A Pseudovirus Nanoparticle-Based Trivalent Rotavirus Vaccine Candidate Elicits High and Cross P Type Immune Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids for Expression of Four Tag-Free S-VP8* Fusion Proteins
2.2. Expression and Purification of Tag-Free S-VP8* Fusion Proteins
2.3. Anion Exchange Chromatography
2.4. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.5. Transmission Electron Microscopy (TEM)
2.6. Cesium Chloride (CsCl) Density Gradient Ultracentrifugation
2.7. Mouse Immunization with the S60-VP8 PVNPs and Controls
2.8. Enzyme Immunoassays (EIAs)
2.9. 50% Blocking Titer (BT50) of Sera against Rotavirus VP8*-Glycan Receptor Attachment
2.10. Rotavirus Neutralization Assays
2.11. Statistical Analyses
3. Results
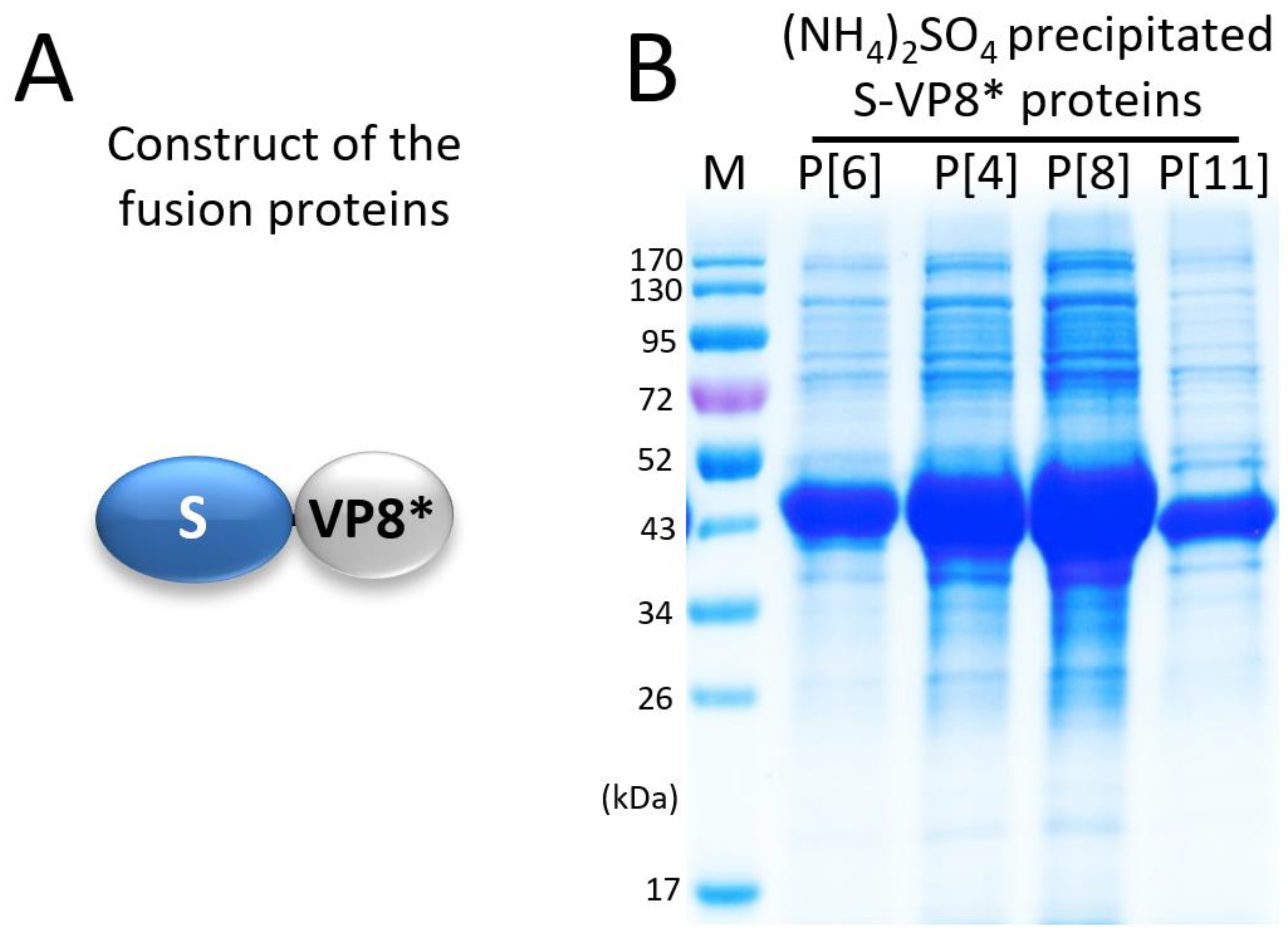
3.1. Expression and Selective Precipitation of the S-VP8* Proteins
3.2. Purification of S-VP8* Proteins by Ion Exchange Chromatography
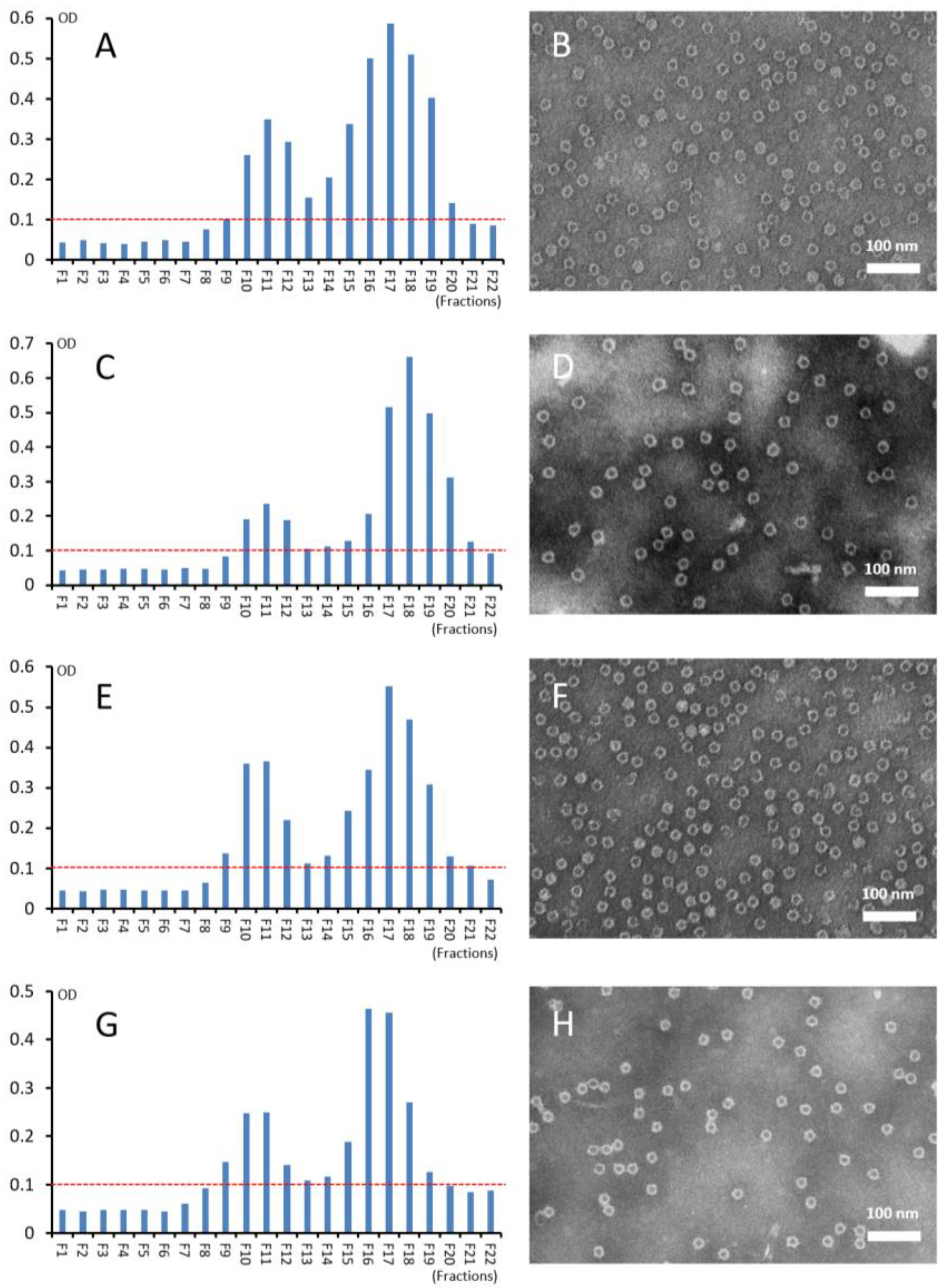
3.3. Self-Formation of Purified S-VP8* Proteins into PVNPs
3.4. CsCl Gradient Ultracentrifugation of the PVNPs
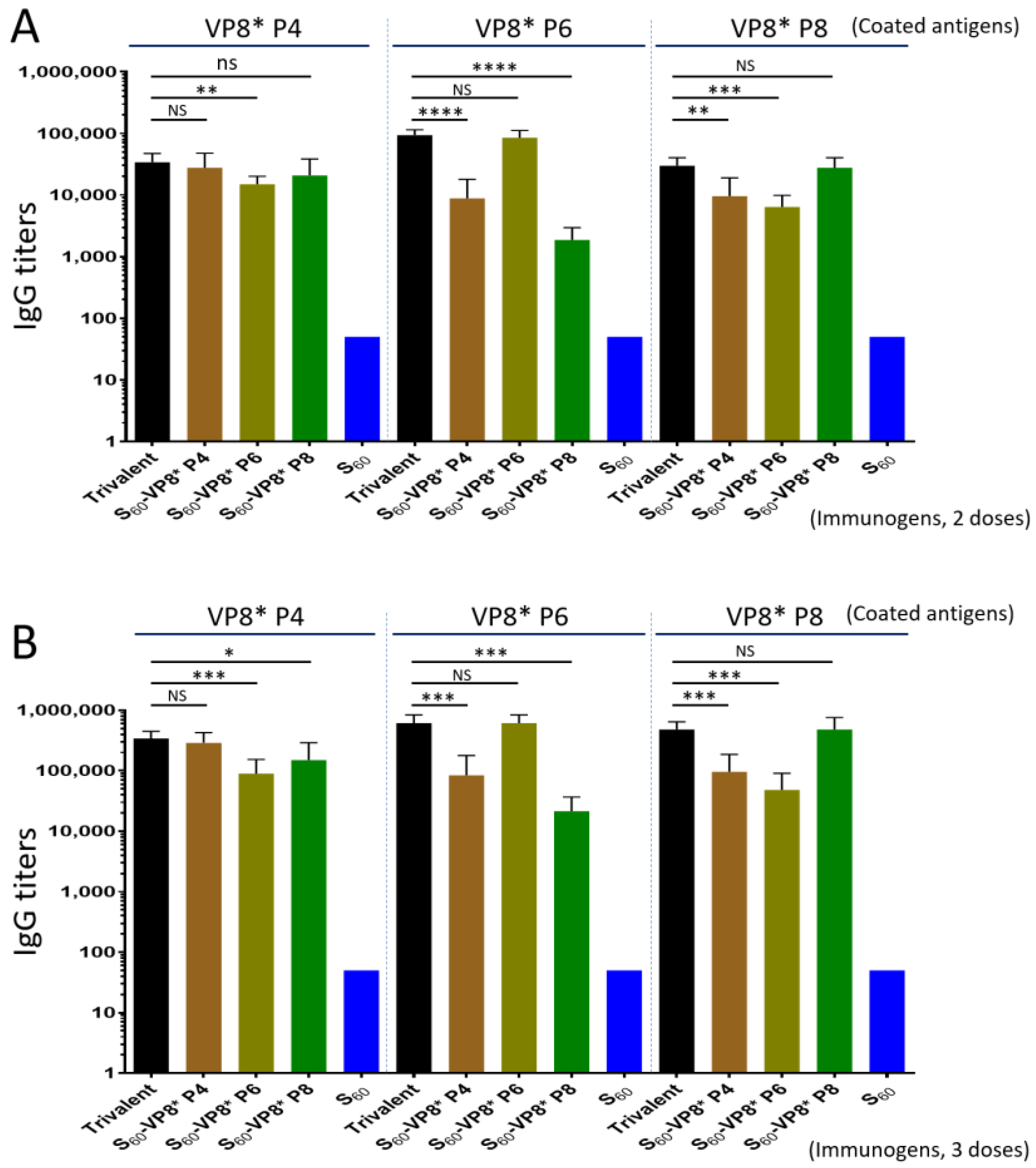
3.5. Trivalent PVNP Vaccine and Its IgG Responses in Mice
3.6. Serum IgA Responses of the Trivalent PVNP Vaccine
3.7. BT50 of the Trivalent Vaccine-Immunized Mouse Sera against VP8*-Glycan Receptor Attachment
3.8. Neutralization of the Trivalent Vaccine-Immunized Mouse Sera
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Parashar, U.D.; Svensson, L.; Hagbom, M.; Franco, M.A.; Greenberg, H.B.; O’Ryan, M.; Kang, G.; et al. Rotavirus infection. Nat. Rev. Dis. Primers 2017, 3, 17083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yen, C.; Tate, J.E.; Patel, M.M.; Cortese, M.M.; Lopman, B.; Fleming, J.; Lewis, K.; Jiang, B.; Gentsch, J.; Steele, D.; et al. Rotavirus vaccines: Update on global impact and future priorities. Hum. Vaccines 2011, 7, 1282–1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vesikari, T.; Karvonen, A.; Prymula, R.; Schuster, V.; Tejedor, J.C.; Cohen, R.; Meurice, F.; Han, H.H.; Damaso, S.; Bouckenooghe, A. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: Randomised, double-blind controlled study. Lancet 2007, 370, 1757–1763. [Google Scholar] [CrossRef]
