A kNGR Peptide-Tethered Lipid–Polymer Hybrid Nanocarrier-Based Synergistic Approach for Effective Tumor Therapy: Development, Characterization, Ex-Vivo, and In-Vivo Assessment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
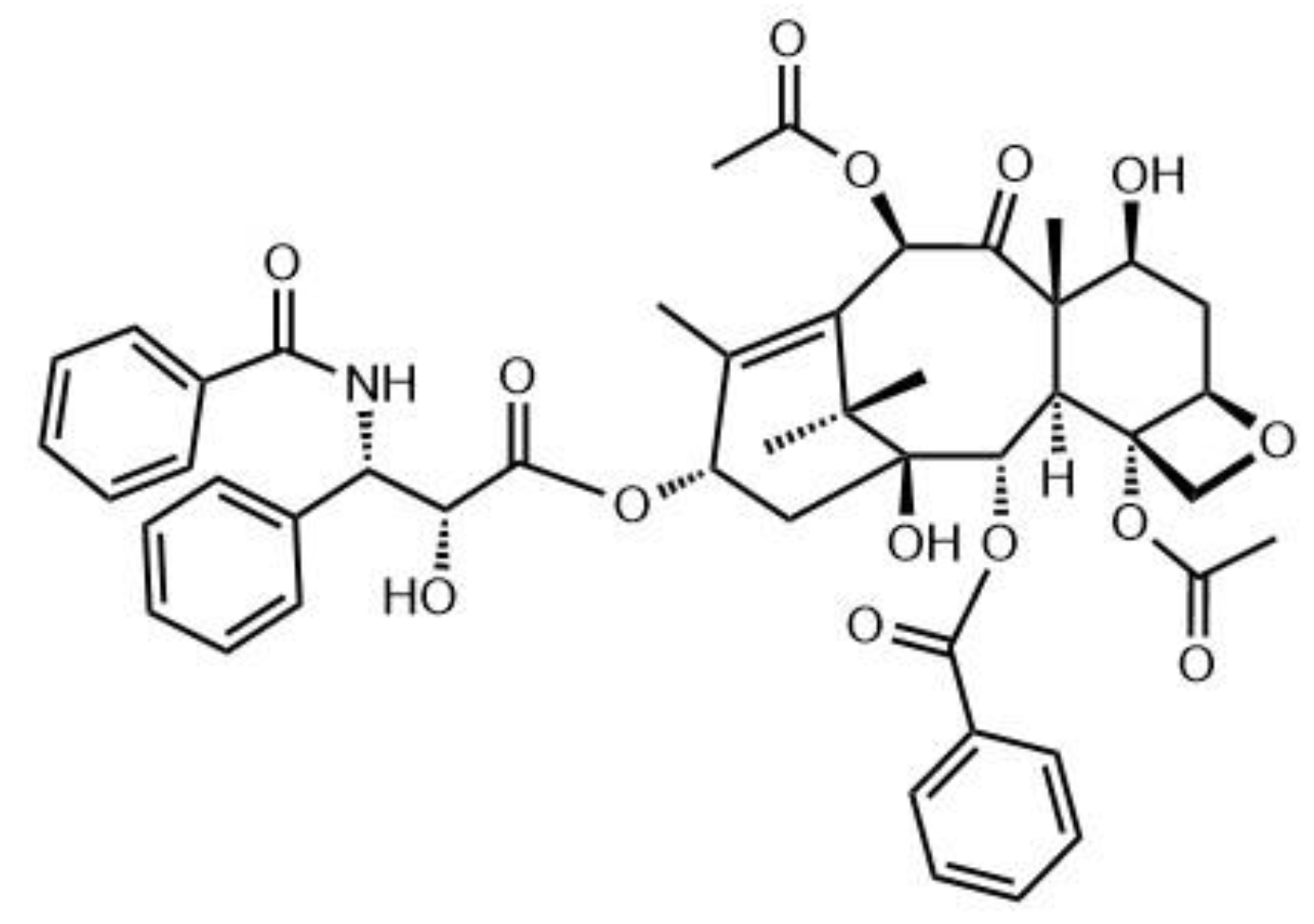
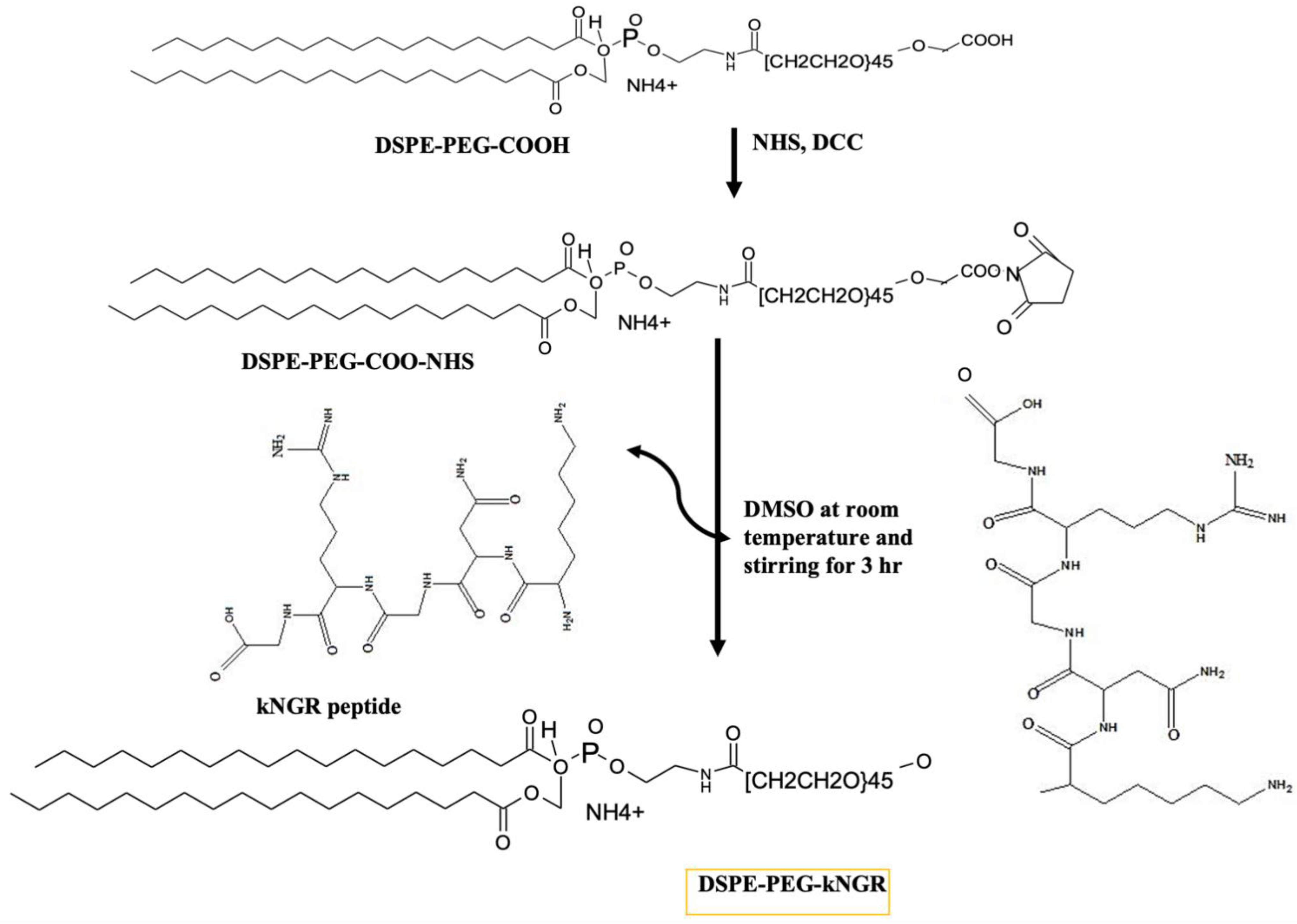
2.2. Synthesis and Characterization of DSPE–PEG–kNGR Conjugate
2.3. Preparation and Physicochemical Characterization of Polymer–Lipid Hybrid N.P.s (PLNs)
2.4. Cell Culture
2.5. Trypan Blue Exclusion Assay and Clonogenic Assay
2.6. Cell Apoptosis and Cytotoxicity Assay
2.7. Cell Cycle Analysis
2.8. Cellular Uptake of PLNs and Competition Assay
2.9. In-Vivo Studies
2.10. In-Vivo Antitumor Activity
2.11. Biodistribution Studies and In-Vivo Toxicological Parameters
2.12. Data Analysis
3. Results
3.1. Synthesis and Characterization of DSPE–PEG–kNGR Conjugate
3.2. Physicochemical Characterization of PLNs
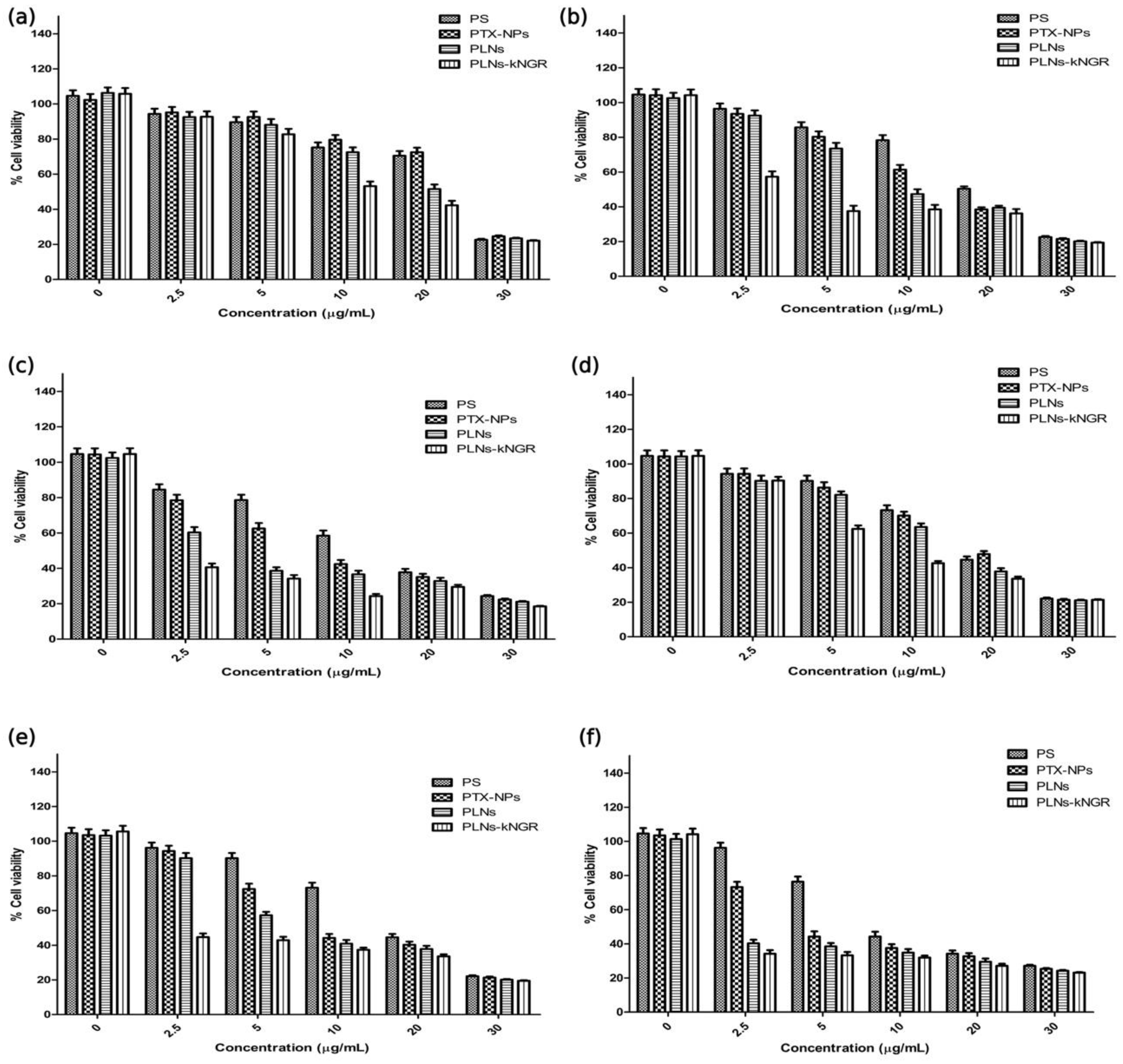
3.3. Cell Membrane Integrity and Anticancer Activity
3.4. Evaluation of Cell Apoptosis Activity
3.5. Cytotoxicity Assay
3.6. Cell Cycle Analysis
3.7. Cellular Uptake of N.P.s and Competition Assay
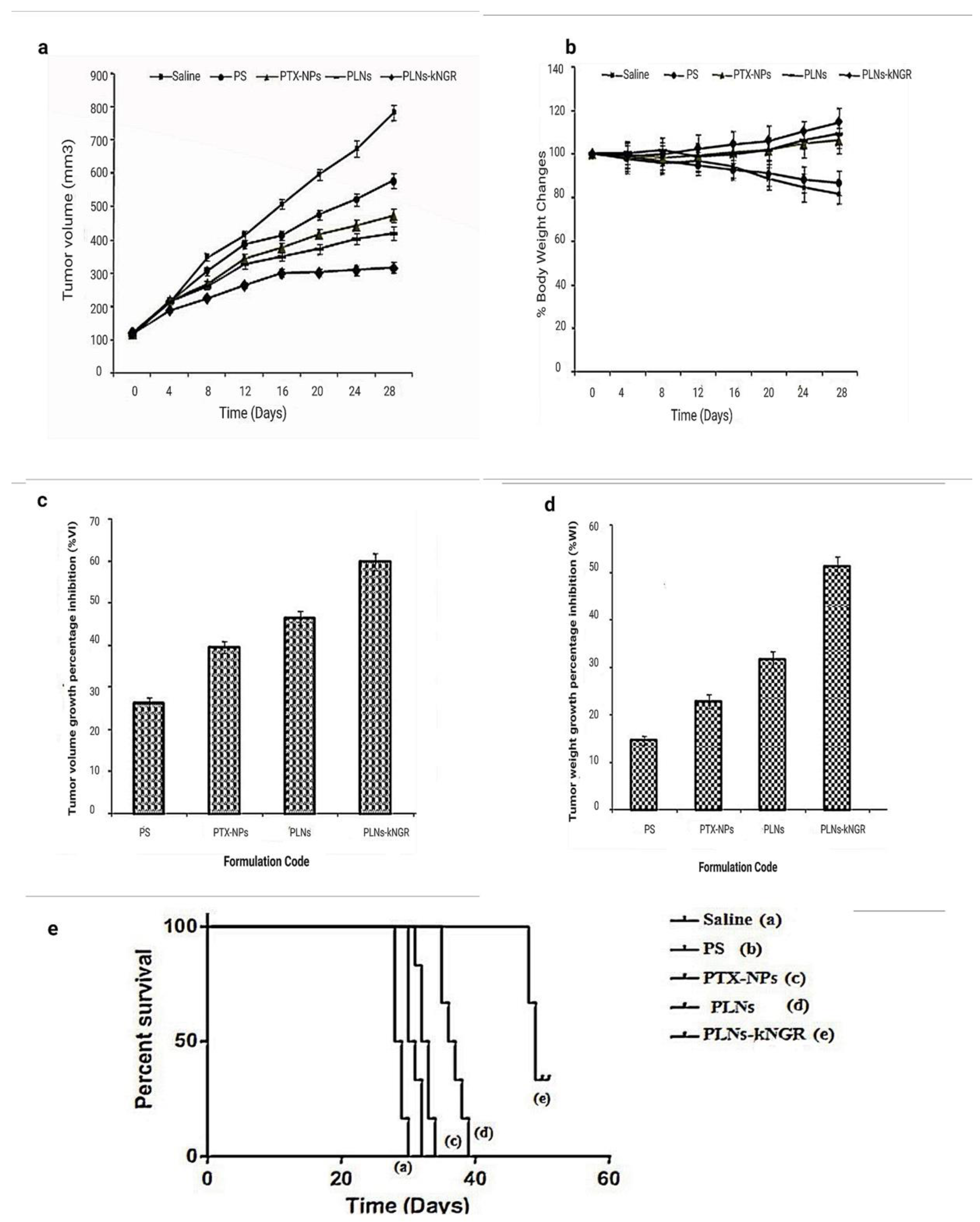
3.8. In-Vivo Antitumor Activity
3.9. Biodistribution Studies and In-Vivo Toxicological Parameters
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Paudel, K.R.; Panth, N.; Pangeni, R.; Awasthi, R.; Chawla, V.; Mehta, M.; Tambuwala, M.M.; Hansbro, P.M. Targeting lung cancer using advanced drug delivery systems. In Targeting Chronic Inflammatory Lung Diseases Using Advanced Drug Delivery Systems; Elsevier: Amsterdam, The Netherlands, 2020; pp. 493–516. [Google Scholar]
- Gupta, M.; Chashoo, G.; Sharma, P.R.; Saxena, A.K.; Gupta, P.N.; Agrawal, G.P.; Vyas, S.P. Dual Targeted Polymeric Nanoparticles Based on Tumor Endothelium and Tumor Cells for Enhanced Antitumor Drug Delivery. Mol. Pharm. 2014, 11, 697–715. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Yue, C.; Ma, Y.; Gong, P.; Zhao, P.; Zheng, C.; Sheng, Z.; Zhang, P.; Wang, Z.; Cai, L. Single-Step Assembly of DOX/ICG Loaded Lipid–Polymer Nanoparticles for Highly Effective Chemo-photothermal Combination Therapy. ACS Nano 2013, 7, 2056–2067. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, K.; Pan, J.; Liu, B.; Feng, S.