Analyzing siRNA Concentration, Complexation and Stability in Cationic Dendriplexes by Stem-Loop Reverse Transcription-qPCR
Abstract
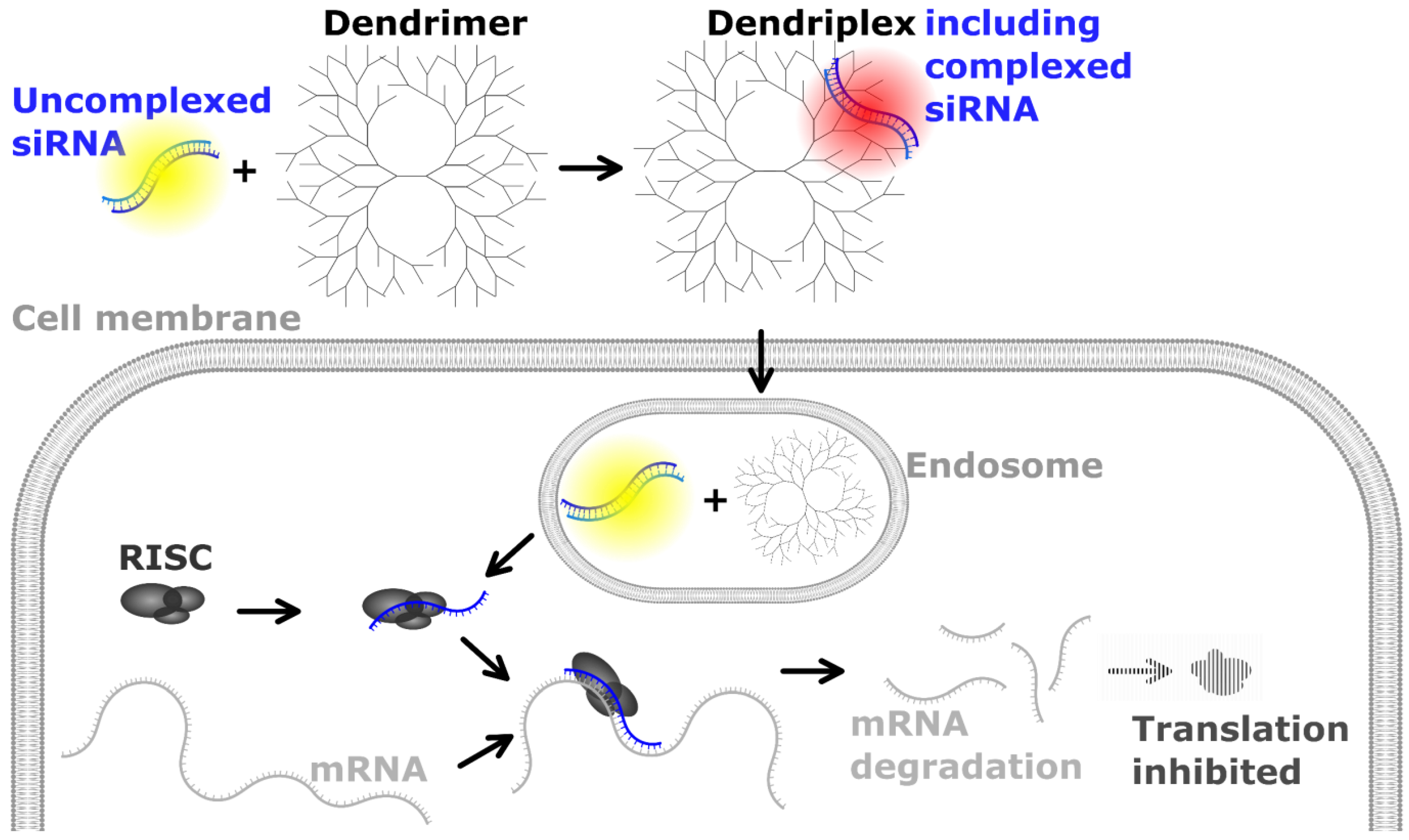
1. Introduction
2. Materials and Methods
2.1. Preparation of Dendriplexes
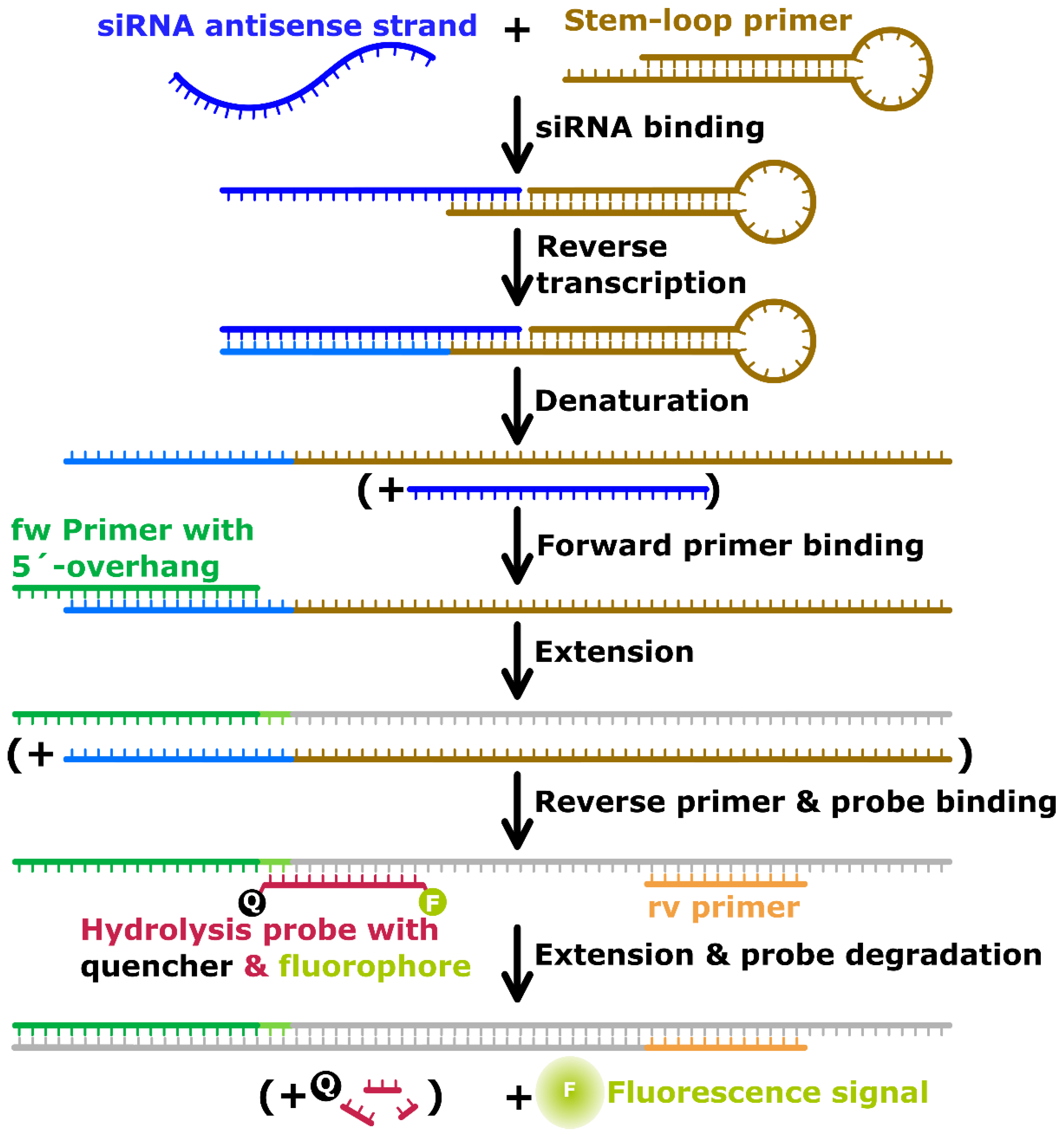
2.2. Targets, Primers and Probes
- Forward primer: 5′-GAGCAAGTCGAACACCTTGA-3′
- Reverse primer: 5′-CCAGTGCAGGGTCCGAGGTA-3′
- Stem-loop primer: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGA TACGACCTCCAT-3′
- Probe: 5′-(6-FAM)-TGGATACGACCTCCATCA-(IBFQ)-3′
- (Anti-Smad6) target siRNA sense strand: 5′-ccaucaagguguucgacuuTT-3′
- (Anti-Smad6) target siRNA antisense strand: 5′-aagucgaacaccuugauggAG-3′
- randomized siRNA: 5′-nnnnnnnnnnnnnnnnnnnNN-3′
2.3. Reverse Transcription-qPCR
2.4. Preparation of siRNA Digest Using RNase A
3. Results
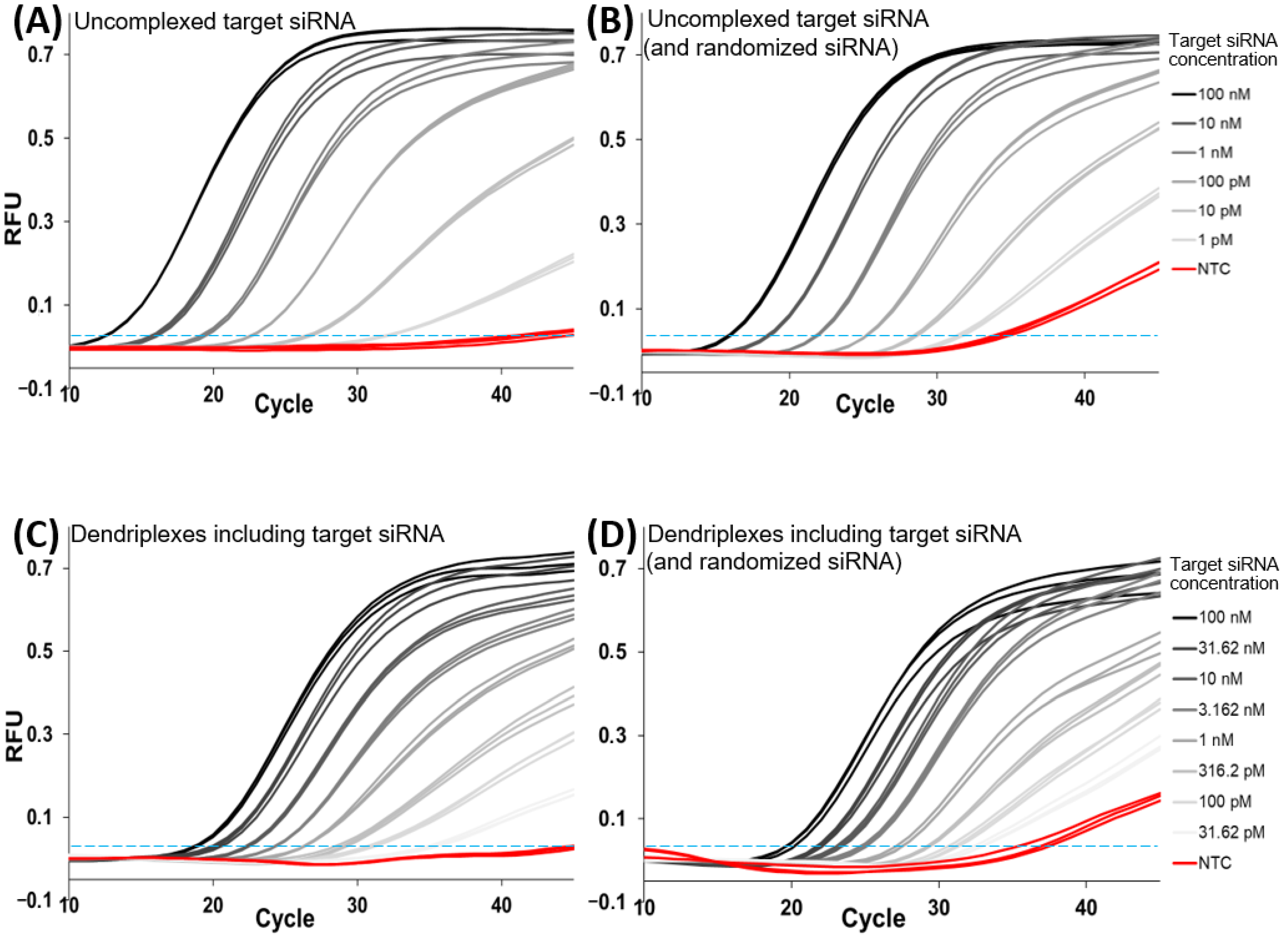
3.1. RT-qPCR Assay Development
3.1.1. Assay Implementation for Uncomplexed Target siRNA
3.1.2. Quantification of Complexed siRNA
3.2. Influence of N/P Ratio on siRNA Accessibility
3.3. Effects of Complexation on siRNA Stability
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.-C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Bahal, R.; Rasmussen, T.P.; Manautou, J.E.; Zhong, X. The growth of siRNA-based therapeutics: Updated clinical studies. Biochem. Pharmacol. 2021, 189, 114432. [Google Scholar] [CrossRef] [PubMed]
- Wittrup, A.; Lieberman, J. Knocking down disease: A progress report on siRNA therapeutics. Nat. Rev. Genet. 2015, 16, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-L.; Chang, W.-T.; Shih, Y.-W. Gene therapy using RNAi. In Gene Therapy—Developments and Future Perspectives; IntechOpen: London, UK, 2011; Volume 1, pp. 31–52. ISBN 978-953-307-617-1. [Google Scholar]
- Dykxhoorn, D.M.; Palliser, D.; Lieberman, J. The silent treatment: siRNAs as small molecule drugs. Gene Ther. 2006, 13, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, Z.; Wientjes, M.G.; Au, J.L.-S. Delivery of siRNA therapeutics: Barriers and carriers. AAPS J. 2010, 12, 492–503. [Google Scholar] [CrossRef]