- Madhi, S.A.; Cunliffe, N.A.; Steele, D.; Witte, D.; Kirsten, M.; Louw, C.; Ngwira, B.; Victor, J.C.; Gillard, P.H.; Cheuvart, B.B.; et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. N. Engl. J. Med. 2010, 362, 289–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armah, G.E.; Sow, S.O.; Breiman, R.F.; Dallas, M.J.; Tapia, M.D.; Feikin, D.R.; Binka, F.N.; Steele, A.D.; Laserson, K.F.; Ansah, N.A.; et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: A randomised, double-blind, placebo-controlled trial. Lancet 2010, 376, 606–614. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Steele, A.D.; Duque, J.; Parashar, U.D.; Network, W.H. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 136–141. [Google Scholar] [CrossRef] [Green Version]
- Parashar, U.D.; Gibson, C.J.; Bresse, J.S.; Glass, R.I. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 2006, 12, 304–306. [Google Scholar] [CrossRef] [Green Version]
- Desselberger, U. Differences of Rotavirus Vaccine Effectiveness by Country: Likely Causes and Contributing Factors. Pathogens 2017, 6, 65. [Google Scholar] [CrossRef] [Green Version]
- Parker, E.P.; Ramani, S.; Lopman, B.A.; Church, J.A.; Iturriza-Gomara, M.; Prendergast, A.J.; Grassly, N.C. Causes of impaired oral vaccine efficacy in developing countries. Future Microbiol. 2018, 13, 97–118. [Google Scholar] [CrossRef] [PubMed]
- Harris, V.C.; Haak, B.W.; Handley, S.A.; Jiang, B.; Velasquez, D.E.; Hykes, B.L., Jr.; Droit, L.; Berbers, G.A.M.; Kemper, E.M.; van Leeuwen, E.M.M.; et al. Effect of Antibiotic-Mediated Microbiome Modulation on Rotavirus Vaccine Immunogenicity: A Human, Randomized-Control Proof-of-Concept Trial. Cell Host Microbe 2018, 24, 197–207 e194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rytter, M.J.; Kolte, L.; Briend, A.; Friis, H.; Christensen, V.B. The immune system in children with malnutrition--a systematic review. PLoS ONE 2014, 9, e105017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniuchi, M.; Platts-Mills, J.A.; Begum, S.; Uddin, M.J.; Sobuz, S.U.; Liu, J.; Kirkpatrick, B.D.; Colgate, E.R.; Carmolli, M.P.; Dickson, D.M.; et