-S. Folic acid conjugated nanoparticles of mixed lipid monolayer shell and biodegradable polymer core for targeted delivery of Docetaxel. Biomaterials 2010, 31, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Khursheed, R.; Dua, K.; Vishwas, S.; Gulati, M.; Jha, N.K.; Aldhafeeri, G.M.; Alanazi, F.G.; Goh, B.H.; Gupta, G.; Paudel, K.R.; et al. Biomedical applications of metallic nanoparticles in cancer: Current status and future perspectives. Biomed. Pharmacother. 2022, 150, 112951. [Google Scholar] [CrossRef]
- Alnuqaydan, A.M.; Almutary, A.G.; Azam, M.; Manandhar, B.; Yin, G.H.S.; Yen, L.L.; Madheswaran, T.; Paudel, K.R.; Hansbro, P.M.; Chellappan, D.K.; et al. Evaluation of the Cytotoxic Activity and Anti-Migratory Effect of Berberine–Phytantriol Liquid Crystalline Nanoparticle Formulation on Non-Small-Cell Lung Cancer In Vitro. Pharmaceutics 2022, 14, 1119. [Google Scholar] [CrossRef]
- Khursheed, R.; Paudel, K.R.; Gulati, M.; Vishwas, S.; Jha, N.K.; Hansbro, P.M.; Oliver, B.G.; Dua, K.; Singh, S.K. Expanding the arsenal against pulmonary diseases using surface-functionalized polymeric micelles: Breakthroughs and bottlenecks. Nanomedicine 2022, 1–31. [Google Scholar] [CrossRef]
- Aravind, A.; Jeyamohan, P.; Nair, R.; Veeranarayanan, S.; Nagaoka, Y.; Yoshida, Y.; Maekawa, T.; Kumar, D.S. AS1411 aptamer tagged PLGA-lecithin-PEG nanoparticles for tumor cell targeting and drug delivery. Biotechnol. Bioeng. 2012, 109, 2920–2931. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, J.J.; Huang, L. Nanoparticles Targeted with NGR Motif Deliver c-myc siRNA and Doxorubicin for Anticancer Therapy. Mol. Ther. 2010, 18, 828–834. [Google Scholar] [CrossRef]
- Zhang, L.; Chan, J.M.; Gu, F.X.; Rhee, J.-W.; Wang, A.Z.; Radovic-Moreno, A.F.; Alexis, F.; Langer, R.; Farokhzad, O.C. Self-Assembled Lipid−Polymer Hybrid Nanoparticles: A Robust Drug Delivery Platform. ACS Nano 2008, 2, 1696–1702. [Google Scholar] [CrossRef] [Green Version]
- Imran, M.; Jha, L.A.; Hasan, N.; Shrestha, J.; Pangeni, R.; Parvez, N.; Mohammed, Y.; Jha, S.K.; Paudel, K.R. “Nanodecoys”-Future of drug delivery by encapsulating nanoparticles in natural cell membranes. Int. J. Pharm. 2022, 621, 121790. [Google Scholar] [CrossRef]
