Abstract
Bacteriophages, viruses that infect and replicate within bacteria, impact bacterial responses to antibiotics in complex ways. Recent studies using lytic bacteriophages to treat bacterial infections (phage therapy) demonstrate that phages can promote susceptibility to chemical antibiotics and that phage/antibiotic synergy is possible. However, both lytic and lysogenic bacteriophages can contribute to antimicrobial resistance. In particular, some phages mediate the horizontal transfer of antibiotic resistance genes between bacteria via transduction and other mechanisms. In addition, chronic infection filamentous phages can promote antimicrobial tolerance, the ability of bacteria to persist in the face of antibiotics. In particular, filamentous phages serve as structural elements in bacterial biofilms and prevent the penetration of antibiotics. Over time, these contributions to antibiotic tolerance favor the selection of resistance clones. Here, we review recent insights into bacteriophage contributions to antibiotic susceptibility, resistance, and tolerance. We discuss the mechanisms involved in these effects and address their impact on bacterial fitness.
1. Introduction
The modern era of antibiotics started with the discovery of penicillin by Sir Alexander Fleming in 1928 [1]. In the following decades, advances in drug screening, chemical synthesis, and manufacturing led to the wide availability of several classes of highly effective antimicrobial agents and a well-developed commercial pharmaceutical industry. Antimicrobial therapy rapidly became a cornerstone of human healthcare.
Bacterial geneticists initially believed that the development of widespread antimicrobial resistance (AMR) was unlikely. However, this view failed to appreciate the facility with which bacteria exchange genetic information [2,3,4], including the horizontal transfer of AMR [5,6]. Researchers also failed to consider the role of antimicrobial tolerance—the ability of metabolically dormant bacteria and bacteria sheltered within biofilms to evade antibiotics—as a gateway for the development of AMR.
Unfortunately, widespread AMR to many classes of antibiotics is now prevalent and is a major threat to human health [7]. In the United States alone, more than 2.8 million infections and 35,000 deaths per year are attributable to AMR bacteria [8]. Globally, at least 1.2 million people died in 2019 because of bacterial AMR infections [9]. The total number of deaths attributable to AMR organisms is expected to reach 10 million globally by 2050 [9].
Despite the advancements in biotechnology, genetic engineering, and synthetic chemistry, antibiotic development has failed to keep pace with the spread of AMR [10]. There is, therefore, great interest in identifying factors that impact antibiotic treatment failures as well as therapies that can complement or substitute for antibiotics [11].
Given this need, there is a resurgence of interest in bacteriophages (phages), viruses that parasitize bacteria [12,13]. Before the discovery of penicillin, phages were discovered independently in 1915 by Frederick Twort, a British pathologist [14], and in 1917 by Félix d’Hérelle, a French-Canadian microbiologist [15]. Despite the great promise of phages as antibacterial agents, penicillin and other antibiotics were more successful. For many decades, research into the clinical applications of phages was largely abandoned in North America and Western Europe [16,17]. The spread of AMR and lack of antibiotic development has led to renewed interest in phages.
Phages employ one of several reproductive strategies. Lytic phages are obligate pathogens of bacteria that lyse their bacterial hosts upon replication [18]. Lysogenic phages can integrate within the bacterial genome and lyse their hosts opportunistically [19]. A sub-set of lysogenic phages (notably inoviruses) emerge from their bacterial hosts without lysis; this is called chronic infection [20,21]. Each of these phage reproductive strategies has distinct impacts on bacterial biology [22] in ways that may influence antimicrobial therapy.
In addition, over the past 15 years, the interest in using bacteriophage therapy has been re-kindled in laboratories and hospitals. Despite mixed results from phage therapy clinical trials [23,24,25], there are multiple instances and case series of successful phage therapy using either conventional or modified phages [26,27,28,29,30]. Phage therapy is generally safe and well-tolerated [31], although many questions remain regarding the optimal dosages and treatment regimens [32].
Bacteriophages impact bacteria in ways that intersect with how conventional antibiotics impact bacteria. Like penicillin and many other early antibiotics, phages have a long evolutionary history with bacteria. Phages exert strong selective pressures on bacteria and have major roles in the transfer of genetic material between bacterial strains and species [33,34,35,36]. This review focuses on the contributions of phages to bacterial tolerance and resistance development to conventional pharmaceutical antibiotics. In particular, we review recent insights into lytic phage contributions to antibiotic susceptibility, lytic and lysogenic phages to overcome antibiotic resistance, and filamentous phages to treat antibiotic tolerance. We discuss the mechanisms involved in these effects and address their impact on bacterial fitness and antimicrobial therapy.
2. Lytic and Lysogenic Phages Contribute to AMR
AMR occurs when inherited mutations in bacteria cause the drugs used to treat infections to become less effective [9,37]. The effects of such mutations are measured by minimum inhibitory concentration (MIC), which is the lowest concentration of antibiotics required to inhibit bacteria growth [38]. As a group, bacteria are not uniformly susceptible or resistant to different antibiotics. The level of susceptibility depends on the composition of bacteria, which leads to a range of MIC for bacterial species. The average MIC is viewed as a convenient metric to evaluate the resistance of tested bacteria.
AMR is heritable and is mediated by the presence of antibiotic resistance genes (ARGs). Bacteria acquire ARGs in multiple ways [39]. One of the ways is that bacteria acquire antibiotic resistance via horizontal transfer of ARGs between individual bacteria or between bacterial species, which can be mediated by bacteriophages, via vertical transfer of ARGs to daughter bacteria, or through de novo chromosomal mutations. There is, therefore, a need to understand the mechanisms, frequency, reservoirs, and vectors governing the horizontal transfer of AMR (Figure 1A).
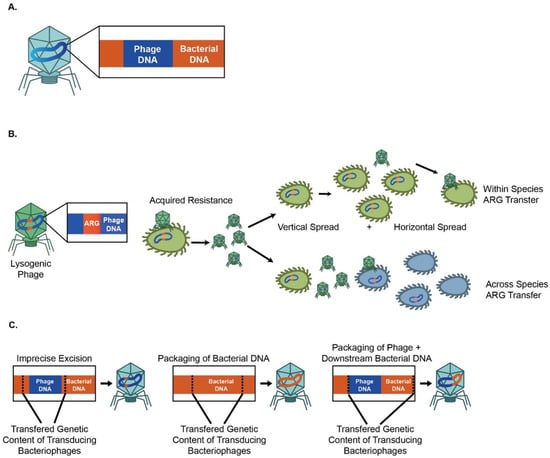
Figure 1.
Lytic and lysogenic phages can contribute to bacterial antimicrobial resistance. (A) Bacteriophages can carry MGEs and mediate ARG movements. (B) Lysogenic bacteriophages contribute to the vertical and horizontal spread of ARGs. (C) Three main mechanisms for the phage-mediated spread of genetic material.
Although most AMR does not originate or spread via phages, there are indications that bacteriophages can be involved in the transfer of AMR. Here we review that literature.
2.1. Horizontal Transfer of Mobile Genetic Elements (MGEs) Promotes the Acquisition and Spread of ARG
Horizontal gene transfer, the movement of genetic material between organisms, is responsible for the dissemination of ARGs [40]. It allows bacteria to acquire new genetic material from outside their clonal lineage. Because of its ability to transfer genetic elements, horizontal gene transfer contributes significantly to the spread of bacterial AMR. Horizontal gene transfer has been extensively covered in several excellent reviews [40,41,42,43]. Horizontal gene transfer is mediated by mobile genetic elements (MGEs), such as bacteriophages and plasmids, which provide an important resource for bacterial genetic diversity as well as bacterial evolution [43,44] (Figure 1B). MGEs mediate the movement of genetic material within genomes or between bacterial hosts [43]. Several comprehensive reviews of MGEs are available [40,43]. The acquisition of ARGs is facilitated by the horizontal gene transfer [45,46] of MGEs, including plasmids [47], transposons, and integrons, through conjugation [48] and viral transduction [48,49]. In a later section, we will further discuss how phages contribute to the acquisition of ARGs. Most evolutionary models consider MGE-mediated horizontal transfer of ARGs from a cost–benefit perspective. Plasmids and other MGEs are an efficient means of exchanging genetic information. However, these elements are still costly as they necessitate the synthesis of proteins (such as conjugation pili), RNA, and DNA, which incur a fitness cost [50,51]. Further, MGEs often integrate into chromosomes, thereby potentially disrupting important genes [52]. However, this cost can be surmounted by other adaptive or addictive traits, such as antibiotic resistance [40,53,54].
2.2. Lysogenic Bacteriophages Can Contribute to the Vertical and Horizontal Spread of ARGs
Lysogenic (temperate) phages can integrate into the bacterial genome as prophages or persist as an extrachromosomal plasmid [55]. However, prophages can also be induced to undergo lytic replication at times of bacterial stress.