al. Impact of enterovirus and other enteric pathogens on oral polio and rotavirus vaccine performance in Bangladeshi infants. Vaccine 2016, 34, 3068–3075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramani, S.; Mamani, N.; Villena, R.; Bandyopadhyay, A.S.; Gast, C.; Sato, A.; Laucirica, D.; Clemens, R.; Estes, M.K.; O’Ryan, M.L. Rotavirus Serum IgA Immune Response in Children Receiving Rotarix Coadministered With bOPV or IPV. Pediatric Infect. Dis. J. 2016, 35, 1137–1139. [Google Scholar] [CrossRef]
- Pitzer, V.E.; Bennett, A.; Bar-Zeev, N.; Jere, K.C.; Lopman, B.A.; Lewnard, J.A.; Parashar, U.D.; Cunliffe, N.A. Evaluating strategies to improve rotavirus vaccine impact during the second year of life in Malawi. Sci. Transl. Med. 2019, 11, eaav6419. [Google Scholar] [CrossRef] [PubMed]
- Desai, R.; Cortese, M.M.; Meltzer, M.I.; Shankar, M.; Tate, J.E.; Yen, C.; Patel, M.M.; Parashar, U.D. Potential intussusception risk versus benefits of rotavirus vaccination in the United States. Pediatric Infect. Dis. J. 2013, 32, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauchau, V.; Van Holle, L.; Mahaux, O.; Holl, K.; Sugiyama, K.; Buyse, H. Post-marketing monitoring of intussusception after rotavirus vaccination in Japan. Pharmacoepidemiol. Drug Saf. 2015, 24, 765–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yung, C.-F.; Chan, S.P.; Soh, S.; Tan, A.; Thoon, K.C. Intussusception and Monovalent Rotavirus Vaccination in Singapore: Self-Controlled Case Series and Risk-Benefit Study. J. Pediatrics 2015, 167, 163–168.e161. [Google Scholar] [CrossRef] [PubMed]
- Rosillon, D.; Buyse, H.; Friedland, L.R.; Ng, S.-P.; Velazquez, F.R.; Breuer, T. Risk of Intussusception After Rotavirus Vaccination: Meta-analysis of Postlicensure Studies. Pediatric Infect. Dis. J. 2015, 34, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Yih, W.K.; Lieu, T.A.; Kulldorff, M.; Martin, D.; McMahill-Walraven, C.N.; Platt, R.; Selvam, N.; Selvan, M.; Lee, G.M.; Nguyen, M. Intussusception risk after rotavirus vaccination in U.S. infants. N. Engl. J. Med. 2014, 370, 503–512. [Google Scholar] [CrossRef] [Green Version]