- Imran, M.; Paudel, K.R.; Jha, S.K.; Hansbro, P.M.; Dua, K.; Mohammed, Y. Dressing multifunctional nanoparticles with natural cell-derived membranes for superior chemotherapy. Nanomedicine 2022, 17, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Sha, X.; Xin, H.; Chen, L.; Gao, X.; Wang, X.; Law, K.; Gu, J.; Chen, Y.; Jiang, Y.; et al. Self-aggregated pegylated poly (trimethylene carbonate) nanoparticles decorated with c(RGDyK) peptide for targeted paclitaxel delivery to integrin-rich tumors. Biomaterials 2011, 32, 9457–9469. [Google Scholar] [CrossRef]
- Mina-Osorio, P. The moonlighting enzyme CD13: Old and new functions to target. Trends Mol. Med. 2008, 14, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Pasqualini, R.; Koivunen, E.; Kain, R.; Lahdenranta, J.; Sakamoto, M.; Stryhn, A.; Ashmun, R.A.; Shapiro, L.H.; Arap, W.; Ruoslahti, E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000, 60, 722–727. [Google Scholar]
- Paudel, K.R.; Panth, N.; Manandhar, B.; Singh, S.K.; Gupta, G.; Wich, P.R.; Nammi, S.; MacLoughlin, R.; Adams, J.; Warkiani, M.E.; et al. Attenuation of Cigarette-Smoke-Induced Oxidative Stress, Senescence, and Inflammation by Berberine-Loaded Liquid Crystalline Nanoparticles: In Vitro Study in 16HBE and RAW264.7 Cells. Antioxidants 2022, 11, 873. [Google Scholar] [CrossRef]
- Curnis, F.; Sacchi, A.; Borgna, L.; Magni, F.; Gasparri, A.; Corti, A. Enhancement of tumor necrosis factor α antitumor immunotherapeutic properties by targeted delivery to aminopeptidase N (CD13). Nat. Biotechnol. 2000, 18, 1185–1190. [Google Scholar] [CrossRef]
- Ellerby, H.M.; Arap, W.; Ellerby, L.M.; Kain, R.; Andrusiak, R.; Del Rio, G.; Krajewski, S.; Lombardo, C.R.; Rao, R.; Ruoslahti, E.; et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat. Med. 1999, 5, 1032–1038. [Google Scholar] [CrossRef]
- Agrawal, U.; Chashoo, G.; Sharma, P.R.; Kumar, A.; Saxena, A.K.; Vyas, S. Tailored polymer–lipid hybrid nanoparticles for the delivery of drug conjugate: Dual strategy for brain targeting. Colloids Surf. B Biointerfaces 2015, 126, 414–425. [Google Scholar] [CrossRef]
