Translating a Thin-Film Rehydration Method to Microfluidics for the Preparation of a SARS-CoV-2 DNA Vaccine: When Manufacturing Method Matters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Plasmid DNA Construct
2.3. Mice Experiments
2.4. Liposomal Formulation and pCMVkan-S Complexation
2.5. Transmission Electron Microscopy (TEM)
2.6. Small Angle X-ray Scattering (SAXS)
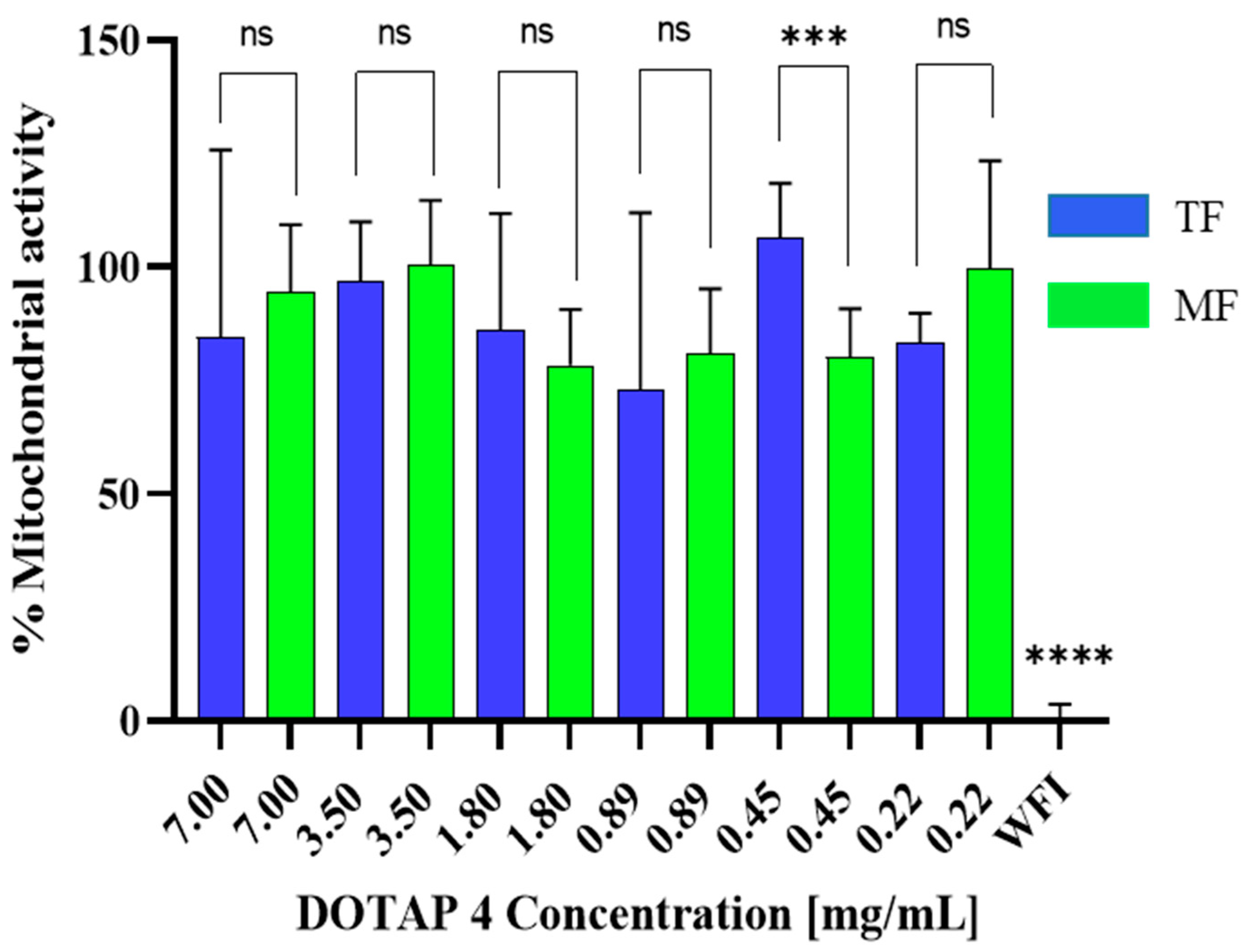
2.7. Mitochondrial Activity (WST-1)
2.8. Liposomal Complex Stability
2.9. Transfection Studies and Immunofluorescence
2.10. Immunogenicity Measurement in Immunized Mice
2.11. Statistical Analysis
3. Results
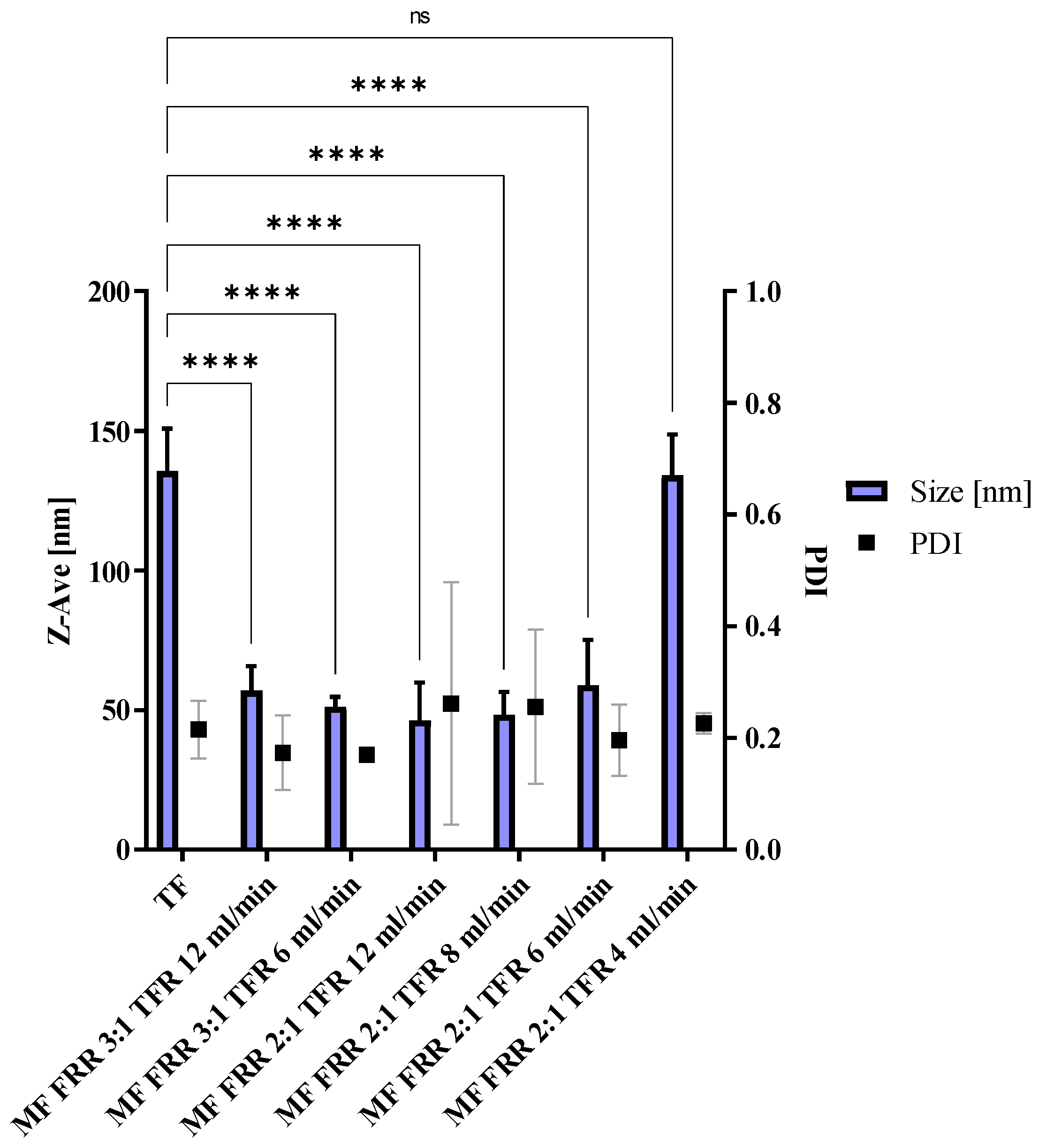
3.1. Microfluidics Method Optimization, Blank Liposomes
3.2. Toxicity of Microfluidics Manufactured Liposomes
3.3. Liposome/pCMVkan-S Complexation: Comparison with Thin-Film Rehydration-Manufactured Liposomes
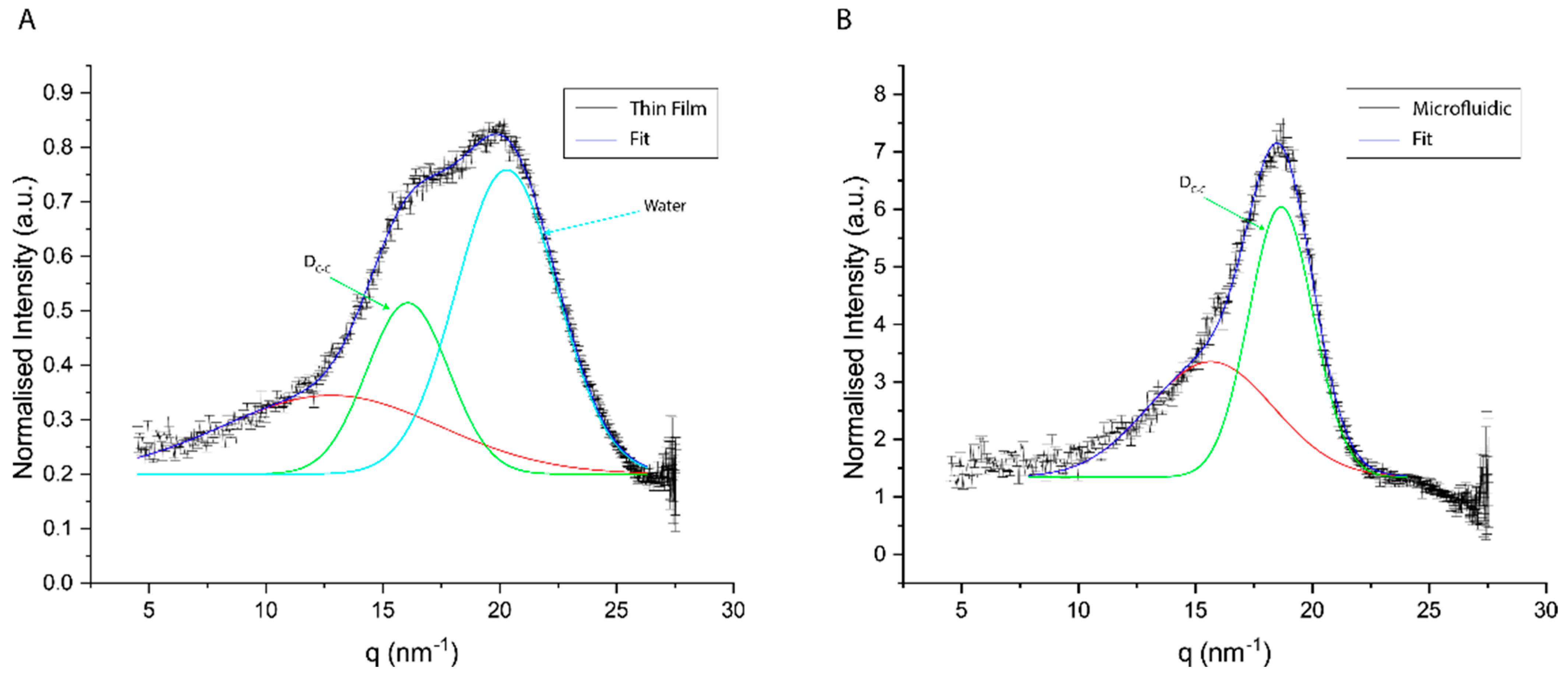
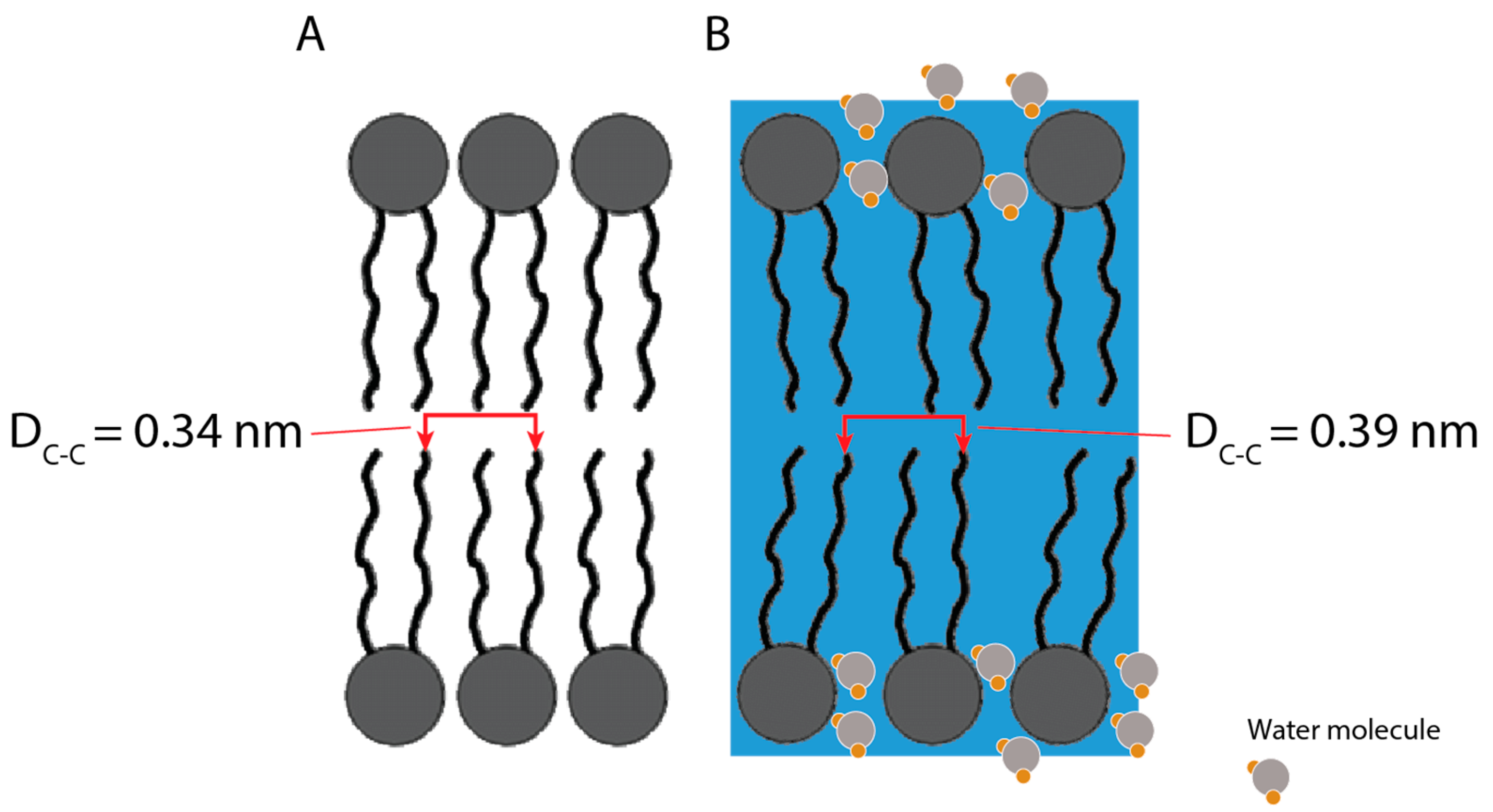
3.4. SAXS Results
3.5. Transfection Efficacy of Microfluidics Manufactured DOTAP 4 on HEK 293 Cells
3.6. Immunogenicity in Mice
3.6.1. Total S-Specific IgG
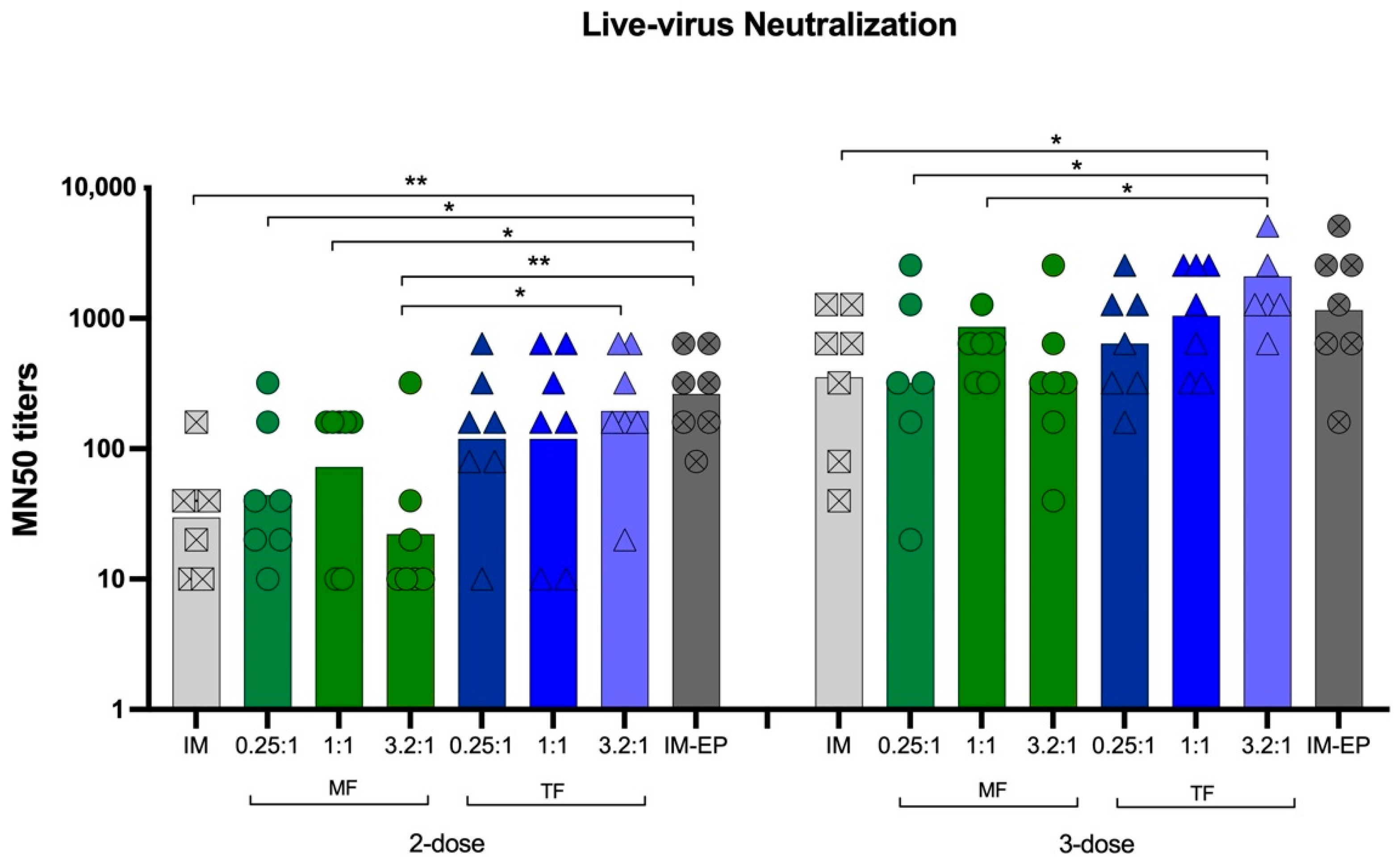
3.6.2. Neutralizing Antibodies against Live Viruses
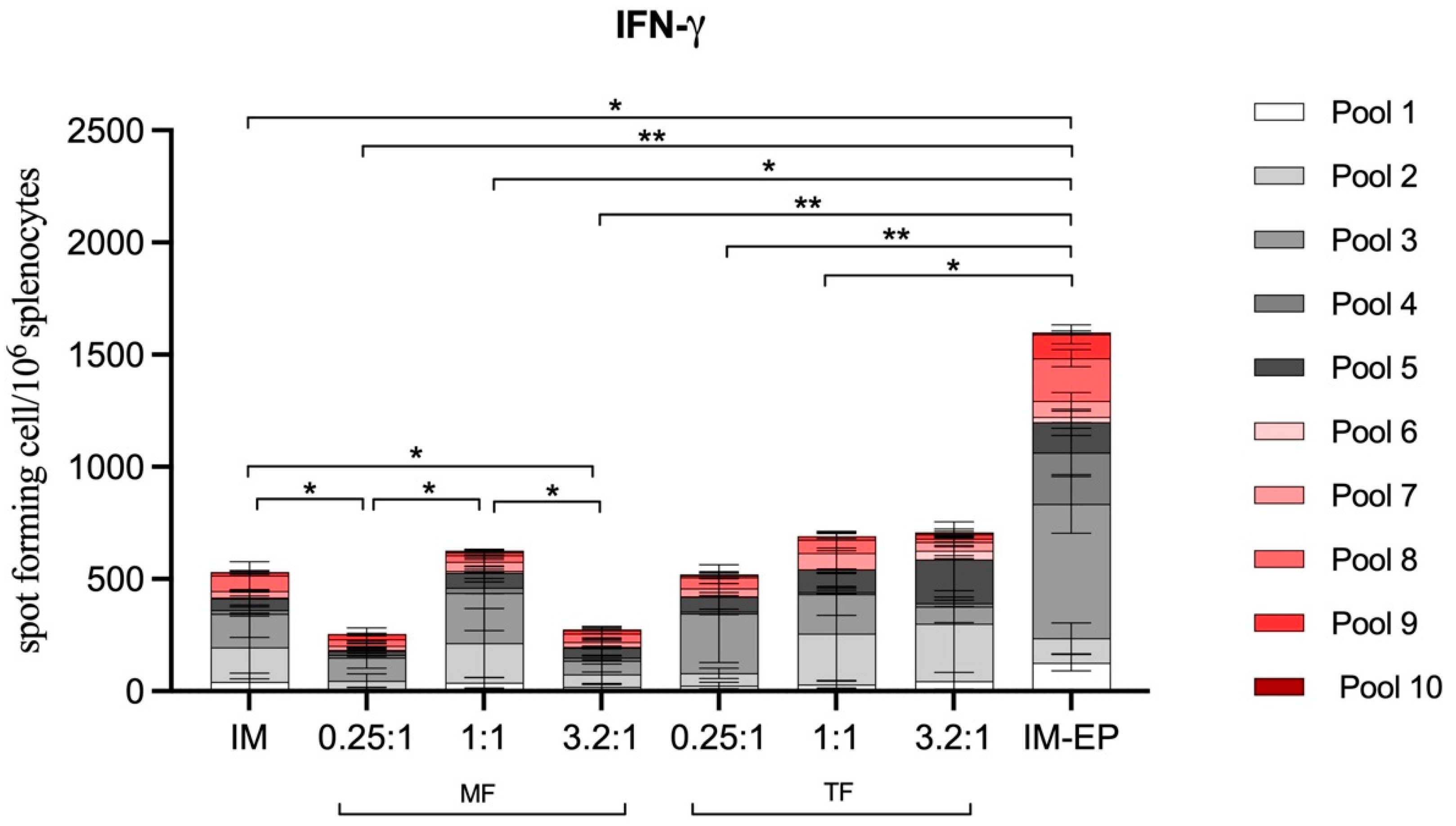
3.6.3. T-Cell Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Calvo Fernández, E.; Zhu, L.Y. Racing to immunity: Journey to a COVID-19 vaccine and lessons for the future. Br. J. Clin. Pharmacol. 2021, 87, 3408–3424. [Google Scholar] [CrossRef] [PubMed]