- Tekade, R.; Maheshwari, R.; Sharma, P.; Tekade, M.; Chauhan, A. siRNA therapy, challenges and underlying perspectives of dendrimer as delivery vector. Curr. Pharm. Des. 2015, 21, 4614–4636. [Google Scholar] [CrossRef]
- Chen, J.; Ellert-Miklaszewska, A.; Garofalo, S.; Dey, A.K.; Tang, J.; Jiang, Y.; Clément, F.; Marche, P.N.; Liu, X.; Kaminska, B.; et al. Synthesis and use of an amphiphilic dendrimer for siRNA delivery into primary immune cells. Nat. Protoc. 2021, 16, 327–351. [Google Scholar] [CrossRef]
- Dhumal, D.; Lan, W.; Ding, L.; Jiang, Y.; Lyu, Z.; Laurini, E.; Marson, D.; Tintaru, A.; Dusetti, N.; Giorgio, S.; et al. An ionizable supramolecular dendrimer nanosystem for effective siRNA delivery with a favorable safety profile. Nano Res. 2021, 14, 2247–2254. [Google Scholar] [CrossRef]
- Wu, J.; Huang, W.; He, Z. Dendrimers as carriers for siRNA delivery and gene silencing: A review. Sci. World J. 2013, 2013, 630654. [Google Scholar] [CrossRef]
- Dykxhoorn, D.M.; Lieberman, J. Knocking down disease with siRNAs. Cell 2006, 126, 231–235. [Google Scholar] [CrossRef]
- Takei, Y.; Kadomatsu, K.; Yuzawa, Y.; Matsuo, S.; Muramatsu, T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004, 64, 3365–3370. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, Y.; Chen, W.L.; Wang, D.D.; Zhou, Y.J.; You, B.G.; Liu, Y.; Qu, C.X.; Yang, S.D.; Chen, M.T.; et al. Co-delivery of VEGF siRNA and etoposide for enhanced anti-angiogenesis and anti-proliferation effect via multi-functional nanoparticles for orthotopic non-small cell lung cancer treatment. Theranostics 2019, 9, 5886–5898. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Torchilin, V. Dendrimers for siRNA delivery. Pharmaceuticals 2013, 6, 161–183. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chen, Q.; Zheng, Y.; Zhang, P.; Chen, X.; Lu, J.; Lv, Y.; Sun, S.; Zeng, W. PAMAM-cRGD mediating efficient siRNA delivery to spermatogonial stem cells. Stem Cell Res. Ther. 2019, 10, 399. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Rocchi, P.; Qu, F.; Zheng, S.; Liang, Z.; Gleave, M.; Iovanna, J.; Peng, L. PAMAM dendrimers mediate siRNA delivery to target Hsp27 and produce potent antiproliferative effects on prostate cancer cells. ChemMedChem 2009, 4, 1302–1310. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, J.; Hafdi, N.; Behr, J.-P.; Erbacher, P.; Peng, L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem. Commun. 2006, 22, 2362–2364. [Google Scholar] [CrossRef]
- Haensler, J.; Szoka, F.C. Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjug. Chem. 1993, 4, 372–379. [Google Scholar] [CrossRef]
- Perche, F.; Yi, Y.; Hespel, L.; Mi, P.; Dirisala, A.; Cabral, H.; Miyata, K.; Kataoka, K. Hydroxychloroquine-conjugated gold nanoparticles for improved siRNA activity. Biomaterials 2016, 90, 62–71. [Google Scholar] [CrossRef]
- Huang, D.; Wu, D. Biodegradable dendrimers for drug delivery. Mater. Sci. Eng. C 2018, 90, 713–727. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Pavan, G.M.; Posocco, P.; Tagliabue, A.; Maly, M.; Malek, A.; Danani, A.; Ragg, E.; Catapano, C.V.; Pricl, S. PAMAM dendrimers for siRNA delivery: Computational and experimental insights. Chem.-A Eur. J. 2010, 16, 7781–7795. [Google Scholar] [CrossRef] [PubMed]
- Sardh, E.; Harper, P.; Balwani, M.; Stein, P.; Rees, D.; Bissell, D.M.; Desnick, R.; Parker, C.; Phillips, J.; Bonkovsky, H.L.; et al. Phase 1 trial of an RNA interference therapy for acute intermittent porphyria. N. Engl. J. Med. 2019, 380, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo, S.; Pérez-Martnez, F.C.; Pérez-Carrin, M.D.; Guerra, J.; Merino, S.; Snchez-Verd, M.P.; Cñea, V. Inhibition of p42 MAPK using a nonviral vector-delivered siRNA potentiates the anti-tumor effect of metformin in prostate cancer cells. Nanomedicine 2012, 7, 493–506. [Google Scholar] [CrossRef]