- Pramual, S.; Lirdprapamongkol, K.; Svasti, J.; Bergkvist, M.; Jouan-Hureaux, V.; Arnoux, P.; Frochot, C.; Barberi-Heyob, M.; Niamsiri, N. Polymer-lipid-PEG hybrid nanoparticles as photosensitizer carrier for photodynamic therapy. J. Photochem. Photobiol. B Biol. 2017, 173, 12–22. [Google Scholar] [CrossRef]
- Garde, S.V.; Forté, A.J.; Ge, M.; Lepekhin, E.A.; Panchal, C.J.; Rabbani, S.; Wu, J.J. Binding and internalization of NGR-peptide-targeted liposomal doxorubicin (TVT-DOX) in CD13-expressing cells and its antitumor effects. Anti-Cancer Drugs 2007, 18, 1189–1200. [Google Scholar] [CrossRef]
- Liu, D.; Liu, Z.; Wang, L.; Zhang, C.; Zhang, N. Nanostructured lipid carriers as novel carrier for parenteral delivery of docetaxel. Colloids Surf. B Biointerfaces 2011, 85, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Lillard, J.W., Jr. Nanoparticle-based targeted drug delivery. Exp. Mol. Pathol. 2009, 86, 215–223. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.-J.; Ke, X.-Y.; Huang, Y.; Chen, X.-M.; Zhao, X.; Zhao, B.-X.; Lu, W.-L.; Lou, J.-N.; Zhang, X.; Zhang, Q. The antiangiogenic efficacy of NGR-modified PEG–DSPE micelles containing paclitaxel (NGR-M-PTX) for the treatment of glioma in rats. J. Drug Target. 2010, 19, 382–390. [Google Scholar] [CrossRef]
- Fahmy, T.M.; Fong, P.M.; Goyal, A.; Saltzman, W.M. Targeted for drug delivery. Mater. Today 2005, 8, 18–26. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Chen, X.; Wang, J.; Zhang, X.; Zhang, Q. NGR-modified micelles enhance their interaction with CD13-overexpressing tumor and endothelial cells. J. Control. Release 2009, 139, 56–62. [Google Scholar] [CrossRef]
- Amiji, M.M.; Lai, P.-K.; Shenoy, D.B.; Rao, M. Intratumoral Administration of Paclitaxel in an in situ Gelling Poloxamer 407 Formulation. Pharm. Dev. Technol. 2002, 7, 195–202. [Google Scholar] [CrossRef]
- Guo, J.; Gao, X.; Su, L.; Xia, H.; Gu, G.; Pang, Z.; Jiang, X.; Yao, L.; Chen, J.; Chen, H. Aptamer-functionalized PEG–PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials 2011, 32, 8010–8020. [Google Scholar] [CrossRef]
- Essa, S.; Rabanel, J.M.; Hildgen, P. Characterization of rhodamine loaded PEG-g-PLA nanoparticles (NPs): Effect of poly(ethylene glycol) grafting density. Int. J. Pharm. 2011, 411, 178–187. [Google Scholar] [CrossRef]