- Karim, S.A. COVID-19 vaccine affordability and accessibility. Lancet 2020, 396, 238. [Google Scholar] [CrossRef]
- Global Dashboard for Vaccine Equity: UNDP. Available online: https://data.undp.org/vaccine-equity/ (accessed on 26 April 2022).
- Huang, C.; Yang, L.; Pan, J.; Xu, X.; Peng, R. Correlation between vaccine coverage and the COVID-19 pandemic throughout the world: Based on real-world data. J. Med. Virol. 2022, 94, 2181–2187. [Google Scholar] [CrossRef] [PubMed]
- Census. Coronavirus (COVID-19) Latest Insights: Deaths 2022. Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/articles/coronaviruscovid19latestinsights/deaths#deaths-by-vaccination-status (accessed on 19 June 2022).
- Funk, P.R.; Yogurtcu, O.N.; Forshee, R.A.; Anderson, S.A.; Marks, P.W.; Yang, H. Benefit-risk assessment of COVID-19 vaccine, mRNA (Comirnaty) for age 16–29 years. Vaccine 2022, 40, 2781–2789. [Google Scholar] [CrossRef]
- Vashi, A.P.; Coiado, O.C. The future of COVID-19: A vaccine review. J. Infect. Public Health 2021, 14, 1461–1465. [Google Scholar] [CrossRef] [PubMed]
- Chatzikleanthous, D.; O’Hagan, D.T.; Adamo, R. Lipid-Based Nanoparticles for Delivery of Vaccine Adjuvants and Antigens: Toward Multicomponent Vaccines. Mol. Pharm. 2021, 18, 2867–2888. [Google Scholar] [CrossRef] [PubMed]