Many prophages carry ARGs [56,57,58,59,60,61]. A longitudinal study of viromes from human fecal samples found that antibiotic resistance genes were highly abundant among phage genomes [62]. In another study, 77% of 80 fecal samples from healthy individuals showed that they harbor at least one ARG [63]. Resistance genes including blaTEM, blaCTX-M-1, mecA, armA, qnrA, and qnrS were identified; blaTEM, qnrA, and blaCTX-M-1 were the most abundant, and armA, qnrS, and mecA were less prevalent [63]. High levels of ARGs were likewise reported in phages from the airways of individuals with cystic fibrosis (CF) [64,65] and feces samples of antibiotic-treated mice [66]. Moreover, in an ex vivo study, phages isolated from antibiotic-treated mice were transferred to aerobically cultured naïve microbiota and found to increase the frequency of drug resistance isolates in naïve microbiota compared to cultures infected with phages from untreated mice [66].
Phages also transfer ARGs to the environment. Lekunberri et al. analyzed 33 viromes sampled from diverse habitats, including human and pig feces, raw sewage, fresh water, and marine environments from public repositories, finding that human-associated viromes do not contain ARGs, while six pig-associated viromes harbored a high abundance of ARGs [59]. Phages from sewage and aquatic environments from around the world have a high diversity of ARGs [59], and studies have shown that aquatic phages serve as reservoirs for ARGs [67,68].
The presence of ARGs in lysogenic phages is consistent with strategies employed by phages to ensure their maintenance within the bacterial genome [55,69,70,71,72,73]. Prophages often express genes that provide competitive advantages to their host, including genes involved in bacterial pathogenicity [74,75,76] or virulence factors [77,78,79,80,81,82]. Prophages also have various mechanisms to prevent infection by other phages [83,84,85,86]. However, the role of phages in the spread of ARGs remains controversial. Enault et al. pointed out that estimates of phage-mediated ARG transfer could be too high due to excessive bacterial DNA content as well as inflated false positives because of the relaxed threshold in in silico detection of ARGs [87]. Instead, Enault et al. suggested that to carefully quantify the bacterial DNA contamination, use a conservative threshold to quantify bona fide ARGs, and apply a discovery-based work process with a manual inspection to remove false positive hits in ARGs. These steps may help properly estimate the role of phages in the spread of ARGs.
The relative importance of phages as a mechanism of horizontal transfer of ARGs is also unclear. The cost/benefit relationship of phages to bacteria is more complex than for plasmids or other MGEs, as the benefits conferred by genes associated with prophages are offset by the threat of bacterial lysis. However, lysis might also help transfer ARGs to neighboring bacterial populations during times of stress, such as during antibiotic treatment. In this way, the lytic portion of the lysogenic phage life cycle might benefit the rest of the bacterial population in some settings [88].
2.3. Both Lytic and Lysogenic Phages Can Promote Dissemination of ARGs via Transduction
Phages can also spread ARGs via transduction, a process in which bacterial DNA is packaged into phage particles during lysis with progeny phages to infect new susceptible bacterial hosts. [89,90]. Phages thereby help ensure the efficient transfer of DNA to appropriate hosts [91,92,93,94]. ARGs can be mobilized by both lytic and lysogenic phages [95].
Three methods of phage-mediated transduction have been identified (Figure 1C). First, specialized transduction is mediated by temperate phages, which inadvertently mobilize host genes adjacent to phage insertion sites as a result of imprecise excision [96]. Second, generalized transduction occurs when bacterial DNA, instead of phage DNA, is packaged into the phage head [96]. Given this ability to package large fragments of DNA, transduction can indirectly mediate the transfer of ARGs associated with other MGE. Zhang et al. showed that T4-like phage misloaded plasmid-borne ARGs by generalized transduction [97]. Transduction can mediate the transfer of ARGs between bacterial species as well. Studies have likewise shown that polyvalent phages disseminate ARGs between several Enterococcus [98] and Staphylococcus [99] species under laboratory conditions. Evidence suggests that phages contribute to the recombination of ARGs such as blaCTX-M, mel, and tetM across multiple bacteria genera, including S. enterica, E. coli, S. pneumoniae, and S. sonnei [100,101]. Although phages can mediate horizontal bacterial DNA exchange via specialized and generalized transduction, these processes are relatively inefficient. The frequencies of these processes are low, and the transfer of ARGs is dependent on antibiotic resistance genes immediately flanking phage insertion sites and imprecise excision in specialized transduction [88].
The third and most recently discovered form of phage-mediated transduction is lateral transduction. Here, newly generated phage capsids package predominantly bacterial DNA downstream of the phage insertion site with high efficiency [102]. Lateral transduction is the most powerful mode of phage-mediated DNA transfer, capable of transferring several hundred kilobases and a large span of the bacterial genome [102]. Instead of using the ppac sites as in generalized transduction, lateral transduction uses embedded pac sites for DNA packaging. Recently, Humphrey et al. used S. aureus and Salmonella spp. as reference organisms and showed that chromosomally encoded bacterial genes could be transferred at up to 1000-fold higher rates by lateral transduction than generalized transduction [103].
Conjugation involving plasmids is perhaps the best-understood route of horizontal gene transfer [104,105]. Studies showed that phages could potentially inhibit bacterial conjugation and potentially reduce ARG dissemination [106]. However, a recent study showed that when phages infect SXT -containing V. cholerae, high-frequency conjugative transfer of SXT ICEs is induced, leading to the dissemination of both phage and antibiotic resistances. Similarly, coliphage could also stimulate higher frequency conjugation of ICEs from an E. coli donor to a V. cholerae recipient [107].
3. Bacteriophage and Antimicrobial Tolerance
Antimicrobial tolerance was first coined by Horne et al. [108]. Later, Kester and Fortune defined antimicrobial tolerance as a population-level phenomenon that enables the population to survive the duration of a transient antibiotic treatment several times above the MIC without a resistance mechanism [38,109]. Unlike AMR, antimicrobial tolerance has a distinct mechanism to escape antibiotic-mediated killing [110]. In addition to occasional mutations, antimicrobial tolerance can result from metabolic adaptations [111] or biofilm production [112] and does not confer a higher MIC to the descendants of bacterial survivors [113]. Moreover, antimicrobial tolerance has been shown to increase antimicrobial resistance [114,115,116]. Hence, it is important to understand how bacteriophages contribute to antimicrobial tolerance to prevent further resistance development.
Filamentous Bacteriophages Contribute to Bacterial Tolerance by Promoting Biofilm Production
A bacterial biofilm is a complex structure that adheres to biological or non-biological surfaces [117]. Biofilms encapsulate bacteria with a matrix that includes polysaccharides (e.g., alginate) and eDNA, as well as bacterial proteins [118].
Bacterial biofilms promote antibiotic tolerance, the ability of bacteria to proliferate despite treatment with antimicrobial agents, by preventing antibiotics from penetrating and then reaching target bacteria [119,120,121,122,123,124,125]. Over time, this tolerance is thought to select for antimicrobial resistance [126,127,128]. Similar effects may also characterize sputum colonized by bacteria. There are reports that the sputum of individuals with CF binds positively charged antibiotics and reduces their efficacy against P. aeruginosa [129,130].
Pf phage harbored by P. aeruginosa contributes to P. aeruginosa biofilm formation by Pf positive strain [131]. Filamentous phages belong to a subgroup of the family Inoviridae, and are long, thin phages ranging from 800 nm to 4 µm in length [20,132]. They are broadly distributed and can infect both Gram-positive and Gram-negative bacteria [133]. Most filamentous phages are lysogenic but extrude progeny phages from the bacterial cell without lysis in a cycle known as chronic infection [20,21]. Hence, when isolated, filamentous phages do not form clear plaques like lytic phages, but instead, form opaque zones of reduced growth that resemble the turbid plaques of lysogenic phages [55]. As with other lysogenic phages, filamentous phages influence the virulence of hosts by transferring genetic material through horizontal and vertical transmission [134,135].
Pf phages are widespread among P. aeruginosa [136]. During biofilm growth, Pf genes are among the most upregulated, with a 100–1000 fold increase in expression relative to the planktonic growth mode [137]. Proteomic studies identified that Pf genes were a major portion of the most upregulated genes during anaerobic growth conditions that mimic those of the lungs of individuals with CF [138]. Rice et al. reported that Pf phages contribute to the P. aeruginosa biofilm formation and virulence. Building on this work, Secor et al. reported that Pf4 promotes the organization of human and microbial biofilm polymers into a liquid crystal [72,139,140,141] (Figure 2A). These effects are mediated by charge-based interactions between phages and polymers, contributing to the adhesivity and viscosity of P. aeruginosa biofilms [142] and promoting bacterial aggregation [143]. Fd phage, a filamentous phage from E. coli, promotes similar structures [142], while a related filamentous phage produced by Neisseria meningitides promotes bacterial colonization to apical surfaces of host epithelial cells [144].
