Combined Transcriptomic and Proteomic Profiling to Unravel Osimertinib, CARP-1 Functional Mimetic (CFM 4.17) Formulation and Telmisartan Combo Treatment in NSCLC Tumor Xenografts
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Formulation of CFM 4.17 Lipid Formulation (CFM-F)
2.3. Cell Culture
2.4. H1975 Xenograft Model of Non-Small Cell Lung Cancer
2.5. RNA Sequencing and Data Analysis
2.6. Proteomics
2.7. RNA Isolation
2.8. Reverse Transcription and RT-qPCR
2.9. TotalSeq Assay for Serum EVs
2.10. Western Blot Analysis
2.11. Statistical Analysis
3. Results
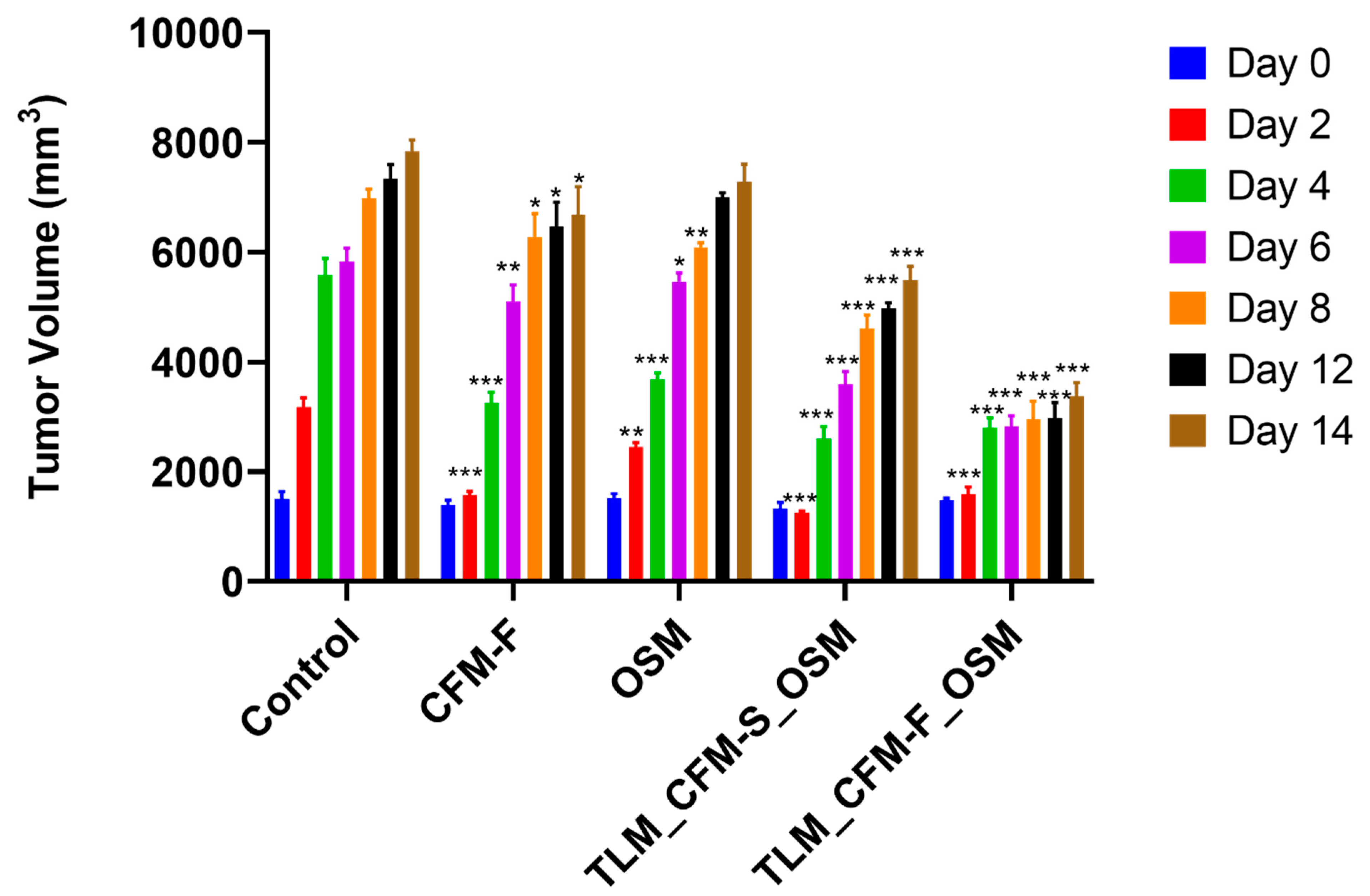
3.1. Effect of TLM_CFM-F_OSM on Tumor Volume in the In Vivo Mouse Model
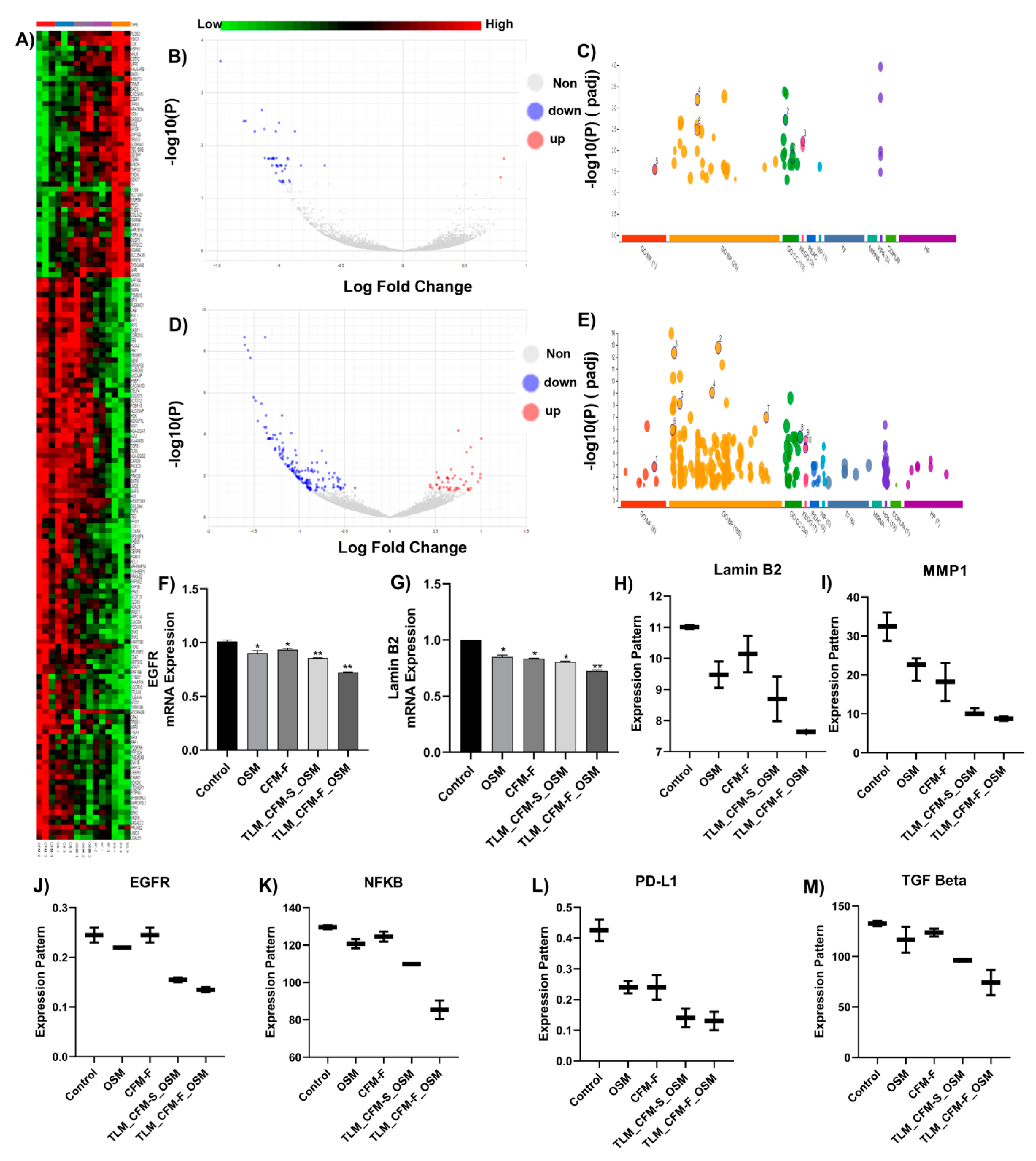
3.2. RNA Sequencing and Differential Gene Expression Analysis in Lung Cancer
3.3. Validation of Differentially Expressed Transcripts via qRT-PCR