- Hoerter, J.A.H.; Krishnan, V.; Lionberger, T.A.; Walter, N.G. siRNA-like double-stranded RNAs are specifically protected against degradation in human cell extract. PLoS ONE 2011, 6, e20359. [Google Scholar] [CrossRef]
- Zhang, C.; Tang, N.; Liu, X.; Liang, W.; Xu, W.; Torchilin, V.P. siRNA-containing liposomes modified with polyarginine effectively silence the targeted gene. J. Control. Release 2006, 112, 229–239. [Google Scholar] [CrossRef]
- Buyens, K.; Lucas, B.; Raemdonck, K.; Braeckmans, K.; Vercammen, J.; Hendrix, J.; Engelborghs, Y.; de Smedt, S.C.; Sanders, N.N. A fast and sensitive method for measuring the integrity of siRNA-carrier complexes in full human serum. J. Control. Release 2008, 126, 67–76. [Google Scholar] [CrossRef]
- Raval, N.; Jogi, H.; Gondaliya, P.; Kalia, K.; Tekade, R.K. Method and its composition for encapsulation, stabilization, and delivery of siRNA in anionic polymeric nanoplex: An in vitro–in vivo assessment. Sci. Rep. 2019, 9, 16047. [Google Scholar] [CrossRef]
- Lee, Y.W.; Hwang, Y.E.; Lee, J.Y.; Sohn, J.H.; Sung, B.H.; Kim, S.C. VEGF siRNA delivery by a cancer-specific cell-penetrating peptide. J. Microbiol. Biotechnol. 2018, 28, 367–374. [Google Scholar] [CrossRef]
- Nguyen, M.K.; Huynh, C.T.; Gilewski, A.; Wilner, S.E.; Maier, K.E.; Kwon, N.; Levy, M.; Alsberg, E. Covalently tethering siRNA to hydrogels for localized, controlled release and gene silencing. Sci. Adv. 2019, 5, eaax0801. [Google Scholar] [CrossRef] [PubMed]
- Oude Blenke, E.; Evers, M.J.W.; Baumann, V.; Winkler, J.; Storm, G.; Mastrobattista, E. Critical evaluation of quantification methods for oligonucleotides formulated in lipid nanoparticles. Int. J. Pharm. 2018, 548, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.-L.; Ali, A.; Chu, C.; Cao, H.; Rana, T.M. Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chem. Biol. 2004, 11, 1165–1175. [Google Scholar] [CrossRef]