- Bangham, A.D.; Standish, M.M.; Watkins, J.C. Diffusion of univalent ions across the lamellae of swollen phospholipids. J. Mol. Biol. 1965, 13, 238–252. [Google Scholar] [CrossRef]
- Mader, C.; Küpcü, S.; Sára, M.; Sleytr, U. Stabilizing effect of an S-layer on liposomes towards thermal or mechanical stress. Biochim. Biophys. Acta (BBA)-Biomembr. 1999, 1418, 106–116. [Google Scholar] [CrossRef] [Green Version]
- Khadke, S.; Roces, C.B.; Cameron, A.; Devitt, A.; Perrie, Y. Formulation and manufacturing of lymphatic targeting liposomes using microfluidics. J. Control. Release 2019, 307, 211–220. [Google Scholar] [CrossRef]
- Roces, C.B.; Lou, G.; Jain, N.; Abraham, S.; Thomas, A.; Halbert, G.W.; Perrie, Y. Manufacturing considerations for the development of lipid nanoparticles using microfluidics. Pharmaceutics 2020, 12, 1095. [Google Scholar] [CrossRef]
- Schmidt, S.T.; Christensen, D.; Perrie, Y. Applying microfluidics for the production of the cationic liposome-based vaccine adjuvant CAF09b. Pharmaceutics 2020, 12, 1237. [Google Scholar] [CrossRef] [PubMed]
- Peletta, A.; Prompetchara, E.; Tharakhet, K.; Kaewpang, P.; Buranapraditkun, S.; Techawiwattanaboon, T.; Jbilou, T.; Krangvichian, P.; Sirivichayakul, S.; Manopwisedjaroen, S.; et al. DNA Vaccine Administered by Cationic Lipoplexes or by In Vivo Electroporation Induces Comparable Antibody Responses against SARS-CoV-2 in Mice. Vaccines 2021, 9, 874. [Google Scholar] [CrossRef] [PubMed]