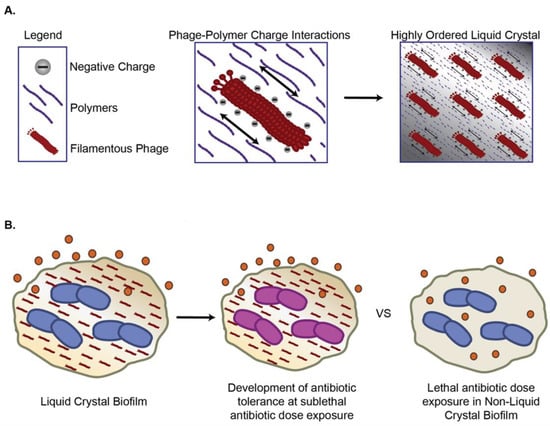
Figure 2.
Filamentous bacteriophages (Inoviruses) increase antimicrobial tolerance via (A) Charged filamentous phages organize polymers into liquid crystal biofilms; (B) Liquid crystal biofilms sequester antibiotics and promote development of antibiotic tolerance at sublethal doses.
Pf phage and liquid crystalline biofilms hinder antibiotic penetration, thereby promoting antibiotic tolerance [72,142] (Figure 2B). In particular, Pf phage increase P. aeruginosa tolerance to tobramycin, gentamicin, and colistin [145]. Tarafder et al. demonstrated that liquid crystalline phage droplets also form occlusive compartments around bacteria that shield them from antibiotics [145]. The potential exclusion could be mediated by either limiting antibiotics diffusion or excluding antibiotics due to thermodynamic forces. However, the underlying mechanisms warrant further investigation.
Filamentous phages may also promote antibiotic tolerance by slowing bacterial growth in ways that diminish the impact of antibiotics targeting cellular division. Pf production comes at a high metabolic cost to P. aeruginosa; Pf+ strains grow more slowly than Pf- strains in vitro [73,131,142,146]. Slow-growing “persister” phenotypes are a major contributor to antibiotic tolerance [147]. These effects may be clinically important. Burgener et al. found that Pf phages were associated with chronic P. aeruginosa infections and worse clinical outcomes in individuals with CF. Moreover, P. aeruginosa strains from patients with Pf phages detected in their sputum show increased antibiotic resistance and, over time, come to dominate the airways of individuals with CF [148]. Similarly, Pf+ strains of P. aeruginosa characterize chronic wound infections [149]. A recent modeling study examined how Pf comes to dominate in the CF lung and other environments. It suggested that antibiotic selection pressure is essential for promoting the dominance of Pf+ strains as in the absence of this, the high energetic cost of producing Pf phage would greatly favor Pf- over Pf+ strains of P. aeruginosa [150]. Together, these results suggest that Pf promotes antibiotic tolerance and may contribute to the selection of antibiotic-resistant mutants over time.
Filamentous phages are associated with drug resistance in other species. For instance, an agricultural pathogen, Ralstonia solanacearum uses an RSM1-like phage to acquire drug resistance at the cost of twitching motility [151,152,153]. Such mechanisms may be common among Inoviridae and their hosts since the production of these bacteriophages without bacterial lysis permits symbiotic relationships to evolve.
4. Lytic Phages and Antibiotic Susceptibility
Lytic or virulent phages infect bacteria and hijack host machinery for genomic replication and virion assembly. Lytic replication results in bacterial lysis, with progeny phages infecting new susceptible bacterial hosts [55].
Bacteria have evolved a myriad of constitutive and inducible defense strategies against lytic phages [154], and these defenses can have direct implications for AMR. Constitutive defense against a phage is achieved by mutation or masking of phage receptors [154]. Surface modifications often come at a fixed but maladaptive and pleiotropic cost to the host bacteria [155], and they can have a direct effect on antibiotic resistance if the phage receptor is involved in AMR mechanisms. Chan et al. showed that Pseudomonas phage OMKO1 binds to the outer membrane porin M (OprM) component of the MexAB and MexXY multi-drug efflux systems of PAO1, which increases bacterial antibiotic susceptibility to drugs exported by this pathway [156] (Figure 3A). This strategy of choosing phages that bind to AMR-involved surface proteins is being exploited by some phage therapy companies. Phage infection poses a selective pressure for the bacteria to lose the AMR transporters and become sensitive to antibiotics. Surface modifications can also indirectly impact bacterial fitness and antibiotic tolerance [157,158]. Westra et al. demonstrated that Pseudomonas aeruginosa PA14 evolves immunity to phage DMS3vir under high-phage conditions through loss of the pilus [159], which can impair biofilm formation and, therefore, may reduce antibiotic tolerance [142,160,161,162] (Figure 3B). In addition, a recent paralleled evolution study suggested a trade-off in bacteria resistant to phage, which leads to a slow growth rate and reduced virulence [163].
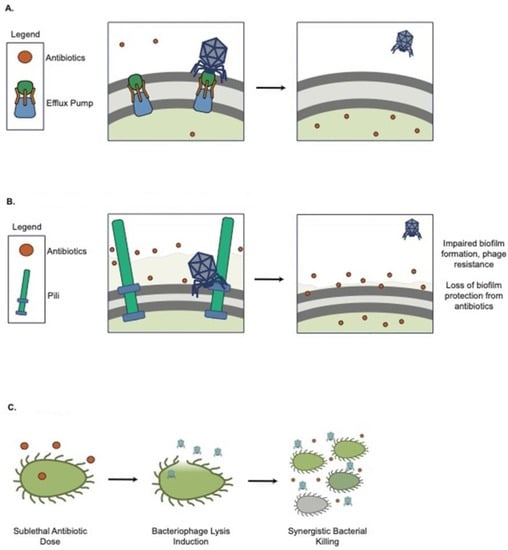
Figure 3.
Lytic phages are used in combination with antimicrobial treatments against bacterial infection. (A) OMKO resistance increases antibiotic susceptibility; (B) DMS3vir resistance (loss of pili) impairs bacterial attachment; (C) Antibiotics trigger phage release and synergistic bacterial killing.
There are indications that lytic phage and conventional antibiotics may act synergistically to kill bacteria (Figure 3C). The phrase phage antibiotic synergy (PAS) was first coined by Comeau et al. [164]. They found that certain antibiotics at sub-lethal concentrations stimulate virulent phage production in vitro, where sub-lethal cefotaxime can enhance uropathogenic E. coli isolate (MFP)’s phage production by 7-fold. Later, many studies, including Tagliaferri et al. [165], showed that various phages could be synergistic with different classes of antibiotics and enhance the eradication of bacteria. Particularly, phage and antibiotic combinations are efficacious in killing P. aeruginosa [166,167,168], E. coli [164,169,170], and S. aureus [171] in both planktonic and biofilm growth modes. Phage/antibiotic interactions can be synergistic, additive, or antagonistic. To facilitate the identification of these patterns, Liu et al. developed a new high-throughput method of screening phage and antibiotic interactions using real-time microtiter plate readouts. Using this approach, they reported that PAS combinations are both phage and antibiotic-specific [172].
PAS has also been efficacious in in vivo and clinical settings [173,174,175,176,177,178]. Various animal models have suggested that phage in combination with antibiotics can enhance the outcomes as well as reduce resistance development. Yilmaz et al. demonstrated a significant effect of PAS in a rat model of implant-associated S. aureus and P. aeruginosa infections. The combination therapy eradicated S. aureus biofilm [174]. Oeschlin et al. demonstrated that phage and ciprofloxacin were effective at rapidly eradicating bacteria and preventing the development of resistance in a mouse model of endocarditis [175]. Khawaldeh et al. described a single case of a 67 y/o woman with a recurrent, multi-drug resistant P. aeruginosa urinary tract infection. The patient was treated with a cocktail of six antipseudomonal phages, meropenem, and colistin. The infection resolved after 21 days, with no recurrence. It is unclear if there was any true synergy between the antibiotic and phage treatments, although phage therapy was successful at reducing bacterial burdens initially prior to the initiation of colistin [176]. Recently, after showing PAS in vitro [156], Chan et al. successfully employed PAS for the treatment of P. aeruginosa aortic graft infection with OMKO1 and continuous treatment of intravenous ceftazidime [177].
Moreover, temperate phages have drawn researchers’ interest due to their abundance in nature. Both natural and engineered temperate phages have been explored for therapy [179]. Recently, Al-Anany et al. demonstrated PAS by co-administration of temperate phage HK97 with sub-MIC ciprofloxacin resulting in an over 8-log bacterial burden reduction in vitro. However, concern has been raised by a recent PAS modeling study using either temperate or chronic phages, which suggests antibiotic resistance would likely develop [21].
Hence, antibiotics must be selected very carefully for PAS to avoid introducing unnecessary phage resistance. Recently, Dimitriu et al. used P. aeruginosa and DMS3vir as a model to demonstrate that bacteriostatic antibiotics, including chloramphenicol, tetracycline, erythromycin, and trimethoprim, can reduce bacterial growth and delay phage development to prompt bacterial acquisition of phage-derived novel spacers into host CRISPR array [178]. Their data suggest the importance of carefully selecting antibiotics in PAS to prevent the development of bacterial CRISPR immunity against lytic phage. Fortunately, studies have shown that bacteria resistant to phage tend to become less virulent and experience loss of fitness in host microenvironments [180,181]. In addition, Salazar et al. used a bioreactor to arise an evolved phage by directed evolution against the bacterial resistant isolates [180].