- Manoharan, M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004, 8, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Overhoff, M. Quantitative detection of siRNA and single-stranded oligonucleotides: Relationship between uptake and biological activity of siRNA. Nucleic Acids Res. 2004, 32, e170. [Google Scholar] [CrossRef]
- Mehrotra, S.; Lee, I.; Chan, C. Multilayer mediated forward and patterned siRNA transfection using linear-PEI at extended N/P ratios. Acta Biomater. 2009, 5, 1474–1488. [Google Scholar] [CrossRef]
- Hecker, K.H.; Green, S.M.; Kobayashi, K. Analysis and purification of nucleic acids by ion-pair reversed-phase high-performance liquid chromatography. J. Biochem. Biophys. Methods 2000, 46, 83–93. [Google Scholar] [CrossRef]
- Andrus, A.; Kuimelis, R.G. Analysis and purification of synthetic nucleic acids using HPLC. Curr. Protoc. Nucleic Acid Chem. 2000, 1, 10-5. [Google Scholar] [CrossRef]
- Sinha, N.D.; Jung, K.E. Analysis and purification of synthetic nucleic acids using HPLC. Curr. Protoc. Nucleic Acid Chem. 2015, 61, 10-5. [Google Scholar] [CrossRef]
- Mouillesseaux, K.P.; Wiley, D.S.; Saunders, L.M.; Wylie, L.A.; Kushner, E.J.; Chong, D.C.; Citrin, K.M.; Barber, A.T.; Park, Y.; Kim, J.-D.; et al. Notch regulates BMP responsiveness and lateral branching in vessel networks via SMAD6. Nat. Commun. 2016, 7, 13247. [Google Scholar] [CrossRef]
- Wylie, L.A.; Mouillesseaux, K.P.; Chong, D.C.; Bautch, V.L. Developmental SMAD6 loss leads to blood vessel hemorrhage and disrupted endothelial cell junctions. Dev. Biol. 2018, 442, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.B.; Pavan, G.M.; Kasimova, M.R.; Rutherford, S.; Danani, A.; Nielsen, H.M.; Foged, C. Elucidating the molecular mechanism of PAMAM–siRNA dendriplex self-assembly: Effect of dendrimer charge density. Int. J. Pharm. 