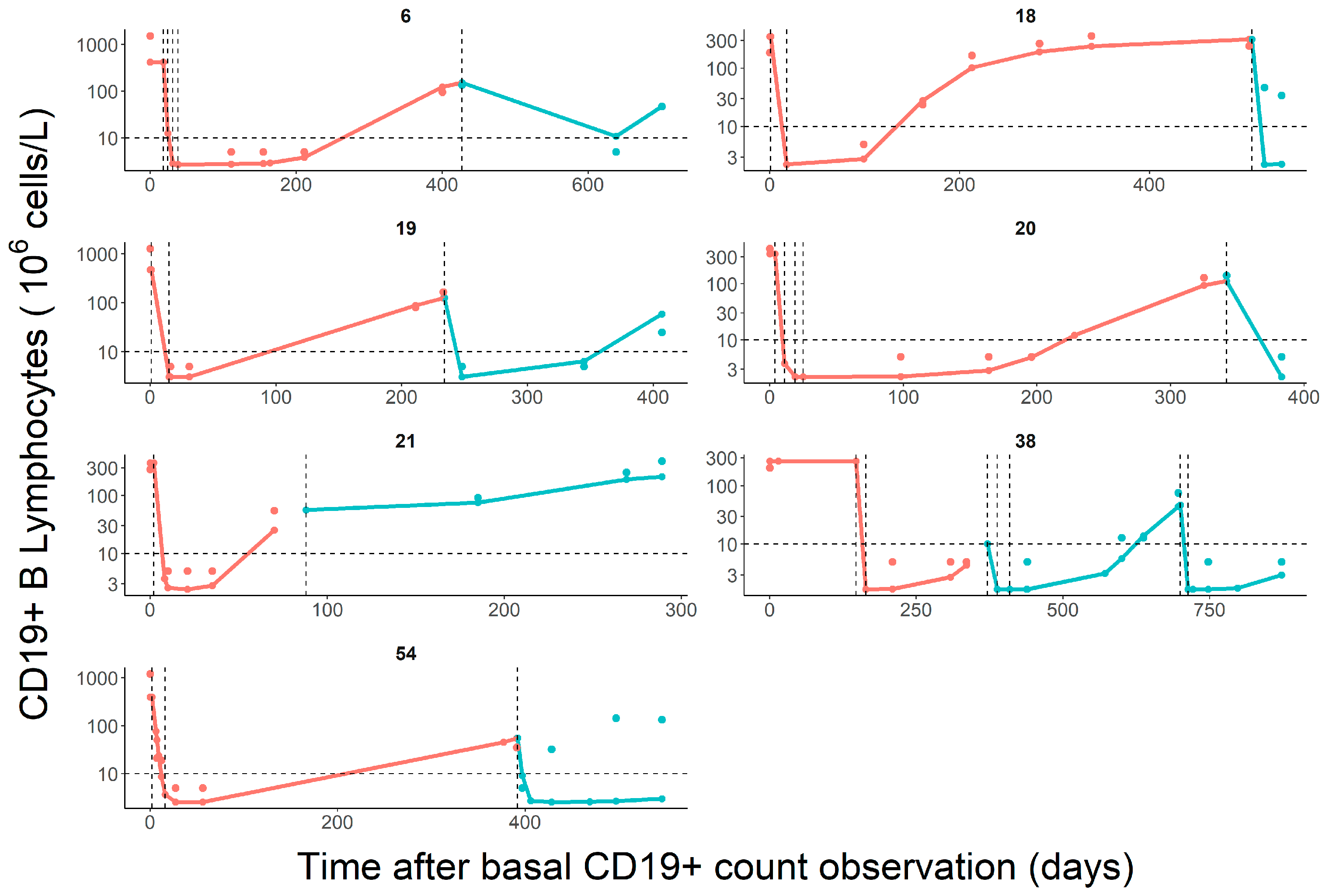
3.1. Population Pharmacodynamic Modelling
3.2. Clinical Response Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kasi, P.M.; Tawbi, H.A.; Oddis, C.V.; Kulkarni, H.S. Clinical review: Serious adverse events associated with the use of rituximab—A critical care perspective. Crit. Care 2012, 16, 231. Available online: https://pubmed.ncbi.nlm.nih.gov/22967460/ (accessed on 16 June 2023). [CrossRef] [PubMed]
- Micromedex-Solutions. Rituximab. 2023. Available online: http://www.micromedexsolutions.com (accessed on 16 August 2023).