5. Conclusions
The data reviewed here suggest that lytic phage therapy may act synergistically with conventional antibiotics in ways that forestall the development or spread of AMR. In the same way that cocktails of antiretrovirals are used to prevent the emergence of resistance in HIV treatment, it is intriguing to imagine that cocktails of phages and conventional antibiotics, with careful selection, could have utility against AMR pathogens. This approach may have particular utility against biofilm infections or other hard-to-treat infections [182,183,184,185].
Most AMR is presumably due to transcriptional adaptation of bacteria against antibiotics. However, there are also indications that phage can contribute to both antibiotic resistance and tolerance. While phages are not the primary mechanism of AMR transfer, lysogenic phages can carry ARGs on prophage. Additionally, both lytic and lysogenic phages can transfer ARGs via general and lateral transductions. Filamentous phages can promote antimicrobial tolerance via antibiotics sequestration and perhaps slow down bacterial growth. Over time, this may promote the development of AMR by selecting for resistant clones/mutants. Currently, with the limited examples of clinical phage therapy, there is yet insufficient evidence to suggest that phage therapy can spread AMR. Nonetheless, care should be taken to avoid the possibility of spreading AMR when designing and testing phage therapy preparations for clinical use.
Many questions remain. It is important to understand the relative contributions of phages versus other MGEs to AMR spread and the evolutionary pressures that mediate these. Similarly, it would be interesting to understand how lytic and lysogenic phages differ with regard to ARG transfer, given their distinct impacts on their bacterial hosts. More work is needed to clarify how such costs influence the development of antibiotic tolerance and resistance.
The intersection between antibiotics and bacteriophages is a frontier in AMR research that is ripe for exploration. The potential dividends of such research are great, with potential benefits for many patients.
Author Contributions
Q.C., T.D., P.C.C., E.B.B., A.J.S. and P.L.B. wrote and revised the manuscript, N.L.H. designed figures. All authors have read and agreed to the published version of the manuscript.
Funding
P.L.B. was supported by grants R01HL148184-01, R01AI12492093, R01DC019965, K24AI166718-01A1, R21GM147838, and grants from Stanford SPARK, The Emerson Collective, and the Cystic Fibrosis Foundation (CFF). Q.C. is supported by the CFF (002572CF221) and the Stanford MCHRI. E.B.B. is supported by the CFF Harry Shwachman Award (BURGEN20Q0).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AMR | Antimicrobial resistance |
| MIC | Minimum inhibitory concentration |
| XDR-TB | Extensively drug-resistant tuberculosis |
| VRSA | Vancomycin resistance genes-resistant Staphylococcus aureus |
| ARG | Antibiotic resistance genes |
| MGE | Mobile genetic elements |
| ICEs | Integrative and conjugative elements |
| CF | Cystic fibrosis |
| PAS | Phage antibiotic synergy |
References
- Dhingra, S.; Rahman, N.A.A.; Peile, E.; Rahman, M.; Sartelli, M.; Hassali, M.A.; Islam, T.; Islam, S.; Haque, M. Microbial Resistance Movements: An Overview of Global Public Health Threats Posed by Antimicrobial Resistance, and How Best to Counter. Front. Public Health 2020, 8, 535668. [Google Scholar] [CrossRef] [PubMed]
- Davies, J. Inactivation of antibiotics and the dissemination of resistance genes. Science 1994, 264, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Lederberg, J.; Tatum, E.L. Gene recombination in Escherichia coli. Nature 1946, 158, 558. [Google Scholar] [CrossRef]
- Zinder, N.D.; Lederberg, J. Genetic exchange in Salmonella. J. Bacteriol. 1952, 64, 679–699. [Google Scholar] [CrossRef]
- Liu, G.; Thomsen, L.E.; Olsen, J.E. Antimicrobial-induced horizontal transfer of antimicrobial resistance genes in bacteria: A mini-review. J. Antimicrob. Chemother. 2022, 77, 556–567. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019.
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global Contributors to Antibiotic Resistance. J. Glob. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [CrossRef]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [CrossRef]
- Suttle, C.A. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Wegrzyn, G. Should Bacteriophages Be Classified as Parasites or Predators? Pol. J. Microbiol. 2022, 71, 3–9. [Google Scholar] [CrossRef]
- Salmond, G.P.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol. 2015, 13, 777–786. [Google Scholar] [CrossRef] [PubMed]
- D’Herelle, F. On an invisible microbe antagonistic toward dysenteric bacilli: Brief note by Mr. F. D’Herelle, presented by Mr. Roux. 1917. Res. Microbiol. 2007, 158, 553–554. [Google Scholar] [CrossRef] [PubMed]
- Summers, W.C. The strange history of phage therapy. Bacteriophage 2012, 2, 130–133. [Google Scholar] [CrossRef]
- Wittebole, X.; De Roock, S.; Opal, S.M. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef]
- Sharma, M. Lytic bacteriophages: Potential interventions against enteric bacterial pathogens on produce. Bacteriophage 2013, 3, e25518. [Google Scholar] [CrossRef]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef]
- Rakonjac, J.; Bennett, N.J.; Spagnuolo, J.; Gagic, D.; Russel, M. Filamentous bacteriophage: Biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 2011, 13, 51–76. [Google Scholar]
- Landa, K.J.; Mossman, L.M.; Whitaker, R.J.; Rapti, Z.; Clifton, S.M. Phage-Antibiotic Synergy Inhibited by Temperate and Chronic Virus Competition. Bull. Math. Biol. 2022, 84, 54. [Google Scholar] [CrossRef]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Gorski, A.; Borysowski, J.; Miedzybrodzki, R. Phage Therapy: Towards a Successful Clinical Trial. Antibiotics 2020, 9, 827. [Google Scholar] [CrossRef] [PubMed]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef]
- Wright, A.; Hawkins, C.H.; Anggard, E.E.; Harper, D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef]
- Simner, P.J.; Cherian, J.; Suh, G.A.; Bergman, Y.; Beisken, S.; Fackler, J.; Lee, M.; Hopkins, R.J.; Tamma, P.D. Combination of phage therapy and cefiderocol to successfully treat Pseudomonas aeruginosa cranial osteomyelitis. JAC Antimicrob. Resist. 2022, 4, dlac046. [Google Scholar] [CrossRef]
- Cano, E.J.; Caflisch, K.M.; Bollyky, P.L.; Van Belleghem, J.D.; Patel, R.; Fackler, J.; Brownstein, M.J.; Horne, B.; Biswas, B.; Henry, M.; et al. Phage Therapy for Limb-threatening Prosthetic Knee Klebsiella pneumoniae Infection: Case Report and In Vitro Characterization of Anti-biofilm Activity. Clin. Infect. Dis. 2021, 73, e144–e151. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Smith, B.E.; Cristinziano, M.; Freeman, K.G.; Jacobs-Sera, D.; Belessis, Y.; Whitney Brown, A.; Cohen, K.A.; Davidson, R.M.; van Duin, D.; et al. Phage Therapy of Mycobacterium Infections: Compassionate-use of Phages in Twenty Patients with Drug-Resistant Mycobacterial Disease. Clin. Infect. Dis. 2022. [Google Scholar] [CrossRef]
- Aslam, S.; Lampley, E.; Wooten, D.; Karris, M.; Benson, C.; Strathdee, S.; Schooley, R.T. Lessons Learned from the First 10 Consecutive Cases of Intravenous Bacteriophage Therapy to Treat Multidrug-Resistant Bacterial Infections at a Single Center in the United States. Open Forum Infect. Dis. 2020, 7, ofaa389. [Google Scholar] [CrossRef]
- Lebeaux, D.; Merabishvili, M.; Caudron, E.; Lannoy, D.; Van Simaey, L.; Duyvejonck, H.; Guillemain, R.; Thumerelle, C.; Podglajen, I.; Compain, F.; et al. A Case of Phage Therapy against Pandrug-Resistant Achromobacter xylosoxidans in a 12-Year-Old Lung-Transplanted Cystic Fibrosis Patient. Viruses 2021, 13, 60. [Google Scholar] [CrossRef]