2011, 416, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, D.; Zhang, H.; Parekh, H.S.; Smith, S.C. The effect of pH on PAMAM dendrimer–siRNA complexation—Endosomal considerations as determined by molecular dynamics simulation. Biophys. Chem. 2011, 158, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Kichler, A.; Mason, A.J.; Bechinger, B. Cationic amphipathic histidine-rich peptides for gene delivery. Biochim. Biophys. Acta-Biomembr. 2006, 1758, 301–307. [Google Scholar] [CrossRef]
- Langlet-Bertin, B.; Leborgne, C.; Scherman, D.; Bechinger, B.; Mason, A.J.; Kichler, A. Design and evaluation of histidine-rich amphipathic peptides for siRNA delivery. Pharm. Res. 2010, 27, 1426–1436. [Google Scholar] [CrossRef]
- Kramer, M.F. Stem-Loop RT-qPCR for miRNAs. Curr. Protoc. Mol. Biol. 2011, 95, 15.10.1–15.10.15. [Google Scholar] [CrossRef]
- Shrivastava, A.; Gupta, V. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron. Young Sci. 2011, 2, 21. [Google Scholar] [CrossRef]
- Zhou, H.; Tsou, J.-H.; Chinthalapally, M.; Liu, H.; Jiang, F. Detection and differentiation of SARS-CoV-2, influenza, and respiratory syncytial viruses by CRISPR. Diagnostics 2021, 11, 823. [Google Scholar] [CrossRef]
- Wolfinger, R.D.; Beedanagari, S.; Boitier, E.; Chen, T.; Couttet, P.; Ellinger-Ziegelbauer, H.; Guillemain, G.; Mariet, C.; Mouritzen, P.; O’Lone, R.; et al. Two approaches for estimating the lower limit of quantitation (LLOQ) of microRNA levels assayed as exploratory biomarkers by RT-qPCR. BMC Biotechnol. 2018, 18, 6. [Google Scholar] [CrossRef]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef]
- Keer, J.T. Chapter 7. Quantitative real-time PCR analysis. In Essentials of Nucleic Acid Analysis; Royal Society of Chemistry: Cambridge, UK, 2008; pp. 132–166. [Google Scholar]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR inhibition in qPCR, dPCR and MPS—Mechanisms and solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef] [PubMed]
- Čepin, U. Understanding qPCR Efficiency and Why It Can Exceed 100%. Available online: https://biosistemika.com/blog/qpcr-efficiency-over-100/ (accessed on 19 November 2021).