- Prompetchara, E.K.C.; Tharakhet, K.; Kaewpang, P.; Buranapraditkun, S.; Techawiwattanaboon, T.; Sathean-anan-kun, S.; Pitakpolrat, P.; Watcharaplueksadee, S.; Phumiamorn, S.; Wijagkanalan, W.; et al. DNA vaccine candidate encoding SARS-CoV-2 spike proteins elicited potent humoral and Th1 cell-mediated immune responses in mice. PLoS ONE 2021, 16, e0248007. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.; Hammel, P. AI334 and AQ806 antibodies recognize the spike S protein from SARS-CoV-2 by immunofluorescence. Antib. Rep. 2020, 3, e192. [Google Scholar] [CrossRef]
- Bonechi, C.; Martini, S.; Ciani, L.; Lamponi, S.; Rebmann, H.; Rossi, C.; Ristori, S. Using liposomes as carriers for polyphenolic compounds: The case of trans-resveratrol. PLoS ONE 2012, 7, e41438. [Google Scholar] [CrossRef] [PubMed]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P.; et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Addetia, A.; Crawford, K.H.D.; Dingens, A.; Zhu, H.; Roychoudhury, P.; Huang, M.L.; Jerome, K.R.; Bloom, J.D.; Greninger, A.L. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol. 2020, 58, e02107-20. [Google Scholar] [CrossRef]