- Liu, D.; Van Belleghem, J.D.; de Vries, C.R.; Burgener, E.; Chen, Q.; Manasherob, R.; Aronson, J.R.; Amanatullah, D.F.; Tamma, P.D.; Suh, G.A. The Safety and Toxicity of Phage Therapy: A Review of Animal and Clinical Studies. Viruses 2021, 13, 1268. [Google Scholar] [CrossRef]
- Suh, G.A.; Lodise, T.P.; Tamma, P.D.; Knisely, J.M.; Alexander, J.; Aslam, S.; Barton, K.D.; Bizzell, E.; Totten, K.M.C.; Campbell, J.L.; et al. Considerations for the Use of Phage Therapy in Clinical Practice. Antimicrob. Agents Chemother. 2022, 66, e0207121. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.N.; Penades, J.R.; Chen, J. Genetic transduction by phages and chromosomal islands: The new and noncanonical. PLoS Pathog. 2019, 15, e1007878. [Google Scholar] [CrossRef] [PubMed]
- Stern, A.; Sorek, R. The phage-host arms race: Shaping the evolution of microbes. Bioessays 2011, 33, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, A.R.; Turner, P.E. Trading-off and trading-up in the world of bacteria-phage evolution. Curr. Biol. 2020, 30, R1120–R1124. [Google Scholar] [CrossRef]
- Koskella, B.; Brockhurst, M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef]
- Micoli, F.; Bagnoli, F.; Rappuoli, R.; Serruto, D. The role of vaccines in combatting antimicrobial resistance. Nat. Microbiol. 2021, 19, 287–302. [Google Scholar] [CrossRef]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Villa, T.G.; Feijoo-Siota, L.; Rama, J.R.; Sánchez-Pérez, A.; Viñas, M. Horizontal Gene Transfer between Bacteriophages and Bacteria: Antibiotic Resistances and Toxin Production. In Horizontal Gene Transfer Breaking Borders between Living Kingdoms; Villa, T.G., Viñas, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 97–142. [Google Scholar]
- van Hoek, A.H.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J. Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Wiedenbeck, J.; Cohan, F.M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 2011, 35, 957–976. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.; Wolffs, P.F. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Xia, Z.J.; Wang, J.; Hu, W.; Liu, H.; Gao, X.Z.; Wu, Z.H.; Zhang, P.Y.; Li, Y.Z. Improving conjugation efficacy of Sorangium cellulosum by the addition of dual selection antibiotics. J. Ind. Microbiol. Biotechnol. 2008, 35, 1157–1163. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 464–473. [Google Scholar] [CrossRef]
- Diaz Ricci, J.C.; Hernandez, M.E. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 2000, 20, 79–108. [Google Scholar] [CrossRef]
- Rasched, I.; Oberer, E. Ff coliphages: Structural and functional relationships. Microbiol. Rev. 1986, 50, 401–427. [Google Scholar] [CrossRef]
- Lerat, E.; Ochman, H. Psi-Phi: Exploring the outer limits of bacterial pseudogenes. Genome Res. 2004, 14, 2273–2278. [Google Scholar] [CrossRef]
- Acman, M.; Wang, R.; van Dorp, L.; Shaw, L.P.; Wang, Q.; Luhmann, N.; Yin, Y.; Sun, S.; Chen, H.; Wang, H.; et al. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene blaNDM. Nat. Commun. 2022, 13, 1131. [Google Scholar] [CrossRef] [PubMed]
- Rankin, D.J.; Rocha, E.P.; Brown, S.P. What traits are carried on mobile genetic elements, and why? Heredity 2011, 106, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Secor, P.R.; Burgener, E.B.; Kinnersley, M.; Jennings, L.K.; Roman-Cruz, V.; Popescu, M.; Van Belleghem, J.D.; Haddock, N.; Copeland, C.; Michaels, L.A.; et al. Pf Bacteriophage and Their Impact on Pseudomonas Virulence, Mammalian Immunity, and Chronic Infections. Front. Immunol. 2020, 11, 244. [Google Scholar] [CrossRef] [PubMed]
- Kobori, H.; Sullivan, C.W.; Shizuya, H. Bacterial plasmids in antarctic natural microbial assemblages. Appl. Environ. Microbiol. 1984, 48, 515–518. [Google Scholar] [CrossRef]
- Calero-Caceres, W.; Melgarejo, A.; Colomer-Lluch, M.; Stoll, C.; Lucena, F.; Jofre, J.; Muniesa, M. Sludge as a potential important source of antibiotic resistance genes in both the bacterial and bacteriophage fractions. Environ. Sci. Technol. 2014, 48, 7602–7611. [Google Scholar] [CrossRef]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS ONE 2011, 6, e17549. [Google Scholar] [CrossRef]
- Lekunberri, I.; Subirats, J.; Borrego, C.M.; Balcazar, J.L. Exploring the contribution of bacteriophages to antibiotic resistance. Environ. Pollut. 2017, 220, 981–984. [Google Scholar] [CrossRef]
- Balcazar, J.L. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 2014, 10, e1004219. [Google Scholar] [CrossRef]
- Kondo, K.; Kawano, M.; Sugai, M. Distribution of Antimicrobial Resistance and Virulence Genes within the Prophage-Associated Regions in Nosocomial Pathogens. Msphere 2021, 6, e0045221. [Google Scholar] [CrossRef]
- Minot, S.; Sinha, R.; Chen, J.; Li, H.; Keilbaugh, S.A.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res. 2011, 21, 1616–1625. [Google Scholar] [CrossRef]
- Quiros, P.; Colomer-Lluch, M.; Martinez-Castillo, A.; Miro, E.; Argente, M.; Jofre, J.; Navarro, F.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples. Antimicrob. Agents Chemother. 2014, 58, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Fancello, L.; Desnues, C.; Raoult, D.; Rolain, J.M. Bacteriophages and diffusion of genes encoding antimicrobial resistance in cystic fibrosis sputum microbiota. J. Antimicrob. Chemother. 2011, 66, 2448–2454. [Google Scholar] [CrossRef]
- Rolain, J.M.; Fancello, L.; Desnues, C.; Raoult, D. Bacteriophages as vehicles of the resistome in cystic fibrosis. J. Antimicrob. Chemother. 2011, 66, 2444–2447. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.R.; Lee, H.H.; Spina, C.S.; Collins, J.J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 2013, 499, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Colombo, S.; Arioli, S.; Neri, E.; Della Scala, G.; Gargari, G.; Mora, D. Viromes as Genetic Reservoir for the Microbial Communities in Aquatic Environments: A Focus on Antimicrobial-Resistance Genes. Front. Microbiol. 2017, 8, 1095. [Google Scholar] [CrossRef]
- Kim, Y.; Van Bonn, W.; Aw, T.G.; Rose, J.B. Aquarium Viromes: Viromes of Human-Managed Aquatic Systems. Front. Microbiol. 2017, 8, 1231. [Google Scholar] [CrossRef]
- Bossi, L.; Fuentes, J.A.; Mora, G.; Figueroa-Bossi, N. Prophage contribution to bacterial population dynamics. J. Bacteriol. 2003, 185, 6467–6471. [Google Scholar] [CrossRef]
- Haaber, J.; Leisner, J.J.; Cohn, M.T.; Catalan-Moreno, A.; Nielsen, J.B.; Westh, H.; Penades, J.R.; Ingmer, H. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat. Commun. 2016, 7, 13333. [Google Scholar] [CrossRef]
- Wang, X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 147. [Google Scholar] [CrossRef]
- Secor, P.R.; Jennings, L.K.; Michaels, L.A.; Sweere, J.M.; Singh, P.K.; Parks, W.C.; Bollyky, P.L. Biofilm assembly becomes crystal clear—Filamentous bacteriophage organize the Pseudomonas aeruginosa biofilm matrix into a liquid crystal. Microb. Cell 2015, 3, 49–52. [Google Scholar] [CrossRef]
- Secor, P.R.; Michaels, L.A.; Smigiel, K.S.; Rohani, M.G.; Jennings, L.K.; Hisert, K.B.; Arrigoni, A.; Braun, K.R.; Birkland, T.P.; Lai, Y.; et al. Filamentous Bacteriophage Produced by Pseudomonas aeruginosa Alters the Inflammatory Response and Promotes Noninvasive Infection In Vivo. Infect. Immun. 2017, 85, e00648-16. [Google Scholar] [CrossRef] [PubMed]
- Vica Pacheco, S.; Garcia Gonzalez, O.; Paniagua Contreras, G.L. The lom gene of bacteriophage lambda is involved in Escherichia coli K12 adhesion to human buccal epithelial cells. FEMS Microbiol. Lett. 1997, 156, 129–132. [Google Scholar] [CrossRef]
- Vaca-Pacheco, S.; Paniagua-Contreras, G.L.; Garcia-Gonzalez, O.; de la Garza, M. The clinically isolated FIZ15 bacteriophage causes lysogenic conversion in Pseudomonas aeruginosa PAO1. Curr. Microbiol. 1999, 38, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.G.; Ing, J.Y.; Cheng, M.K.; Flitter, B.A.; Moe, G.R. Identification of a phage-encoded Ig-binding protein from invasive Neisseria meningitidis. J. Immunol. 2013, 191, 3287–3296. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, A.D.; Newland, J.W.; Miller, S.F.; Holmes, R.K.; Smith, H.W.; Formal, S.B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 1984, 226, 694–696. [Google Scholar] [CrossRef]