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Jensen, S.G.; Lamy, P.; Rasmussen, M.H.; Ostenfeld, M.S.; Dyrskjøt, L.; Ørntoft, T.F.; Andersen, C.L. Evaluation of two commercial global miRNA expression profiling platforms for detection of less abundant miRNAs. BMC Genom. 2011, 12, 435. [Google Scholar] [CrossRef]
- Benes, V.; Castoldi, M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods 2010, 50, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Vermeer, L.S.; Hamon, L.; Schirer, A.; Schoup, M.; Cosette, J.; Majdoul, S.; Pastré, D.; Stockholm, D.; Holic, N.; Hellwig, P.; et al. Vectofusin-1, a potent peptidic enhancer of viral gene transfer forms pH-dependent α-helical nanofibrils, concentrating viral particles. Acta Biomater. 2017, 64, 259–268. [Google Scholar] [CrossRef]
- Lointier, M.; Dussouillez, C.; Glattard, E.; Kichler, A.; Bechinger, B. Different biological activities of histidine-rich peptides are favored by variations in their design. Toxins 2021, 13, 363. [Google Scholar] [CrossRef]
- Singh, T.; Murthy, A.S.N.; Yang, H.-J.; Im, J. Versatility of cell-penetrating peptides for intracellular delivery of siRNA. Drug Deliv. 2018, 25, 1996–2006. [Google Scholar] [CrossRef]
- Spyropoulos-Antonakakis, N.; Sarantopoulou, E.; Trohopoulos, P.N.; Stefi, A.L.; Kollia, Z.; Gavriil, V.E.; Bourkoula, A.; Petrou, P.S.; Kakabakos, S.; Semashko, V.V.; et al. Selective aggregation of PAMAM dendrimer nanocarriers and PAMAM/ZnPc nanodrugs on human atheromatous carotid tissues: A photodynamic therapy for atherosclerosis. Nanoscale Res. Lett. 2015, 10, 210. [Google Scholar] [CrossRef]
- Cahill, B.P.; Papastavrou, G.; Koper, G.J.M.; Borkovec, M. Adsorption of poly(amido amine) (PAMAM) dendrimers on silica: Importance of electrostatic three-body attraction. Langmuir 2008, 24, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Yablon, D.G.; Karchner, R.; Knapp, D.; Kleinman, M.H.; Fang, H.; Durning, C.J.; Tomalia, D.A.; Turro, N.J.; Flynn, G.W. AFM Studies of high-generation PAMAM dendrimers at the liquid/solid interface. Langmuir 2002, 18, 7452–7455. [Google Scholar] [CrossRef]
- Veiman, K.-L.; Mäger, I.; Ezzat, K.; Margus, H.; Lehto, T.; Langel, K.; Kurrikoff, K.; Arukuusk, P.; Suhorutšenko, J.; Padari, K.; et al. PepFect14 peptide vector for efficient gene delivery in cell cultures. Mol. Pharm. 2013, 10, 199–210. [Google Scholar] [CrossRef]
- Perez, A.P.; Cosaka, M.L.; Romero, E.L.; Morilla, M.J. Uptake and intracellular traffic of siRNA dendriplexes in glioblastoma cells and macrophages. Int. J. Nanomed. 2011, 6, 2715. [Google Scholar] [CrossRef]
- Tang, Y.; Li, Y.-B.; Wang, B.; Lin, R.-Y.; van Dongen, M.; Zurcher, D.M.; Gu, X.-Y.; Banaszak Holl, M.M.; Liu, G.; Qi, R. Efficient in vitro siRNA delivery and intramuscular gene silencing using PEG-modified PAMAM dendrimers. Mol. Pharm. 2012, 9, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Ihnatsyeu-Kachan, A.; Dzmitruk, V.; Apartsin, E.; Krasheninina, O.; Ionov, M.; Loznikova, S.; Venyaminova, A.; Miłowska, K.; Shcharbin, D.; Mignani, S.; et al. Multi-target inhibition of cancer cell growth by SiRNA cocktails and 5-fluorouracil using effective piperidine-terminated phosphorus dendrimers. Colloids Interfaces 2017, 1, 6. [Google Scholar] [CrossRef]
- Lazniewska, J.; Milowska, K.; Zablocka, M.; Mignani, S.; Caminade, A.-M.; Majoral, J.-P.; Bryszewska, M.; Gabryelak, T. Mechanism of cationic phosphorus dendrimer toxicity against murine neural cell lines. Mol. Pharm. 2013, 10, 3484–3496. [Google Scholar] [CrossRef]
- Nolan, T.; Huggett, J.; Sanchez, E. Good Practice Guide for the Application of Quantitative PCR (qPCR); LGC: Teddington, UK, 2013. [Google Scholar]
- Roberts, C.A.; Dietzgen, R.G.; Heelan, L.A.; Maclean, D.J. Real-time RT-PCR fluorescent detection of tomato spotted wilt virus. J. Virol. Methods 2000, 88, 1–8. [Google Scholar] [CrossRef]