- Wang, L.; Ho, P.; Lee, H.; Vaddi, H.; Chan, Y.; Yung, C.S. Quantitation of paclitaxel in micro-sample rat plasma by a sensitive reversed-phase HPLC assay. J. Pharm. Biomed. Anal. 2003, 31, 283–289. [Google Scholar] [CrossRef]
- Yu, D.-H.; Lu, Q.; Xie, J.; Fang, C.; Chen, H.-Z. Peptide-conjugated biodegradable nanoparticles as a carrier to target paclitaxel to tumor neovasculature. Biomaterials 2010, 31, 2278–2292. [Google Scholar] [CrossRef]
- Liu, C.; Yu, W.; Chen, Z.; Zhang, J.; Zhang, N. Enhanced gene transfection efficiency in CD13-positive vascular endothelial cells with targeted poly(lactic acid)–poly(ethylene glycol) nanoparticles through caveolae-mediated endocytosis. J. Control. Release 2011, 151, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Paudel, K.R.; Mehta, M.; Yin, G.H.S.; Yen, L.L.; Malyla, V.; Patel, V.K.; Panneerselvam, J.; Madheswaran, T.; MacLoughlin, R.; Jha, N.K.; et al. Berberine-loaded liquid crystalline nanoparticles inhibit non-small cell lung cancer proliferation and migration in vitro. Environ. Sci. Pollut. Res. 2022, 29, 46830–46847. [Google Scholar] [CrossRef] [PubMed]
- Paudel, K.R.; Kim, D.-W. Microparticles-Mediated Vascular Inflammation and its Amelioration by Antioxidant Activity of Baicalin. Antioxidants 2020, 9, 890. [Google Scholar] [CrossRef]
- Wong, H.L.; Rauth, A.M.; Bendayan, R.; Manias, J.L.; Ramaswamy, M.; Liu, Z.; Erhan, S.Z.; Wu, X.Y. A New Polymer–Lipid Hybrid Nanoparticle System Increases Cytotoxicity of Doxorubicin against Multidrug-Resistant Human Breast Cancer Cells. Pharm. Res. 2006, 23, 1574–1585. [Google Scholar] [CrossRef]
- Liu, B.; Yang, M.; Li, X.; Qian, X.; Shen, Z.; Ding, Y.; Yu, L. Enhanced Efficiency of Thermally Targeted Taxanes Delivery in a Human Xenograft Model of Gastric Cancer. J. Pharm. Sci. 2008, 97, 3170–3181. [Google Scholar] [CrossRef] [PubMed]
- Teow, H.M.; Zhou, Z.; Najlah, M.; Yusof, S.R.; Abbott, N.J.; D’Emanuele, A. Delivery of paclitaxel across cellular barriers using a dendrimer-based nanocarrier. Int. J. Pharm. 2013, 441, 701–711. [Google Scholar] [CrossRef]