- Tyler, J.S.; Mills, M.J.; Friedman, D.I. The operator and early promoter region of the Shiga toxin type 2-encoding bacteriophage 933W and control of toxin expression. J. Bacteriol. 2004, 186, 7670–7679. [Google Scholar] [CrossRef]
- Karaolis, D.K.; Somara, S.; Maneval, D.R., Jr.; Johnson, J.A.; Kaper, J.B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 1999, 399, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Mirold, S.; Rabsch, W.; Rohde, M.; Stender, S.; Tschape, H.; Russmann, H.; Igwe, E.; Hardt, W.D. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 1999, 96, 9845–9850. [Google Scholar] [CrossRef]
- Figueroa-Bossi, N.; Bossi, L. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 1999, 33, 167–176. [Google Scholar] [CrossRef]
- Nakayama, K.; Kanaya, S.; Ohnishi, M.; Terawaki, Y.; Hayashi, T. The complete nucleotide sequence of phi CTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: Implications for phage evolution and horizontal gene transfer via bacteriophages. Mol. Microbiol. 1999, 31, 399–419. [Google Scholar] [CrossRef]
- Vostrov, A.A.; Vostrukhina, O.A.; Svarchevsky, A.N.; Rybchin, V.N. Proteins responsible for lysogenic conversion caused by coliphages N15 and phi80 are highly homologous. J. Bacteriol. 1996, 178, 1484–1486. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.K.; Fitzpatrick, A.D.; Schwartzkopf, C.M.; Faith, D.R.; Jennings, L.K.; Coluccio, A.; Hunt, D.J.; Michaels, L.A.; Hargil, A.; Chen, Q.; et al. A Filamentous Bacteriophage Protein Inhibits Type IV Pili to Prevent Superinfection of Pseudomonas aeruginosa. mBio 2022, 13, e02441-21. [Google Scholar] [CrossRef]
- Newton, G.J.; Daniels, C.; Burrows, L.L.; Kropinski, A.M.; Clarke, A.J.; Lam, J.S. Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol. Microbiol. 2001, 39, 1237–1247. [Google Scholar] [CrossRef]
- Cumby, N.; Edwards, A.M.; Davidson, A.R.; Maxwell, K.L. The bacteriophage HK97 gp15 moron element encodes a novel superinfection exclusion protein. J. Bacteriol. 2012, 194, 5012–5019. [Google Scholar] [CrossRef] [PubMed]
- Enault, F.; Briet, A.; Bouteille, L.; Roux, S.; Sullivan, M.B.; Petit, M.A. Phages rarely encode antibiotic resistance genes: A cautionary tale for virome analyses. ISME J. 2017, 11, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Torres-Barcelo, C. The disparate effects of bacteriophages on antibiotic-resistant bacteria. Emerg. Microbes Infect. 2018, 7, 168. [Google Scholar] [CrossRef]
- Willi, K.; Sandmeier, H.; Kulik, E.M.; Meyer, J. Transduction of antibiotic resistance markers among Actinobacillus actinomycetemcomitans strains by temperate bacteriophages Aa phi 23. Cell. Mol. Life Sci. 1997, 53, 904–910. [Google Scholar] [CrossRef]
- Del Grosso, M.; Camilli, R.; Barbabella, G.; Blackman Northwood, J.; Farrell, D.J.; Pantosti, A. Genetic resistance elements carrying mef subclasses other than mef(A) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 2011, 55, 3226–3230. [Google Scholar] [CrossRef]
- Conway, J.F.; Wikoff, W.R.; Cheng, N.; Duda, R.L.; Hendrix, R.W.; Johnson, J.E.; Steven, A.C. Virus maturation involving large subunit rotations and local refolding. Science 2001, 292, 744–748. [Google Scholar] [CrossRef]
- Kanamaru, S.; Leiman, P.G.; Kostyuchenko, V.A.; Chipman, P.R.; Mesyanzhinov, V.V.; Arisaka, F.; Rossmann, M.G. Structure of the cell-puncturing device of bacteriophage T4. Nature 2002, 415, 553–557. [Google Scholar] [CrossRef]
- Molineux, I.J. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 2001, 40, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Greene, B.; Thuman-Commike, P.A.; Jakana, J.; Prevelige, P.E., Jr.; King, J.; Chiu, W. Visualization of the maturation transition in bacteriophage P22 by electron cryomicroscopy. J. Mol. Biol. 2000, 297, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Popescu, M.; Van Belleghem, J.D.; Khosravi, A.; Bollyky, P.L. Bacteriophages and the Immune System. Annu. Rev. Virol. 2021, 8, 415–435. [Google Scholar] [CrossRef] [PubMed]
- Thierauf, A.; Perez, G.; Maloy, A.S. Generalized transduction. Methods Mol. Biol. 2009, 501, 267–286. [Google Scholar] [CrossRef]
- Zhang, J.; He, X.; Shen, S.; Shi, M.; Zhou, Q.; Liu, J.; Wang, M.; Sun, Y. Effects of the Newly Isolated T4-like Phage on Transmission of Plasmid-Borne Antibiotic Resistance Genes via Generalized Transduction. Viruses 2021, 13, 2070. [Google Scholar] [CrossRef]
- Mazaheri Nezhad Fard, R.; Barton, M.D.; Heuzenroeder, M.W. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett. Appl. Microbiol. 2011, 52, 559–564. [Google Scholar] [CrossRef]
- Zeman, M.; Maslanova, I.; Indrakova, A.; Siborova, M.; Mikulasek, K.; Bendickova, K.; Plevka, P.; Vrbovska, V.; Zdrahal, Z.; Doskar, J.; et al. Staphylococcus sciuri bacteriophages double-convert for staphylokinase and phospholipase, mediate interspecies plasmid transduction, and package mecA gene. Sci. Rep. 2017, 7, 46319. [Google Scholar] [CrossRef]
- Gabashvili, E.; Osepashvili, M.; Koulouris, S.; Ujmajuridze, L.; Tskhitishvili, Z.; Kotetishvili, M. Phage Transduction is Involved in the Intergeneric Spread of Antibiotic Resistance-Associated blaCTX-M, mel, and tetM Loci in Natural Populations of Some Human and Animal Bacterial Pathogens. Curr. Microbiol. 2020, 77, 185–193. [Google Scholar] [CrossRef]
- Gabashvili, E.; Kobakhidze, S.; Koulouris, S.; Robinson, T.; Kotetishvili, M. Bi- and Multi-directional Gene Transfer in the Natural Populations of Polyvalent Bacteriophages, and Their Host Species Spectrum Representing Foodborne Versus Other Human and/or Animal Pathogens. Food Environ. Virol. 2021, 13, 179–202. [Google Scholar] [CrossRef]
- Chen, J.; Quiles-Puchalt, N.; Chiang, Y.N.; Bacigalupe, R.; Fillol-Salom, A.; Chee, M.S.J.; Fitzgerald, J.R.; Penades, J.R. Genome hypermobility by lateral transduction. Science 2018, 362, 207–212. [Google Scholar] [CrossRef]
- Humphrey, S.; Fillol-Salom, A.; Quiles-Puchalt, N.; Ibarra-Chavez, R.; Haag, A.F.; Chen, J.; Penades, J.R. Bacterial chromosomal mobility via lateral transduction exceeds that of classical mobile genetic elements. Nat. Commun. 2021, 12, 6509. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.P.; Prior, S.E.; Barstow, D.A.; Minton, N.P. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 1988, 68, 139–149. [Google Scholar] [CrossRef]
- Smillie, C.; Garcillan-Barcia, M.P.; Francia, M.V.; Rocha, E.P.; de la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Jimenez, J.; Derr, J.; Vera, P.; Manapat, M.L.; Esvelt, K.M.; Villanueva, L.; Liu, D.R.; Chen, I.A. Inhibition of bacterial conjugation by phage M13 and its protein g3p: Quantitative analysis and model. PLoS ONE 2011, 6, e19991. [Google Scholar] [CrossRef] [PubMed]
- LeGault, K.N.; Hays, S.G.; Angermeyer, A.; McKitterick, A.C.; Johura, F.T.; Sultana, M.; Ahmed, T.; Alam, M.; Seed, K.D. Temporal shifts in antibiotic resistance elements govern phage-pathogen conflicts. Science 2021, 373, 6554. [Google Scholar] [CrossRef] [PubMed]
- Horne, D.; Tomasz, A. Tolerant response of Streptococcus sanguis to beta-lactams and other cell wall inhibitors. Antimicrob. Agents Chemother. 1977, 11, 888–896. [Google Scholar] [CrossRef][Green Version]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef]
- Levin-Reisman, I.; Brauner, A.; Ronin, I.; Balaban, N.Q. Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc. Natl. Acad. Sci. USA 2019, 116, 14734–14739. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, K.; Zhang, H.; Jia, Y.; Wang, Z. Combating Antibiotic Tolerance Through Activating Bacterial Metabolism. Front. Microbiol. 2020, 11, 577564. [Google Scholar] [CrossRef]