- Babiuk, S.; Baca-Estrada, M.E.; Foldvari, M.; Middleton, D.M.; Rabussay, D.; Widera, G.; Babiuk, L.A. Increased gene expression and inflammatory cell infiltration caused by electroporation are both important for improving the efficacy of DNA vaccines. J. Biotechnol. 2004, 110, 1–10. [Google Scholar] [CrossRef]
- Chiarella, P.; Massi, E.; De Robertis, M.; Sibilio, A.; Parrella, P.; Fazio, V.M.; Signori, E. Electroporation of skeletal muscle induces danger signal release and antigen-presenting cell recruitment independently of DNA vaccine administration. Expert Opin. Biol. Ther. 2008, 8, 1645–1657. [Google Scholar] [CrossRef]
- Plotkin, S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010, 17, 1055–1065. [Google Scholar] [CrossRef] [Green Version]
- Mills, T.T.; Toombes, G.E.; Tristram-Nagle, S.; Smilgies, D.-M.; Feigenson, G.W.; Nagle, J.F. Order parameters and areas in fluid-phase oriented lipid membranes using wide angle X-ray scattering. Biophys. J. 2008, 95, 669–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitter, J.; Lechner, R.; Dencher, N. Interactions of hydration water and biological membranes studied by neutron scattering. ACS Publ. 1999, 103, 8036–8050. [Google Scholar] [CrossRef]
- Zaccai, G.; Blasie, J.; Schoenborn, B. Neutron diffraction studies on the location of water in lecithin bilayer model membranes. Proc. Natl. Acad. Sci. USA 1975, 72, 376–380. [Google Scholar] [CrossRef] [Green Version]