- Tenembaum, S.; Yeh, E.A. Pediatric NMOSD: A Review and Position Statement on Approach to Work-Up and Diagnosis. Front. Pediatr. 2020, 8, 339. Available online: https://pubmed.ncbi.nlm.nih.gov/32671002/ (accessed on 16 June 2023). [CrossRef] [PubMed]
- Genberg, H.; Hansson, A.; Wernerson, A.; Wennberg, L.; Tydén, G. Pharmacodynamics of rituximab in kidney allotransplantation. Am. J. Transplant. 2006, 6, 2418–2428. Available online: https://pubmed.ncbi.nlm.nih.gov/16925569/ (accessed on 16 June 2023). [CrossRef]
- Ellrichmann, G.; Bolz, J.; Peschke, M.; Duscha, A.; Hellwig, K.; Lee, D.H.; Linker, R.A.; Gold, R.; Haghikia, A. Peripheral CD19+ B-cell counts and infusion intervals as a surrogate for long-term B-cell depleting therapy in multiple sclerosis and neuromyelitis optica/neuromyelitis optica spectrum disorders. J. Neurol. 2019, 266, 57–67. Available online: https://pubmed.ncbi.nlm.nih.gov/30377816/ (accessed on 16 June 2023). [CrossRef]
- Tedder, T.F. CD19: A promising B cell target for rheumatoid arthritis. Nat. Rev. Rheumatol. 2009, 5, 572–577. Available online: https://pubmed.ncbi.nlm.nih.gov/19798033/ (accessed on 16 June 2023). [CrossRef] [PubMed]
- Marks, S.D.; Patey, S.; Brogan, P.A.; Hasson, N.; Pilkington, C.; Woo, P.; Tullus, K. B lymphocyte depletion therapy in children with refractory systemic lupus erythematosus. Arthritis Rheum. 2005, 52, 3168–3174. Available online: https://pubmed.ncbi.nlm.nih.gov/16200620/ (accessed on 16 June 2023). [CrossRef]
- Naegelin, Y.; Naegelin, P.; von Felten, S.; Lorscheider, J.; Sonder, J.; Uitdehaag, B.M.; Scotti, B.; Zecca, C.; Gobbi, C.; Kappos, L.; et al. Association of Rituximab Treatment with Disability Progression among Patients with Secondary Progressive Multiple Sclerosis. JAMA Neurol. 2019, 76, 274–281. Available online: https://pubmed.ncbi.nlm.nih.gov/30615019/ (accessed on 16 June 2023). [CrossRef]
- Kurtzke, J.F. Rating neurologic impairment in multiple sclerosis: An expanded disability status scale (EDSS). Neurology 1983, 33, 1444–1452. Available online: https://pubmed.ncbi.nlm.nih.gov/6685237/ (accessed on 16 June 2023). [CrossRef]
- Bombardier, C.; Gladman, D.D.; Urowitz, M.B.; Caron, D.; Chang, C.H.; Austin, A.; Bell, A.; Bloch, D.A.; Corey, P.N.; Decker, J.L.; et al. Derivation of the SLEDAI. A disease activity index for lupus patients. Comm. Progn. Stud. SLE Arthritis Rheum. 1992, 35, 630–640. Available online: https://pubmed.ncbi.nlm.nih.gov/1599520/ (accessed on 16 June 2023). [CrossRef]
- Uribe, A.G.; Vilá, L.M.; McGwin, G.; Sanchez, M.L.; Reveille, J.D.; Alarcón, G.S. The Systemic Lupus Activity Measure-revised, the Mexican Systemic Lupus Erythematosus Disease Activity Index (SLEDAI), and a modified SLEDAI-2K are adequate instruments to measure disease activity in systemic lupus erythematosus. J. Rheumatol. 2004, 31, 1934–1940. Available online: http://www.ncbi.nlm.nih.gov/pubmed/15468356 (accessed on 16 August 2023).