- Ganesh, T.; Yang, C.; Norris, A.; Glass, T.; Bane, S.; Ravindra, R.; Banerjee, A.; Metaferia, B.; Thomas, S.L.; Giannakakou, P.; et al. Evaluation of the Tubulin-Bound Paclitaxel Conformation: Synthesis, Biology, and SAR Studies of C-4 to C-3′ Bridged Paclitaxel Analogues. J. Med. Chem. 2007, 50, 713–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Shi, Y.; Chen, Y.; Yu, S.; Hao, J.; Luo, J.; Sha, X.; Fang, X. Enhanced antitumor efficacy by Paclitaxel-loaded Pluronic P123/F127 mixed micelles against non-small cell lung cancer based on passive tumor targeting and modulation of drug resistance. Eur. J. Pharm. Biopharm. 2010, 75, 341–353. [Google Scholar] [CrossRef]
- Son, S.; Singha, K.; Kim, W.J. Bioreducible BPEI-SS-PEG-cNGR polymer as a tumor targeted nonviral gene carrier. Biomaterials 2010, 31, 6344–6354. [Google Scholar] [CrossRef]
- Farokhzad, O.C.; Cheng, J.; Teply, B.A.; Sherifi, I.; Jon, S.; Kantoff, P.W.; Richie, J.P.; Langer, R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 6315–6320. [Google Scholar] [CrossRef] [Green Version]
- Min, K.H.; Kim, J.-H.; Bae, S.M.; Shin, H.; Kim, M.S.; Park, S.; Lee, H.; Park, R.-W.; Kim, K.; Kwon, I.C.; et al. Tumoral acidic pH-responsive MPEG-poly(β-amino ester) polymeric micelles for cancer targeting therapy. J. Control. Release 2010, 144, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.K.; Labhasetwar, V. Enhanced Antiproliferative Activity of Transferrin-Conjugated Paclitaxel-Loaded Nanoparticles Is Mediated via Sustained Intracellular Drug Retention. Mol. Pharm. 2005, 2, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Zhang, L.; Teply, B.A.; Mann, N.; Wang, A.; Radovic-Moreno, A.F.; Langer, R.; Farokhzad, O.C. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc. Natl. Acad. Sci. USA 2008, 105, 2586–2591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Hensbergen, Y.; Broxterman, H.J.; Elderkamp, Y.W.; Lankelma, J.; Beers, J.C.; Heijn, M.; Boven, E.; Hoekman, K.; Pinedo, H.M. A doxorubicin–CNGRC-peptide conjugate with prodrug properties. Biochem. Pharmacol. 2002, 63, 897–908. [Google Scholar] [CrossRef]
- Yokoyama, Y.; Ramakrishnan, S. Addition of an aminopeptidase N-binding sequence to human endostatin improves inhibition of ovarian carcinoma growth. Cancer 2005, 104, 321–331. [Google Scholar] [CrossRef]
- Naik, S.; Patel, D.; Chuttani, K.; Mishra, A.K.; Misra, A. In vitro mechanistic study of cell death and in vivo performance evaluation of RGD grafted PEGylated docetaxel liposomes in breast cancer. Nanomed. Nanotechnol. Biol. Med. 2012, 8, 951–962. [Google Scholar] [CrossRef]