- Weintraub, E.S.; Baggs, J.; Duffy, J.; Vellozzi, C.; Belongia, E.A.; Irving, S.; Klein, N.P.; Glanz, J.M.; Jacobsen, S.J.; Naleway, A.; et al. Risk of intussusception after monovalent rotavirus vaccination. N. Engl. J. Med. 2014, 370, 513–519. [Google Scholar] [CrossRef]
- Glass, R.I.; Parashar, U.D. Rotavirus vaccines--balancing intussusception risks and health benefits. N. Engl. J. Med. 2014, 370, 568–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, M.; Huang, P.; Jiang, X.; Tan, M. A Nanoparticle-Based Trivalent Vaccine Targeting the Glycan Binding VP8* Domains of Rotaviruses. Viruses 2021, 13, 72. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Huang, P.; Xia, M.; Fang, P.A.; Zhong, W.; McNeal, M.; Wei, C.; Jiang, W.; Jiang, X. Norovirus P particle, a novel platform for vaccine development and antibody production. J. Virol. 2011, 85, 753–764. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Jiang, X. Norovirus Capsid Protein-Derived Nanoparticles and Polymers as Versatile Platforms for Antigen Presentation and Vaccine Development. Pharmaceutics 2019, 11, 472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, M.; Jiang, X. The p domain of norovirus capsid protein forms a subviral particle that binds to histo-blood group antigen receptors. J. Virol. 2005, 79, 14017–14030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, M.; Fang, P.; Chachiyo, T.; Xia, M.; Huang, P.; Fang, Z.; Jiang, W.; Jiang, X. Noroviral P particle: Structure, function and applications in virus-host interaction. Virology 2008, 382, 115–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dormitzer, P.R.; Sun, Z.Y.; Blixt, O.; Paulson, J.C.; Wagner, G.; Harrison, S.C. Specificity and affinity of sialic acid binding by the rhesus rotavirus VP8* core. J. Virol. 2002, 76, 10512–10517. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Crawford, S.E.; Czako, R.; Cortes-Penfield, N.W.; Smith, D.F.; Le Pendu, J.; Estes, M.K.; Prasad, B.V. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 2012, 485, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Xia, M.; Tan, M.; Zhong, W.; Wei, C.; Wang, L.; Morrow, A.; Jiang, X. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J. Virol. 2012, 86, 4833–4843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramani, S.; Cortes-Penfield, N.W.; Hu, L.; Crawford, S.E.; Czako, R.; Smith, D.F.; Kang, G.; Ramig, R.F.; Le Pendu, J.; Prasad, B.V.; et al. The VP8* Domain of Neonatal Rotavirus Strain G10P[11] Binds to Type II Precursor Glycans. J. Virol. 2013, 87, 7255–7264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Huang, P.; Tan, M.; Liu, Y.; Biesiada, J.; Meller, J.; Castello, A.A.; Jiang, B.; Jiang, X. Rotavirus VP8*: Phylogeny, host range, and interaction with histo-blood group antigens. J. Virol. 2012, 86, 9899–9910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, M.; Huang, P.; Jiang, X.; Tan, M. Immune response and protective efficacy of the S particle presented rotavirus VP8* vaccine in mice. Vaccine 2019, 37, 4103–4110. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Yu, L.; Jia, L.; Li, Y.; Zeng, Y.; Li, T.; Ge, S.; Xia, N. Immunogenicity and protective efficacy of rotavirus VP8* fused to cholera toxin B subunit in a mouse model. Hum. Vaccines Immunother. 2016, 12, 2959–2968. [Google Scholar] [CrossRef]
- Ramesh, A.; Mao, J.; Lei, S.; Twitchell, E.; Shiraz, A.; Jiang, X.; Tan, M.; Yuan, A.L. Parenterally Administered P24-VP8* Nanoparticle Vaccine Conferred Strong Protection against Rotavirus Diarrhea and Virus Shedding in Gnotobiotic Pigs. Vaccines 2019, 7, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, M.P.; Vlasova, A.N.; Saif, L.J. Human rotavirus virus-like particle vaccines evaluated in a neonatal gnotobiotic pig model of human rotavirus disease. Expert Rev. Vaccines 2013, 12, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cui, K.; Wang, H.; Liu, F.; Huang, K.; Duan, Z.; Wang, F.; Shi, D.; Liu, Q. A milk-based self-assemble rotavirus VP6-ferritin nanoparticle vaccine elicited protection against the viral infection. J. Nanobiotech. 2019, 17, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groome, M.J.; Fairlie, L.; Morrison, J.; Fix, A.; Koen, A.; Masenya, M.; Jose, L.; Madhi, S.A.; Page, N.; McNeal, M.; et al. Safety and immunogenicity of a parenteral trivalent P2-VP8 subunit rotavirus vaccine: A multisite, randomised, double-blind, placebo-controlled trial. Lancet Infect. Dis. 2020, 20, 851–863. [Google Scholar] [CrossRef]