- Wolfe, G.I.; Herbelin, L.; Nations, S.P.; Foster, B.; Bryan, W.W.; Barohn, R.J. Myasthenia gravis activities of daily living profile. Neurology 1999, 52, 1487–1489. Available online: https://pubmed.ncbi.nlm.nih.gov/10227640/ (accessed on 16 June 2023). [CrossRef]
- Pichler, W.J. Adverse side-effects to biological agents. Allergy 2006, 61, 912–920. Available online: http://www.ncbi.nlm.nih.gov/pubmed/16867042 (accessed on 28 August 2023). [CrossRef]
- World Health Organization. Guidelines on Evaluation of Similar Biotherapeutic Products (SBPs). WHO Tech. Rep. Ser. 2017, 2, 34. Available online: https://www.who.int/publications/m/item/mAbs-trs-no-1004-a2 (accessed on 28 August 2023).
- Cohen, D. European Medicines Agency tightens rules on conflict of interest. BMJ 2010, 341, c5902. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20959306 (accessed on 28 August 2023). [CrossRef] [PubMed]
- U.S. Food and Drug Administration. Scientific Considerations in Demonstrating Biosimilarity to a Reference Product. FDA Guid. Ind. 2012. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/scientific-considerations-demonstrating-biosimilarity-reference-product (accessed on 28 August 2023).
- Riva, N.; Molina, M.; Cornaló, B.L.; Salvador, M.V.; Savransky, A.; Tenembaum, S.; Katsicas, M.M.; Monteverde, M.; Cáceres Guido, P.; Rousseau, M.; et al. Intensive Safety Monitoring of Rituximab (Biosimilar Novex® and the Innovator) in Pediatric Patients with Complex Diseases. Front. Pharmacol. 2022, 12, 785770. Available online: https://pubmed.ncbi.nlm.nih.gov/35153748/ (accessed on 16 June 2023). [CrossRef]
- Pan, S.; Yu, H.; Surti, A.; Cheng, I.; Marks, S.D.; Brogan, P.A.; Eleftheriou, D.; Standing, J.F. Pharmacodynamics of rituximab on B lymphocytes in paediatric patients with autoimmune diseases. Br. J. Clin. Pharmacol. 2019, 85, 1790–1797. Available online: https://pubmed.ncbi.nlm.nih.gov/31026092/ (accessed on 16 June 2023). [CrossRef] [PubMed]
- Adeli, M.M.G.; Eichner, B.H.; Thornburg, C.; Williams, L. Persistent antibody depletion after rituximab in three children with autoimmune cytopenias. Pediatr. Hematol. Oncol. 2009, 26, 566–572. Available online: https://pubmed.ncbi.nlm.nih.gov/19954366/ (accessed on 16 June 2023). [CrossRef]
- Quartier, P.; Brethon, B.; Philippet, P.; Landman-Parker, J.; Le Deist, F.; Fischer, A. Treatment of childhood autoimmune haemolytic anaemia with rituximab. Lancet 2001, 358, 1511–1513. Available online: https://pubmed.ncbi.nlm.nih.gov/11705566/ (accessed on 16 June 2023). [CrossRef] [PubMed]
- Bauer, R.J. NONMEM Tutorial Part II: Estimation Methods and Advanced Examples. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 538–556. Available online: https://pubmed.ncbi.nlm.nih.gov/31044558/ (accessed on 21 June 2023). [CrossRef]
- Beal, S.L. Ways to fit a PK model with some data below the quantification limit. J. Pharmacokinet. Pharmacodyn. 2001, 28, 481–504. Available online: http://www.ncbi.nlm.nih.gov/pubmed/11768292 (accessed on 28 August 2023). [CrossRef] [PubMed]
- ICON. NONMEM®. Available online: https://www.iconplc.com/solutions/technologies/nonmem/ (accessed on 21 June 2023).