- Nickels, J.D.; Katsaras, J. Water and lipid bilayers. Membr. Hydration 2015, 71, 45–67. [Google Scholar]
- Caracciolo, G.; Pozzi, D.; Amenitsch, H.; Caminiti, R. Multicomponent cationic lipid−DNA complex formation: Role of lipid mixing. Langmuir 2005, 21, 11582–11587. [Google Scholar] [CrossRef] [PubMed]
- Koltover, I.; Salditt, T.; Safinya, C. Phase diagram, stability, and overcharging of lamellar cationic lipid–DNA self-assembled complexes. Biophys. J. 1999, 77, 915–924. [Google Scholar] [CrossRef] [Green Version]
- Zasadzinski, J. Transmission electron microscopy observations of sonication-induced changes in liposome structure. Biophys. J. 1986, 49, 1119–1130. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Luo, L.; Liang, S.; Long, M.; Xu, H. Influence of probe-sonication process on drug entrapment efficiency of liposomes loaded with a hydrophobic drug. Int. J. Polym. Mater. Polym. Biomater. 2019, 68, 193–197. [Google Scholar] [CrossRef]
- Modena, M.M.; Rühle, B.; Burg, T.P.; Wuttke, S. Nanoparticle characterization: What to measure? Adv. Mater. 2019, 31, 1901556. [Google Scholar] [CrossRef]
- O’Sullivan, C.; Rutten, P.; Schatz, C. Why tech transfer may be critical to beating COVID-19. Pharm. Med. Prod. Pract. 2020, 23, 2020. [Google Scholar]