- Groome, M.J.; Koen, A.; Fix, A.; Page, N.; Jose, L.; Madhi, S.A.; McNeal, M.; Dally, L.; Cho, I.; Power, M.; et al. Safety and immunogenicity of a parenteral P2-VP8-P[8] subunit rotavirus vaccine in toddlers and infants in South Africa: A randomised, double-blind, placebo-controlled trial. Lancet Infect. Dis. 2017, 17, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xue, M.; Yu, L.; Luo, G.; Yang, H.; Jia, L.; Zeng, Y.; Li, T.; Ge, S.; Xia, N. Expression and characterization of a novel truncated rotavirus VP4 for the development of a recombinant rotavirus vaccine. Vaccine 2018, 36, 2086–2092. [Google Scholar] [CrossRef] [PubMed]
- Todd, S.; Page, N.A.; Steele, A.D.; Peenze, I.; Cunliffe, N.A. Rotavirus Strain Types Circulating in Africa: Review of Studies Published during 1997-2006. J. Infect. Dis. 2010, 202, S34–S42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, N.; Hoshino, Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 2005, 15, 29–56. [Google Scholar] [CrossRef] [PubMed]
- Ouermi, D.; Soubeiga, D.; Nadembega, W.M.C.; Sawadogo, P.M.; Zohoncon, T.M.; Obiri-Yeboah, D.; Djigma, F.W.; Nordgren, J.; Simpore, J. Molecular Epidemiology of Rotavirus in Children under Five in Africa (2006–2016): A Systematic Review. Pak. J. Biol. Sci. 2017, 20, 59–69. [Google Scholar] [CrossRef]
- Jain, V.; Parashar, U.D.; Glass, R.I.; Bhan, M.K. Epidemiology of rotavirus in India. Indian J. Pediatr. 2001, 68, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Iturriza Gomara, M.; Kang, G.; Mammen, A.; Jana, A.K.; Abraham, M.; Desselberger, U.; Brown, D.; Gray, J. Characterization of G10P[11] rotaviruses causing acute gastroenteritis in neonates and infants in Vellore, India. J. Clin. Microbiol. 2004, 42, 2541–2547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Libonati, M.H.; Dennis, A.F.; Ramani, S.; McDonald, S.M.; Akopov, A.; Kirkness, E.F.; Kang, G.; Patton, J.T. Absence of Genetic Differences among G10P[11] Rotaviruses Associated with Asymptomatic and Symptomatic Neonatal Infections in Vellore, India. J. Virol. 2014, 88, 9060–9071. [Google Scholar] [CrossRef] [Green Version]