- Steimer, J.L.; Mallet, A.; Golmard, J.L.; Boisvieux, J.F. Alternative approaches to estimation of population pharmacokinetic parameters: Comparison with the nonlinear mixed-effect model. Drug Metab. Rev. 1984, 15, 265–292. Available online: https://pubmed.ncbi.nlm.nih.gov/6745083/ (accessed on 21 June 2023). [CrossRef] [PubMed]
- Bauer, R.J.; Guzy, S.; Ng, C. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J. 2007, 9, E60–E83. Available online: https://pubmed.ncbi.nlm.nih.gov/17408237/ (accessed on 21 June 2023). [CrossRef] [PubMed]
- Keizer, R.J.; Karlsson, M.O.; Hooker, A. Modeling and Simulation Workbench for NONMEM: Tutorial on Pirana, PsN, and Xpose. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, e50. Available online: http://www.ncbi.nlm.nih.gov/pubmed/23836189 (accessed on 28 August 2023). [CrossRef]
- Lindbom, L.; Pihlgren, P.; Jonsson, N. PsN-Toolkit—A collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput. Methods Programs Biomed. 2005, 79, 241–257. Available online: https://pubmed.ncbi.nlm.nih.gov/16023764/ (accessed on 16 June 2023). [CrossRef]
- Lindbom, L.; Ribbing, J.; Jonsson, E.N. Perl-speaks-NONMEM (PsN)—A Perl module for NONMEM related programming. Comput. Methods Programs Biomed. 2004, 75, 85–94. Available online: https://pubmed.ncbi.nlm.nih.gov/15212851/ (accessed on 16 June 2023). [CrossRef]
- Nguyen, T.H.T.; Mouksassi, M.S.; Holford, N.; Al-Huniti, N.; Freedman, I.; Hooker, A.C.; John, J.; Karlsson, M.O.; Mould, D.R.; Pérez Ruixo, J.J.; et al. Model Evaluation of Continuous Data Pharmacometric Models: Metrics and Graphics. CPT Pharmacomet. Syst. Pharmacol. 2017, 6, 87–109. Available online: https://pubmed.ncbi.nlm.nih.gov/27884052/ (accessed on 16 June 2023). [CrossRef]
- Broeker, A.; Nardecchia, M.; Klinker, K.P.; Derendorf, H.; Day, R.O.; Marriott, D.J.; Carland, J.E.; Stocker, S.L.; Wicha, S.G. Towards precision dosing of vancomycin: A systematic evaluation of pharmacometric models for Bayesian forecasting. Clin. Microbiol. Infect. 2019, 25, e1–e1286. Available online: https://linkinghub.elsevier.com/retrieve/pii/S1198743X19300977 (accessed on 28 August 2023). [CrossRef]
- Sheiner, L.B.; Beal, S.L. Some suggestions for measuring predictive performance. J. Pharmacokinet. Biopharm. 1981, 9, 503–512. [Google Scholar] [CrossRef]
- Chen, Y.; Shen, Q.; Dong, M.; Xiong, Y.; Xu, H.; Li, Z. Population Pharmacokinetics of Rituximab in Pediatric Patients With Frequent-Relapsing or Steroid-Dependent Nephrotic Syndrome. Front. Pharmacol. 2021, 12, 725665. Available online: https://pubmed.ncbi.nlm.nih.gov/34539407/ (accessed on 16 June 2023). [CrossRef]
- Svensson, R.J.; Ooi, Q.X.; Friberg, L.E.; Maharaj, N.; Reddy, P.K.; López-Lázaro, L.; Hansson, E. Rituximab pharmacokinetic and pharmacokinetic–pharmacodynamic evaluation based on a study in diffuse large B-cell lymphoma: Influence of tumor size on pharmacokinetic and assessment of pharmacokinetic similarity. CPT Pharmacomet. Syst. Pharmacol. 2023, 12, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Du, W.; Zhang, X.; Li, P. The Influence of Different Disease States on Rituximab Pharmacokinetics. Curr. Drug Metab. 2020, 21, 938–946. [Google Scholar] [CrossRef]
- Lioger, B.; Edupuganti, S.R.; Mulleman, D.; Passot, C.; Desvignes, C.; Bejan-Angoulvant, T.; Thibault, G.; Gouilleux-Gruart, V.; Mélet, J.; Paintaud, G.; et al. Antigenic burden and serum IgG concentrations influence rituximab pharmacokinetics in rheumatoid arthritis patients. Br. J. Clin. Pharmacol. 2017, 83, 1773–1781. [Google Scholar] [CrossRef]
- Bensalem, A.; Cartron, G.; Specks, U.; Mulleman, D.; Gyan, E.; Cornec, D.; Desvignes, C.; Casasnovas, O.; Lamy, T.; Leprêtre, S.; et al. The Influence of Underlying Disease on Rituximab Pharmacokinetics May be Explained by Target-Mediated Drug Disposition. Clin. Pharmacokinet. 2022, 61, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Perinparajah, S.; Booth, J.; Shah, M.; Amrolia, P.; Amy Cheung, S.Y.; James, W.T.; Klein, N.Y.; Joseph, F.S. PAGE 31: Abstr 10656—Modelling the Pharmacodynamics of Rituximab Biosimilar in Children with Rheumatological Conditions. 2023. Available online: https://www.page-meeting.org/default.asp?abstract=10656 (accessed on 28 August 2023).