- Molina-Quiroz, R.C.; Silva-Valenzuela, C.; Brewster, J.; Castro-Nallar, E.; Levy, S.B.; Camilli, A. Cyclic AMP Regulates Bacterial Persistence through Repression of the Oxidative Stress Response and SOS-Dependent DNA Repair in Uropathogenic Escherichia coli. mBio 2018, 9, e02144-17. [Google Scholar] [CrossRef]
- Crabbe, A.; Jensen, P.O.; Bjarnsholt, T.; Coenye, T. Antimicrobial Tolerance and Metabolic Adaptations in Microbial Biofilms. Trends Microbiol. 2019, 27, 850–863. [Google Scholar] [CrossRef] [PubMed]
- Windels, E.M.; Michiels, J.E.; Fauvart, M.; Wenseleers, T.; Van den Bergh, B.; Michiels, J. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 2019, 13, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Levin-Reisman, I.; Ronin, I.; Gefen, O.; Braniss, I.; Shoresh, N.; Balaban, N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355, 826–830. [Google Scholar] [CrossRef]
- Liu, J.; Gefen, O.; Ronin, I.; Bar-Meir, M.; Balaban, N.Q. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 2020, 367, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Vestby, L.K.; Gronseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013, 121, 1–51. [Google Scholar] [CrossRef]
- Chiang, W.C.; Nilsson, M.; Jensen, P.O.; Hoiby, N.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsen, T. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2013, 57, 2352–2361. [Google Scholar] [CrossRef]
- Ishida, H.; Ishida, Y.; Kurosaka, Y.; Otani, T.; Sato, K.; Kobayashi, H. In vitro and in vivo activities of levofloxacin against biofilm-producing Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1998, 42, 1641–1645. [Google Scholar] [CrossRef]
- Starkey, M.; Hickman, J.H.; Ma, L.; Zhang, N.; De Long, S.; Hinz, A.; Palacios, S.; Manoil, C.; Kirisits, M.J.; Starner, T.D.; et al. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 2009, 191, 3492–3503. [Google Scholar] [CrossRef]
- Hoiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Meers, P.; Neville, M.; Malinin, V.; Scotto, A.W.; Sardaryan, G.; Kurumunda, R.; Mackinson, C.; James, G.; Fisher, S.; Perkins, W.R. Biofilm penetration, triggered release and in vivo activity of inhaled liposomal amikacin in chronic Pseudomonas aeruginosa lung infections. J. Antimicrob. Chemother. 2008, 61, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.M.; Neill, D.R.; Kaman, B.; Sahota, J.S.; Clokie, M.R.J.; Winstanley, C.; Kadioglu, A. Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax 2017, 72, 666–667. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, E.B.; Tam, V.H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharm. Outcomes Res. 2010, 10, 441–451. [Google Scholar] [CrossRef]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, R.S.; Christie, C.D.; Smith, G.J. Immunohistopathologic localization of Pseudomonas aeruginosa in lungs from patients with cystic fibrosis. Implications for the pathogenesis of progressive lung deterioration. Am. Rev. Respir. Dis. 1989, 140, 1650–1661. [Google Scholar] [CrossRef]
- Samad, T.; Co, J.Y.; Witten, J.; Ribbeck, K. Mucus and Mucin Environments Reduce the Efficacy of Polymyxin and Fluoroquinolone Antibiotics against Pseudomonas aeruginosa. ACS Biomater. Sci. Eng. 2019, 5, 1189–1194. [Google Scholar] [CrossRef]
- Huang, J.X.; Blaskovich, M.A.; Pelingon, R.; Ramu, S.; Kavanagh, A.; Elliott, A.G.; Butler, M.S.; Montgomery, A.B.; Cooper, M.A. Mucin Binding Reduces Colistin Antimicrobial Activity. Antimicrob. Agents Chemother. 2015, 59, 5925–5931. [Google Scholar] [CrossRef]
- Rice, S.A.; Tan, C.H.; Mikkelsen, P.J.; Kung, V.; Woo, J.; Tay, M.; Hauser, A.; McDougald, D.; Webb, J.S.; Kjelleberg, S. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009, 3, 271–282. [Google Scholar] [CrossRef]
- Hay, I.D.; Lithgow, T. Filamentous phages: Masters of a microbial sharing economy. EMBO Rep. 2019, 20, e47427. [Google Scholar] [CrossRef]
- Roux, S.; Krupovic, M.; Daly, R.A.; Borges, A.L.; Nayfach, S.; Schulz, F.; Sharrar, A.; Matheus Carnevali, P.B.; Cheng, J.F.; Ivanova, N.N.; et al. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat. Microbiol. 2019, 4, 1895–1906. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Kan, B.; Wang, R. Isolation and characterization of the new mosaic filamentous phage VFJ Phi of Vibrio cholerae. PLoS ONE 2013, 8, e70934. [Google Scholar] [CrossRef]
- Chevallereau, A.; Pons, B.J.; van Houte, S.; Westra, E.R. Interactions between bacterial and phage communities in natural environments. Nat. Rev. Microbiol. 2022, 20, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, P.; Voet, M.; Lavigne, R. Prevalence of Pf1-like (pro)phage genetic elements among Pseudomonas aeruginosa isolates. Virology 2015, 483, 64–71. [Google Scholar] [CrossRef]
- Whiteley, M.; Bangera, M.G.; Bumgarner, R.E.; Parsek, M.R.; Teitzel, G.M.; Lory, S.; Greenberg, E.P. Gene expression in Pseudomonas aeruginosa biofilms. Nature 2001, 413, 860–864. [Google Scholar] [CrossRef]
- Platt, M.D.; Schurr, M.J.; Sauer, K.; Vazquez, G.; Kukavica-Ibrulj, I.; Potvin, E.; Levesque, R.C.; Fedynak, A.; Brinkman, F.S.; Schurr, J.; et al. Proteomic, microarray, and signature-tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late-stage cystic fibrosis airway conditions. J. Bacteriol. 2008, 190, 2739–2758. [Google Scholar] [CrossRef]
- Lieleg, O.; Vladescu, I.; Ribbeck, K. Characterization of particle translocation through mucin hydrogels. Biophys. J. 2010, 98, 1782–1789. [Google Scholar] [CrossRef]
- Park, S.H.; Marassi, F.M.; Black, D.; Opella, S.J. Structure and dynamics of the membrane-bound form of Pf1 coat protein: Implications of structural rearrangement for virus assembly. Biophys. J. 2010, 99, 1465–1474. [Google Scholar] [CrossRef]
- Janmey, P.A.; Slochower, D.R.; Wang, Y.H.; Wen, Q.; Cebers, A. Polyelectrolyte properties of filamentous biopolymers and their consequences in biological fluids. Soft. Matter. 2014, 10, 1439–1449. [Google Scholar] [CrossRef]
- Secor, P.R.; Sweere, J.M.; Michaels, L.A.; Malkovskiy, A.V.; Lazzareschi, D.; Katznelson, E.; Rajadas, J.; Birnbaum, M.E.; Arrigoni, A.; Braun, K.R.; et al. Filamentous Bacteriophage Promote Biofilm Assembly and Function. Cell Host Microbe 2015, 18, 549–559. [Google Scholar] [CrossRef]
- Secor, P.R.; Michaels, L.A.; Ratjen, A.; Jennings, L.K.; Singh, P.K. Entropically driven aggregation of bacteria by host polymers promotes antibiotic tolerance in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2018, 115, 10780–10785. [Google Scholar] [CrossRef]
- Bille, E.; Meyer, J.; Jamet, A.; Euphrasie, D.; Barnier, J.P.; Brissac, T.; Larsen, A.; Pelissier, P.; Nassif, X. A virulence-associated filamentous bacteriophage of Neisseria meningitidis increases host-cell colonisation. PLoS Pathog. 2017, 13, e1006495. [Google Scholar] [CrossRef] [PubMed]
- Tarafder, A.K.; von Kugelgen, A.; Mellul, A.J.; Schulze, U.; Aarts, D.; Bharat, T.A.M. Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 4724–4731. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.S.; Lau, M.; Kjelleberg, S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2004, 186, 8066–8073. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Multidrug tolerance of biofilms and persister cells. Curr. Top Microbiol. Immunol. 2008, 322, 107–131. [Google Scholar] [CrossRef]
- Burgener, E.B.; Sweere, J.M.; Bach, M.S.; Secor, P.R.; Haddock, N.; Jennings, L.K.; Marvig, R.L.; Johansen, H.K.; Rossi, E.; Cao, X.; et al. Filamentous bacteriophages are associated with chronic Pseudomonas lung infections and antibiotic resistance in cystic fibrosis. Sci. Transl. Med. 2019, 11, eaau9748. [Google Scholar] [CrossRef]
- Sweere, J.M.; Van Belleghem, J.D.; Ishak, H.; Bach, M.S.; Popescu, M.; Sunkari, V.; Kaber, G.; Manasherob, R.; Suh, G.A.; Cao, X.; et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 2019, 363, eaat9691. [Google Scholar] [CrossRef]