- Gazal, S.; Taku, A.K.; Kumar, B. Predominance of rotavirus genotype G6P[11] in diarrhoeic lambs. Vet. J. 2012, 193, 299–300. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Huang, P.; Sun, C.; Han, L.; Vago, F.S.; Li, K.; Zhong, W.; Jiang, W.; Klassen, J.S.; Jiang, X.; et al. Bioengineered Norovirus S60 Nanoparticles as a Multifunctional Vaccine Platform. ACS Nano. 2018, 12, 10665–10682. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Huang, P.; Zhao, D.; Xia, M.; Zhong, W.; Jiang, X.; Tan, M. Effects of rotavirus NSP4 protein on the immune response and protection of the SR69A-VP8* nanoparticle rotavirus vaccine. Vaccine 2020, 39, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Ahmed, L.U.; Stuckert, M.R.; McGinnis, K.R.; Liu, Y.; Tan, M.; Huang, P.; Zhong, W.; Zhao, D.; Jiang, X.; et al. Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens. PLoS Pathog. 2020, 16, e1008386. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Hegde, R.S.; Jiang, X. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J. Virol. 2004, 78, 6233–6242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.; Farkas, T.; Zhong, W.; Tan, M.; Thornton, S.; Morrow, A.L.; Jiang, X. Norovirus and histo-blood group antigens: Demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 2005, 79, 6714–6722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, M.; Wei, C.; Wang, L.; Cao, D.; Meng, X.J.; Jiang, X.; Tan, M. Development and evaluation of two subunit vaccine candidates containing antigens of hepatitis E virus, rotavirus, and astrovirus. Sci. Rep. 2016, 6, 25735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Liu, Y.; Tan, M. Histo-blood group antigens as receptors for rotavirus, new understanding on rotavirus epidemiology and vaccine strategy. Emerg. Microbes Infect. 2017, 6, e22. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; McGinnis, K.R.; Liu, Y.; Huang, P.; Tan, M.; Stuckert, M.R.; Burnside, R.E.; Jacob, E.G.; Ni, S.; Jiang, X.; et al. Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens. Proc. Natl. Acad. Sci. USA 2021, 118, e2107963118. [Google Scholar] [CrossRef]