- Dunne, M.; Zheng, J.; Rosenblat, J.; Jaffray, D.A.; Allen, C. APN/CD13-targeting as a strategy to alter the tumor accumulation of liposomes. J. Control. Release 2011, 154, 298–305. [Google Scholar] [CrossRef]
- Murase, Y.; Asai, T.; Katanasaka, Y.; Sugiyama, T.; Shimizu, K.; Maeda, N.; Oku, N. A novel DDS strategy, “dual-targeting”, and its application for antineovascular therapy. Cancer Lett. 2010, 287, 165–171. [Google Scholar] [CrossRef]
- Pastorino, F.; Brignole, C.; Marimpietri, D.; Cilli, M.; Gambini, C.; Ribatti, D.; Longhi, R.; Allen, T.M.; Corti, A.; Ponzoni, M. Vascular damage and anti-angiogenic effects of tumor vessel-targeted liposomal chemotherapy. Cancer Res. 2003, 63, 7400–7409. [Google Scholar]




| Formulation Code | Size (nm) | PI (Polydispersity Index) | Zeta Potential (mV) | %EE (Entrapment Efficiency) | CE% (Conjugation Efficiency) | Surface Density (P) | (Average Distancein nm) D |
|---|---|---|---|---|---|---|---|
| PTX-NPs (Polymer-based nanoparticles) | 163.5 ± 5.52 | 0.128 ± 0.011 | −22.4 ± 1.8 | 72.24 ± 4.43 | --- | --- | --- |
| PLNs (Polymer–lipid hybrid nanoprtaicles without ligand) | 178.8 ± 8.41 | 0.126 ± 0.012 | −26.6 ± 1.9 | 78.88 ± 5.38 | --- | --- | --- |
| PLNs-kNGR (Ligand-conjugated Polymer–lipid hybrid nanoprtaicles) | 205.1 ± 9.1 | 0.117 ± 0.011 | −31.3 ± 2.3 | 82.21 ± 3.75 | 34.7 | 198 ± 6.8 | 16 + 1.4 |
| Time (h) | HUVEC Cell Line, IC50 μg/mL) | HT-1080 Cell Line, IC50 (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|
| PS | PTX-NPs | PLNs | PLNs-kNGR | PS | PTX-NPs | PLNs | PLNs-kNGR | |
| 24 | 22.5 | 23.5 | 18.8 | 9.6 | 18.5 | 19.8 | 12.9 | 7.6 |
| 48 | 19.3 | 13.8 | 8.9 | 3.3 | 17.2 | 9.0 | 5.6 | 2.4 |
| 72 | 12.1 | 7.2 | 2.72 | 0.98 | 9.25 | 4.8 | 2.2 | 0.85 |
| Formulation Code | Dose (mg/kg) | %VI | %WI | Median Survival Time (days) | Mean Survival Time (days) | Standard Error | 95% Confidence in Interval | Increase in Survival Time (% IST) | Log Rank Test | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Saline | PS | PTX-NPs | PLNs | |||||||||
| Saline | --- | --- | --- | 28 | 28.3 | 0.516 | 27.791–28.875 | --- | --- | --- | --- | --- |