- Oomen, I.; Bouts, A.H.M.; Gouw, S.C.; Kuijpers, T.W.; Rispens, T.; de Vries, A.; Wolbink, G.; van den Berg, J.M.; Schonenberg-Meinema, D. Anti-rituximab antibodies affect pharmacokinetics and pharmacodynamics of rituximab in children with immune-mediated diseases. Clin. Exp. Rheumatol. 2022, 40, 183–190. Available online: http://www.ncbi.nlm.nih.gov/pubmed/34251329 (accessed on 28 August 2023). [CrossRef]
- Reddy, V.R.; Pepper, R.J.; Shah, K.; Cambridge, G.; Henderson, S.R.; Klein, C.; Kell, L.; Taylor, S.J.; Isenberg, D.A.; Cragg, M.S.; et al. Disparity in peripheral and renal B-cell depletion with rituximab in systemic lupus erythematosus: An opportunity for obinutuzumab? Rheumatology 2022, 61, 2894–2904. Available online: http://www.ncbi.nlm.nih.gov/pubmed/34788412 (accessed on 28 August 2023). [CrossRef]
- Cain, D.; Kondo, M.; Chen, H.; Kelsoe, G. Effects of acute and chronic inflammation on B-cell development and differentiation. J. Investig. Dermatol. 2009, 129, 266–277. Available online: http://www.ncbi.nlm.nih.gov/pubmed/19148216 (accessed on 28 August 2023). [CrossRef] [PubMed]
- Generics and Biosimilars Initiative (GaBI). Biosimilars of Rituximab. 2021. Available online: https://www.gabionline.net/biosimilars/general/Biosimilars-of-rituximab (accessed on 28 August 2023).
- Mitchell, C.; Crayne, C.B.; Cron, R.Q. Patterns of B Cell Repletion Following Rituximab Therapy in a Pediatric Rheumatology Cohort. ACR Open Rheumatol. 2019, 1, 527–532. [Google Scholar] [CrossRef]
- Reddy, V.; Cambridge, G.; Isenberg, D.A.; Glennie, M.J.; Cragg, M.S.; Leandro, M. Internalization of rituximab and the efficiency of B Cell depletion in rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheumatol. 2015, 67, 2046–2055. Available online: http://www.ncbi.nlm.nih.gov/pubmed/25916583 (accessed on 28 August 2023). [CrossRef]
- Reddy, V.; Croca, S.; Gerona, D.; De La Torre, I.; Isenberg, D.; McDonald, V.; Leandro, M.; Cambridge, G. Serum rituximab levels and efficiency of B cell depletion: Differences between patients with rheumatoid arthritis and systemic lupus erythematosus. Rheumatology 2013, 52, 951–952. Available online: http://www.ncbi.nlm.nih.gov/pubmed/23424262 (accessed on 28 August 2023). [CrossRef]
- Walport, M.J. Complement and systemic lupus erythematosus. Arthritis Res. 2002, 4 (Suppl. 3), S279–S293. Available online: http://www.ncbi.nlm.nih.gov/pubmed/12110148 (accessed on 28 August 2023). [CrossRef]
- Ahuja, A.; Teichmann, L.L.; Wang, H.; Dunn, R.; Kehry, M.R.; Shlomchik, M.J. An acquired defect in IgG-dependent phagocytosis explains the impairment in antibody-mediated cellular depletion in Lupus. J. Immunol. 2011, 187, 3888–3894. Available online: http://www.ncbi.nlm.nih.gov/pubmed/21873531 (accessed on 28 August 2023). [CrossRef] [PubMed]