- Pourtois, J.D.; Kratochvil, M.J.; Chen, Q.; Haddock, N.L.; Burgener, E.B.; De Leo, G.A.; Bollyky, P.L. Filamentous Bacteriophages and the Competitive Interaction between Pseudomonas aeruginosa Strains under Antibiotic Treatment: A Modeling Study. mSystems 2021, 6, e00193-21. [Google Scholar] [CrossRef]
- Addy, H.S.; Askora, A.; Kawasaki, T.; Fujie, M.; Yamada, T. Loss of virulence of the phytopathogen Ralstonia solanacearum through infection by phiRSM filamentous phages. Phytopathology 2012, 102, 469–477. [Google Scholar] [CrossRef][Green Version]
- Ahmad, A.A.; Stulberg, M.J.; Huang, Q. Prophage Rs551 and Its Repressor Gene orf14 Reduce Virulence and Increase Competitive Fitness of Its Ralstonia solanacearum Carrier Strain UW551. Front. Microbiol. 2017, 8, 2480. [Google Scholar] [CrossRef]
- Narulita, E.; Addy, H.S.; Kawasaki, T.; Fujie, M.; Yamada, T. The involvement of the PilQ secretin of type IV pili in phage infection in Ralstonia solanacearum. Biochem. Biophys. Res. Commun. 2016, 469, 868–872. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Lenski, R.E. Experimental Studies of Pleiotropy and Epistasis in Escherichia coli. I. Variation in Competitive Fitness among Mutants Resistant to Virus T4. Evolution 1988, 42, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Hossain, M.J.; Rahman, K.S.; Terhune, J.S.; Liles, M.R. An outer membrane porin protein modulates phage susceptibility in Edwardsiella ictaluri. Microbiology 2012, 158, 474–487. [Google Scholar] [CrossRef][Green Version]
- Sjahriani, T.; Wasito, E.B.; Tyasningsih, W. The Analysis of OmpA and Rz/Rz1 of Lytic Bacteriophage from Surabaya, Indonesia. Scientifica 2021, 2021, 7494144. [Google Scholar] [CrossRef]
- Westra, E.R.; van Houte, S.; Oyesiku-Blakemore, S.; Makin, B.; Broniewski, J.M.; Best, A.; Bondy-Denomy, J.; Davidson, A.; Boots, M.; Buckling, A. Parasite Exposure Drives Selective Evolution of Constitutive versus Inducible Defense. Curr. Biol. 2015, 25, 1043–1049. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef]
- Webster, S.S.; Lee, C.K.; Schmidt, W.C.; Wong, G.C.L.; O’Toole, G.A. Interaction between the type 4 pili machinery and a diguanylate cyclase fine-tune c-di-GMP levels during early biofilm formation. Proc. Natl. Acad. Sci. USA 2021, 118, e2105566118. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- Castledine, M.; Padfield, D.; Sierocinski, P.; Soria Pascual, J.; Hughes, A.; Makinen, L.; Friman, V.P.; Pirnay, J.P.; Merabishvili, M.; de Vos, D.; et al. Parallel evolution of Pseudomonas aeruginosa phage resistance and virulence loss in response to phage treatment in vivo and in vitro. eLife 2022, 11, e73679. [Google Scholar] [CrossRef]
- Comeau, A.M.; Tetart, F.; Trojet, S.N.; Prere, M.F.; Krisch, H.M. Phage-Antibiotic Synergy (PAS): Beta-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS ONE 2007, 2, e799. [Google Scholar] [CrossRef] [PubMed]
- Tagliaferri, T.L.; Jansen, M.; Horz, H.P. Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy. Front. Cell. Infect. Microbiol. 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, W.N.; Concepcion-Acevedo, J.; Park, T.; Andleeb, S.; Bull, J.J.; Levin, B.R. Synergy and Order Effects of Antibiotics and Phages in Killing Pseudomonas aeruginosa Biofilms. PLoS ONE 2017, 12, e0168615. [Google Scholar] [CrossRef] [PubMed]
- Torres-Barcelo, C.; Arias-Sanchez, F.I.; Vasse, M.; Ramsayer, J.; Kaltz, O.; Hochberg, M.E. A window of opportunity to control the bacterial pathogen Pseudomonas aeruginosa combining antibiotics and phages. PLoS ONE 2014, 9, e106628. [Google Scholar] [CrossRef]
- Uchiyama, J.; Shigehisa, R.; Nasukawa, T.; Mizukami, K.; Takemura-Uchiyama, I.; Ujihara, T.; Murakami, H.; Imanishi, I.; Nishifuji, K.; Sakaguchi, M.; et al. Piperacillin and ceftazidime produce the strongest synergistic phage-antibiotic effect in Pseudomonas aeruginosa. Arch. Virol. 2018, 163, 1941–1948. [Google Scholar] [CrossRef]
- Ryan, E.M.; Alkawareek, M.Y.; Donnelly, R.F.; Gilmore, B.F. Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol. Med. Microbiol. 2012, 65, 395–398. [Google Scholar] [CrossRef]
- Coulter, L.B.; McLean, R.J.; Rohde, R.E.; Aron, G.M. Effect of bacteriophage infection in combination with tobramycin on the emergence of resistance in Escherichia coli and Pseudomonas aeruginosa biofilms. Viruses 2014, 6, 3778–3786. [Google Scholar] [CrossRef]
- Rahman, M.; Kim, S.; Kim, S.M.; Seol, S.Y.; Kim, J. Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling 2011, 27, 1087–1093. [Google Scholar] [CrossRef]
- Gu Liu, C.; Green, S.I.; Min, L.; Clark, J.R.; Salazar, K.C.; Terwilliger, A.L.; Kaplan, H.B.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio 2020, 11, e01462-20. [Google Scholar] [CrossRef]
- Hagens, S.; Habel, A.; Blasi, U. Augmentation of the antimicrobial efficacy of antibiotics by filamentous phage. Microb. Drug Resist. 2006, 12, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, C.; Colak, M.; Yilmaz, B.C.; Ersoz, G.; Kutateladze, M.; Gozlugol, M. Bacteriophage therapy in implant-related infections: An experimental study. J. Bone. Jt. Surg. Am. 2013, 95, 117–125. [Google Scholar] [CrossRef]
- Oechslin, F.; Piccardi, P.; Mancini, S.; Gabard, J.; Moreillon, P.; Entenza, J.M.; Resch, G.; Que, Y.A. Synergistic Interaction Between Phage Therapy and Antibiotics Clears Pseudomonas aeruginosa Infection in Endocarditis and Reduces Virulence. J. Infect. Dis. 2017, 215, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Khawaldeh, A.; Morales, S.; Dillon, B.; Alavidze, Z.; Ginn, A.N.; Thomas, L.; Chapman, S.J.; Dublanchet, A.; Smithyman, A.; Iredell, J.R. Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J. Med. Microbiol. 2011, 60, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef]
- Dimitriu, T.; Kurilovich, E.; Lapinska, U.; Severinov, K.; Pagliara, S.; Szczelkun, M.D.; Westra, E.R. Bacteriostatic antibiotics promote CRISPR-Cas adaptive immunity by enabling increased spacer acquisition. Cell Host Microbe 2022, 30, 31–40.e35. [Google Scholar] [CrossRef]
- Monteiro, R.; Pires, D.P.; Costa, A.R.; Azeredo, J. Phage Therapy: Going Temperate? Trends Microbiol. 2019, 27, 368–378. [Google Scholar] [CrossRef]
- Salazar, K.C.; Ma, L.; Green, S.I.; Zulk, J.J.; Trautner, B.W.; Ramig, R.F.; Clark, J.R.; Terwilliger, A.L.; Maresso, A.W. Antiviral Resistance and Phage Counter Adaptation to Antibiotic-Resistant Extraintestinal Pathogenic Escherichia coli. mBio 2021, 12, e00211-21. [Google Scholar] [CrossRef]
- Canfield, G.S.; Chatterjee, A.; Espinosa, J.; Mangalea, M.R.; Sheriff, E.K.; Keidan, M.; McBride, S.W.; McCollister, B.D.; Hang, H.C.; Duerkop, B.A. Lytic bacteriophages facilitate antibiotic sensitization of Enterococcus faecium. Antimicrob. Agents Chemother. 2021, 65, e00143-21. [Google Scholar] [CrossRef]
- Akturk, E.; Oliveira, H.; Santos, S.B.; Costa, S.; Kuyumcu, S.; Melo, L.D.R.; Azeredo, J. Synergistic Action of Phage and Antibiotics: Parameters to Enhance the Killing Efficacy Against Mono and Dual-Species Biofilms. Antibiotics 2019, 8, 103. [Google Scholar] [CrossRef]
- Chang, R.Y.K.; Das, T.; Manos, J.; Kutter, E.; Morales, S.; Chan, H.K. Bacteriophage PEV20 and Ciprofloxacin Combination Treatment Enhances Removal of Pseudomonas aeruginosa Biofilm Isolated from Cystic Fibrosis and Wound Patients. AAPS J. 2019, 21, 49. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tkhilaishvili, T.; Bernal Andres, B.; Trampuz, A.; Gonzalez Moreno, M. Bacteriophage-antibiotic combinations against ciprofloxacin/ceftriaxone-resistant Escherichia coli in vitro and in an experimental Galleria mellonella model. Int. J. Antimicrob. Agents 2020, 56, 106200. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Xie, L.; Liu, M.; Li, Q.; Wang, P.; Luo, C. Bactericidal Synergism between Phage YC#06 and Antibiotics: A Combination Strategy to Target Multidrug-Resistant Acinetobacter baumannii In Vitro and In Vivo. Microbiol. Spectr. 2022, e0009622. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).