| PS | 10 | 26.2 ± 1.2 | 14.6 ± 1.0 | 32 | 31.7 | 0.422 | 30.583–32.751 | 14.3 | ** p < 0.01 | --- | --- | --- |
| PTX-NPs | 10 | 39.5 ± 1.5 | 22.8 ± 1.4 | 33 | 32.7 | 0.422 | 31.583–33.751 | 17.9 | *** p < 0.001 | ns (p >0.05) | --- | --- |
| PLNs | 10 | 46.4 ± 1.7 | 31.6 ± 1.7 | 37 | 36.7 | 0.76 | 34.712–38.621 | 32.1 | *** p < 0.001 | *** p < 0.001 | *** p < 0.001 | --- |
| PLNs-kNGR | 10 | 59.7 ± 2.1 | 51.3 ± 1.9 | 49 | 49.2 | 0.477 | 47.940–50.394 | 75 | *** p < 0.001 | *** p < 0.001 | *** p < 0.001 | *** p < 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, M.; Sharma, V.; Sharma, K.; Kumar, A.; Sharma, A.; Kazmi, I.; Al-Abbasi, F.A.; Alzarea, S.I.; Afzal, O.; Altamimi, A.S.A.; et al. A kNGR Peptide-Tethered Lipid–Polymer Hybrid Nanocarrier-Based Synergistic Approach for Effective Tumor Therapy: Development, Characterization, Ex-Vivo, and In-Vivo Assessment. Pharmaceutics 2022, 14, 1401. https://doi.org/10.3390/pharmaceutics14071401
Gupta M, Sharma V, Sharma K, Kumar A, Sharma A, Kazmi I, Al-Abbasi FA, Alzarea SI, Afzal O, Altamimi ASA, et al. A kNGR Peptide-Tethered Lipid–Polymer Hybrid Nanocarrier-Based Synergistic Approach for Effective Tumor Therapy: Development, Characterization, Ex-Vivo, and In-Vivo Assessment. Pharmaceutics. 2022; 14(7):1401. https://doi.org/10.3390/pharmaceutics14071401
Chicago/Turabian StyleGupta, Madhu, Vikas Sharma, Kalicharan Sharma, Anoop Kumar, Ajay Sharma, Imran Kazmi, Fahad A. Al-Abbasi, Sami I. Alzarea, Obaid Afzal, Abdulmalik Saleh Alfawaz Altamimi, and et al. 2022. "A kNGR Peptide-Tethered Lipid–Polymer Hybrid Nanocarrier-Based Synergistic Approach for Effective Tumor Therapy: Development, Characterization, Ex-Vivo, and In-Vivo Assessment" Pharmaceutics 14, no. 7: 1401. https://doi.org/10.3390/pharmaceutics14071401
APA StyleGupta, M., Sharma, V., Sharma, K., Kumar, A., Sharma, A., Kazmi, I., Al-Abbasi, F. A., Alzarea, S. I., Afzal, O., Altamimi, A. S. A., Singh, S. K., Gupta, G., Paudel, K. R., Hansbro, P. M., & Dua, K. (2022). A kNGR Peptide-Tethered Lipid–Polymer Hybrid Nanocarrier-Based Synergistic Approach for Effective Tumor Therapy: Development, Characterization, Ex-Vivo, and In-Vivo Assessment. Pharmaceutics, 14(7), 1401. https://doi.org/10.3390/pharmaceutics14071401