- Carter, L.M.; Isenberg, D.A.; Ehrenstein, M.R. Elevated serum BAFF levels are associated with rising anti-double-stranded DNA antibody levels and disease flare following B cell depletion therapy in systemic lupus erythematosus. Arthritis Rheum. 2013, 65, 2672–2679. Available online: http://www.ncbi.nlm.nih.gov/pubmed/23839909 (accessed on 28 August 2023). [CrossRef] [PubMed]
- Albert, D.; Dunham, J.; Khan, S.; Stansberry, J.; Kolasinski, S.; Tsai, D.; Pullman-Mooar, S.; Barnack, F.; Striebich, C.; Looney, R.J.; et al. Variability in the biological response to anti-CD20 B cell depletion in systemic lupus erythaematosus. Ann. Rheum. Dis. 2008, 67, 1724–1731. Available online: http://www.ncbi.nlm.nih.gov/pubmed/18250115 (accessed on 28 August 2023). [CrossRef] [PubMed]
- Pierpont, T.M.; Limper, C.B.; Richards, K.L. Past, present, and future of Rituximab-The world’s first oncology monoclonal antibody therapy. Front. Oncol. 2018, 8, 163. [Google Scholar] [CrossRef]
- Breedveld, F.; Agarwal, S.; Yin, M.; Ren, S.; Li, N.F.; Shaw, T.M.; Davies, B.E. Rituximab pharmacokinetics in patients with rheumatoid arthritis: B-cell levels do not correlate with clinical response. J. Clin. Pharmacol. 2007, 47, 1119–1128. Available online: http://www.ncbi.nlm.nih.gov/pubmed/17766699 (accessed on 28 August 2023). [CrossRef] [PubMed]
- Tavakolpour, S.; Alesaeidi, S.; Darvishi, M.; GhasemiAdl, M.; Darabi-Monadi, S.; Akhlaghdoust, M.; Elikaei Behjati, S.; Jafarieh, A. A comprehensive review of rituximab therapy in rheumatoid arthritis patients. Clin. Rheumatol. 2019, 38, 2977–2994. Available online: http://www.ncbi.nlm.nih.gov/pubmed/31367943 (accessed on 28 August 2023). [CrossRef]
- Thurlings, R.M.; Teng, O.; Vos, K.; Gerlag, D.M.; Aarden, L.; Stapel, S.O.; van Laar, J.M.; Tak, P.P.; Wolbink, G.J. Clinical response, pharmacokinetics, development of human anti-chimaeric antibodies, and synovial tissue response to rituximab treatment in patients with rheumatoid arthritis. Ann. Rheum. Dis. 2010, 69, 409–412. Available online: http://www.ncbi.nlm.nih.gov/pubmed/19596693 (accessed on 28 August 2023). [CrossRef]

| Training Cohort | Predictive Performance Evaluation Cohort | |||||
|---|---|---|---|---|---|---|
| Variable | Neurologic | IHR | Total | Neurologic | IHR | Total |
| No. patients | 30 | 22 | 52 | 6 | 5 | 11 |
| No. of CD19+ B cell levels | 107 | 88 | 195 | 68 | 18 | 86 |
| Age (y.o.) | 11.1 (0.1–15.4) | 11.5 (1.6–18.7) | 11.1 (0.1–18.7) | 12.1 (7.0–15.2) | 13.9 (7.4–17.5) | 13.9 (7.0–17.5) |
| BSA (m2) | 1.3 (0.2–2.0) | 1.2 (0.4–1.8) | 1.2 (0.2–2.0) | 1.3 (0.9–1.8) | 1.4 (0.9–1.7) | 1.3 (0.9–1.8) |
| Body weight (kg) | 40 (4–87) | 37 (7–75) | 38 (4–87) | 42 (26–75) | 48 (23–75) | 43 (23–75) |
| Sex (Fem/Male) a | 18/12 | 18/4 | 36/16 | 5/1 | 5/0 | 10/1 |
| CD19+B0 (×106cel/L) | 437 (5–1953) | 315 (5–2513) | 544 (5–2513) | 83 (34–317) | 235 (67–652) | 162 (34–652) |