- Lemoine, C.; Nidom, R.; Ventura, R.; Indrasari, S.; Normalina, I.; Santoso, K.; Derouet, F.; Barnier-Quer, C.; Borchard, G.; Collin, N.; et al. Better Pandemic Influenza Preparedness through Adjuvant Technology Transfer: Challenges and Lessons Learned. Vaccines 2021, 9, 461. [Google Scholar] [CrossRef] [PubMed]







| Sample | Z-Ave (nm) ± SD TF | Z-Ave (nm) ± SD MF | p-Value Summary 1 |
|---|---|---|---|
| Blank Liposomes | 135.4 ± 15.3 | 134.0 ± 14.7 | ns |
| pCMVkan-S 0.25:1 | 490.7 ± 149.7 | 829.8 ± 26.7 | **** |
| pCMVkan-S 1:1 | 515.8 ± 35.9 | 925.8 ± 68.5 | **** |
| pCMVkan-S 3.2:1 | 565.9 ± 174.4 | 329.9 ± 35.9 | ** |
| pCMVkan-S 10:1 | 6444 ± 2853 | 757.2 ± 252.3 | *** |
| pCMVkan-S 25:1 | 8974 ± 6056 | 8294 ± 7483 | ns |
| pCMVkan-S 50:1 | 5776 ± 2788 | 6112 ± 4514 | ns |
| pCMVkan-S 100:1 | 434.5 ± 11.9 | 636.8 ± 283.3 | ns |
| Thin Film | Microfluidic | ||
|---|---|---|---|
| Chain–Chain Spacing (DC-C) | Water Peak | Chain–Chain Spacing (DC-C) | |
| q0 (nm−1) | 16.06 ± 0.04 | 20.29 ± 0.03 | 18.66 ± 0.01 |
| D (nm) | 0.39 ± < 0.01 | 0.31 ± < 0.01 | 0.34 ± < 0.01 |
| HWHM (nm−1) | 1.57 ± 0.07 | 4.32 ± 0.05 | 1.37 ± 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peletta, A.; Prompetchara, E.; Tharakhet, K.; Kaewpang, P.; Buranapraditkun, S.; Yostrerat, N.; Manopwisedjaroen, S.; Thitithanyanont, A.; Avaro, J.; Krupnik, L.; et al. Translating a Thin-Film Rehydration Method to Microfluidics for the Preparation of a SARS-CoV-2 DNA Vaccine: When Manufacturing Method Matters. Pharmaceutics 2022, 14, 1427. https://doi.org/10.3390/pharmaceutics14071427