- Aigner, A. Perspectives, issues and solutions in RNAi therapy: The expected and the less expected. Nanomedicine 2019, 14, 2777–2782. [Google Scholar] [CrossRef]
- Fattal, E.; Fay, F. Nanomedicine-based delivery strategies for nucleic acid gene inhibitors in inflammatory diseases. Adv. Drug Deliv. Rev. 2021, 175, 113809. [Google Scholar] [CrossRef]
- Edy, V.G.; Szekely, M.; Loviny, T.; Dreyer, C. Action of nucleases on double-stranded RNA. Eur. J. Biochem. 1976, 61, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, S.; Naddeo, M.; Russo, A.; D’Alessio, G. Degradation of double-stranded RNA by human pancreatic ribonuclease: Crucial role of noncatalytic basic amino acid residues. Biochemistry 2003, 42, 10182–10190. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip. Rev. RNA 2014, 5, 31–48. [Google Scholar] [CrossRef]
- Ohtani, N. Cleavage of double-stranded RNA by RNase HI from a thermoacidophilic archaeon, Sulfolobus tokodaii 7. Nucleic Acids Res. 2004, 32, 5809–5819. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hickerson, R.P.; Vlassov, A.V.; Wang, Q.; Leake, D.; Ilves, H.; Gonzalez-Gonzalez, E.; Contag, C.H.; Johnston, B.H.; Kaspar, R.L. Stability study of unmodified siRNA and relevance to clinical use. Oligonucleotides 2008, 18, 345–354. [Google Scholar] [CrossRef]
- Wang, Q.; Carmichael, G.G. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 2004, 68, 432–452. [Google Scholar] [CrossRef]
- Elmén, J.; Thonberg, H.; Ljungberg, K.; Frieden, M.; Westergaard, M.; Xu, Y.; Wahren, B.; Liang, Z.; Ørum, H.; Koch, T.; et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005, 33, 439–447. [Google Scholar] [CrossRef] [PubMed]



| Randomized siRNA | Sample Type | Calibration Curve Replicate No | Coefficient of Determination | Slope | PCR Efficiency | LOD [pM] | LOQ [pM] |
|---|---|---|---|---|---|---|---|
| − | Uncomplexed target siRNA | 1 | 0.99 | −3.83 | 82.5% | 4.1 | 70.3 |
| 2 | 0.99 | −3.82 | 82.6% | 4.0 | 66.4 | ||
| 3 | 0.988 | −3.83 | 82.4% | 4.5 | 96.6 | ||
| + | Uncomplexed target siRNA | 1 | 0.997 | −3.31 | 100.4% | 10.0 | 10.0 |
| 2 | 0.998 | −3.57 | 90.6% | 10.0 | 10.0 | ||
| 3 | 0.993 | −3.49 | 93.5% | 10.0 | 10.0 | ||
| − | Dendriplexes | 1 | 0.98 | −4.47 | 67.5% | 31.6 | 50.2 |
| 2 | 0.98 | −4.50 | 66.8% | 31.6 | 51.3 | ||
| 3 | 0.987 | −4.78 | 61.9% | 31.6 | 31.6 | ||
| + | Dendriplexes | 1 | 0.998 | −3.52 | 92.4% | 31.6 | 31.6 |
| 2 | 0.995 | −3.71 | 85.9% | 31.6 | 31.6 | ||
| 3 | 0.993 | −3.15 | 107.8% | 31.6 | 31.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neugebauer, M.; Grundmann, C.E.; Lehnert, M.; von Stetten, F.; Früh, S.M.; Süss, R. Analyzing siRNA Concentration, Complexation and Stability in Cationic Dendriplexes by Stem-Loop Reverse Transcription-qPCR. Pharmaceutics 2022, 14, 1348. https://doi.org/10.3390/pharmaceutics14071348
Neugebauer M, Grundmann CE, Lehnert M, von Stetten F, Früh SM, Süss R. Analyzing siRNA Concentration, Complexation and Stability in Cationic Dendriplexes by Stem-Loop Reverse Transcription-qPCR. Pharmaceutics. 2022; 14(7):1348. https://doi.org/10.3390/pharmaceutics14071348
Chicago/Turabian StyleNeugebauer, Maximilian, Clara E. Grundmann, Michael Lehnert, Felix von Stetten, Susanna M. Früh, and Regine Süss. 2022. "Analyzing siRNA Concentration, Complexation and Stability in Cationic Dendriplexes by Stem-Loop Reverse Transcription-qPCR" Pharmaceutics 14, no. 7: 1348. https://doi.org/10.3390/pharmaceutics14071348
APA StyleNeugebauer, M., Grundmann, C. E., Lehnert, M., von Stetten, F., Früh, S. M., & Süss, R. (2022). Analyzing siRNA Concentration, Complexation and Stability in Cationic Dendriplexes by Stem-Loop Reverse Transcription-qPCR. Pharmaceutics, 14(7), 1348. https://doi.org/10.3390/pharmaceutics14071348