- Patel, M.; Glass, R.I.; Jiang, B.; Santosham, M.; Lopman, B.; Parashar, U. A systematic review of anti-rotavirus serum IgA antibody titer as a potential correlate of rotavirus vaccine efficacy. J. Infect. Dis. 2013, 208, 284–294. [Google Scholar] [CrossRef] [Green Version]
- Clarke, E.; Desselberger, U. Correlates of protection against human rotavirus disease and the factors influencing protection in low-income settings. Mucosal. Immunol. 2015, 8, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Ward, R.L.; Bernstein, D.I.; Shukla, R.; Young, E.C.; Sherwood, J.R.; McNeal, M.M.; Walker, M.C.; Schiff, G.M. Effects of antibody to rotavirus on protection of adults challenged with a human rotavirus. J. Infect. Dis. 1989, 159, 79–88. [Google Scholar] [CrossRef]
- Azevedo, M.S.; Yuan, L.; Iosef, C.; Chang, K.O.; Kim, Y.; Nguyen, T.V.; Saif, L.J. Magnitude of serum and intestinal antibody responses induced by sequential replicating and nonreplicating rotavirus vaccines in gnotobiotic pigs and correlation with protection. Clin. Diagn. Lab. Immunol. 2004, 11, 12–20. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, M.; Huang, P.; Tan, M. A Pseudovirus Nanoparticle-Based Trivalent Rotavirus Vaccine Candidate Elicits High and Cross P Type Immune Response. Pharmaceutics 2022, 14, 1597. https://doi.org/10.3390/pharmaceutics14081597
Xia M, Huang P, Tan M. A Pseudovirus Nanoparticle-Based Trivalent Rotavirus Vaccine Candidate Elicits High and Cross P Type Immune Response. Pharmaceutics. 2022; 14(8):1597. https://doi.org/10.3390/pharmaceutics14081597
Chicago/Turabian StyleXia, Ming, Pengwei Huang, and Ming Tan. 2022. "A Pseudovirus Nanoparticle-Based Trivalent Rotavirus Vaccine Candidate Elicits High and Cross P Type Immune Response" Pharmaceutics 14, no. 8: 1597. https://doi.org/10.3390/pharmaceutics14081597
APA StyleXia, M., Huang, P., & Tan, M. (2022). A Pseudovirus Nanoparticle-Based Trivalent Rotavirus Vaccine Candidate Elicits High and Cross P Type Immune Response. Pharmaceutics, 14(8), 1597. https://doi.org/10.3390/pharmaceutics14081597