Peletta A, Prompetchara E, Tharakhet K, Kaewpang P, Buranapraditkun S, Yostrerat N, Manopwisedjaroen S, Thitithanyanont A, Avaro J, Krupnik L, et al. Translating a Thin-Film Rehydration Method to Microfluidics for the Preparation of a SARS-CoV-2 DNA Vaccine: When Manufacturing Method Matters. Pharmaceutics. 2022; 14(7):1427. https://doi.org/10.3390/pharmaceutics14071427
Chicago/Turabian StylePeletta, Allegra, Eakachai Prompetchara, Kittipan Tharakhet, Papatsara Kaewpang, Supranee Buranapraditkun, Nongnaphat Yostrerat, Suwimon Manopwisedjaroen, Arunee Thitithanyanont, Jonathan Avaro, Leonard Krupnik, and et al. 2022. "Translating a Thin-Film Rehydration Method to Microfluidics for the Preparation of a SARS-CoV-2 DNA Vaccine: When Manufacturing Method Matters" Pharmaceutics 14, no. 7: 1427. https://doi.org/10.3390/pharmaceutics14071427
APA StylePeletta, A., Prompetchara, E., Tharakhet, K., Kaewpang, P., Buranapraditkun, S., Yostrerat, N., Manopwisedjaroen, S., Thitithanyanont, A., Avaro, J., Krupnik, L., Neels, A., Ruxrungtham, K., Ketloy, C., & Borchard, G. (2022). Translating a Thin-Film Rehydration Method to Microfluidics for the Preparation of a SARS-CoV-2 DNA Vaccine: When Manufacturing Method Matters. Pharmaceutics, 14(7), 1427. https://doi.org/10.3390/pharmaceutics14071427