| Switch between innovator and biosimilar (yes/no/NA) a | 8/15/7 | 6/12/4 | 14/27/11 | 0/3/3 | 0/3/2 | 0/6/5 |
| Rituximab dose (mg) | 500 (41.5–1000) | 500 (100–1000) | 500 (41.5–1000) | 700 (500–1000) | 449 (130–1000) | 700 (130–1000) |
| Follow-up time (days) | 128 (27–701) | 190 (30–872) | 199 (27–872) | 837 (303–973) | 243 (22–688) | 688 (22–973) |
| No. of patients with Co-medication (%) | ||||||
| Cyclophosphamide | 3 (10) | 2 (9) | 5 (10) | - | - | - |
| Methotrexate | 2 (6.7) | - | 2 (4) | - | - | - |
| Steroids | 24 (80) | 17 (77.3) | 41 (79) | 3 (50) | 5 (100) | 8 (73) |
| Intravenous immunoglobulin | 5 (16.7) | 1 (4.5) | 6 (11.5) | 3 (50) | - | 3 (27) |
| Parameter | Estimate | RSE (%) | Shrinkage (%) | Bootstrap a Median, 95% CI |
|---|---|---|---|---|
| KE (days−1) | 0.06 | 17 | - | 0.06 (0.049–0.09) |
| CD19+0 (106 cells/L) | 353 | 25 | - | 352 (248–516) |
| Kdeg (days−1) | 0.004 | 22 | - | 0.004 (0.0008–0.005) |
| EMAX | 155 | 23 | - | 173 (153–713) |
| ED50 (mg) | 0.692 | 61 | - | 0.692 (0.177–2.259) |
| IIV KE (%) b | 55.4 | 27 | 47 | 59.7 (34.8–92.9) |
| IIV CD19+0 (%) b | 70.1 | 32 | 35 | 60.5 (8.6–183.0) |
| IIV Kdeg (%) b | 80.7 | 43 | 57 | 122.2 (39.6–323.3) |
| Residual error (Ln(106 cells/L)) | 0.94 | 9.5 | 24 | 0.93 (0.68–1.03) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riva, N.; Brstilo, L.; Sancho-Araiz, A.; Molina, M.; Savransky, A.; Roffé, G.; Sanz, M.; Tenembaum, S.; Katsicas, M.M.; Trocóniz, I.F.; et al. Population Pharmacodynamic Modelling of the CD19+ Suppression Effects of Rituximab in Paediatric Patients with Neurological and Autoimmune Diseases. Pharmaceutics 2023, 15, 2534. https://doi.org/10.3390/pharmaceutics15112534
Riva N, Brstilo L, Sancho-Araiz A, Molina M, Savransky A, Roffé G, Sanz M, Tenembaum S, Katsicas MM, Trocóniz IF, et al. Population Pharmacodynamic Modelling of the CD19+ Suppression Effects of Rituximab in Paediatric Patients with Neurological and Autoimmune Diseases. Pharmaceutics. 2023; 15(11):2534. https://doi.org/10.3390/pharmaceutics15112534
Chicago/Turabian StyleRiva, Natalia, Lucas Brstilo, Aymara Sancho-Araiz, Manuel Molina, Andrea Savransky, Georgina Roffé, Marianela Sanz, Silvia Tenembaum, Maria M. Katsicas, Iñaki F. Trocóniz, and et al. 2023. "Population Pharmacodynamic Modelling of the CD19+ Suppression Effects of Rituximab in Paediatric Patients with Neurological and Autoimmune Diseases" Pharmaceutics 15, no. 11: 2534. https://doi.org/10.3390/pharmaceutics15112534
APA StyleRiva, N., Brstilo, L., Sancho-Araiz, A., Molina, M., Savransky, A., Roffé, G., Sanz, M., Tenembaum, S., Katsicas, M. M., Trocóniz, I. F., & Schaiquevich, P. (2023). Population Pharmacodynamic Modelling of the CD19+ Suppression Effects of Rituximab in Paediatric Patients with Neurological and Autoimmune Diseases. Pharmaceutics, 15(11), 2534. https://doi.org/10.3390/pharmaceutics15112534