3.4. Proteomics and Differential Gene Expression Analysis in Drug-Treated H1975 Tumors
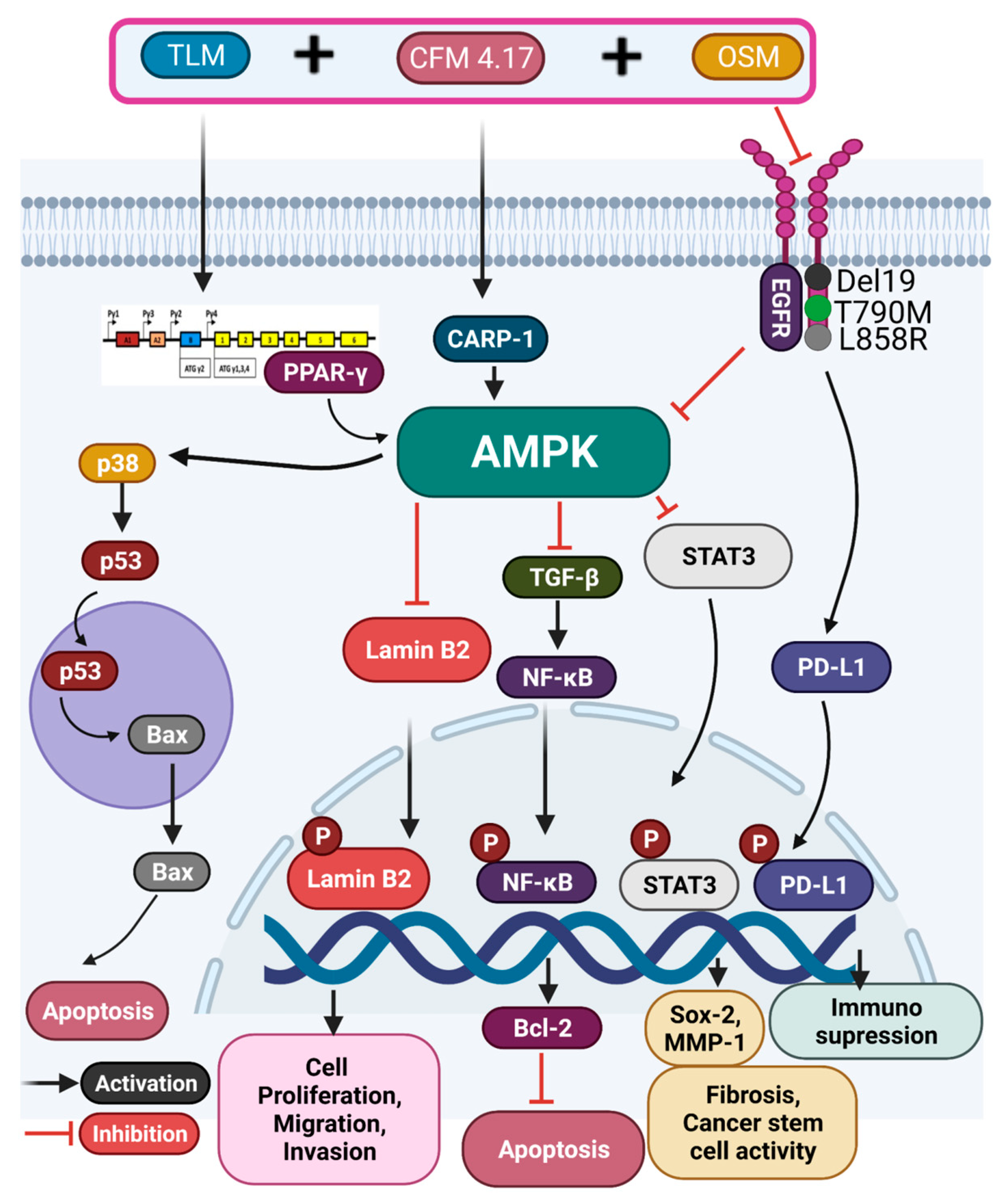
3.5. Combination of TLM_CFM-F_OSM Induces Anti-Cancer Effect via the LAMIN B2/STAT3/NF-κB Signaling Pathways in Lung Cancer
3.6. Combination of TLM_CFM-F_OSM Reduced the Protein Expression of Lung Cancer Stem Cells, Fibrosis, and Migration
3.7. Combination of TLM_CFM-F_OSM Effects on the Protein Expression of Tumor Suppressor Proteins and Apoptotic Proteins
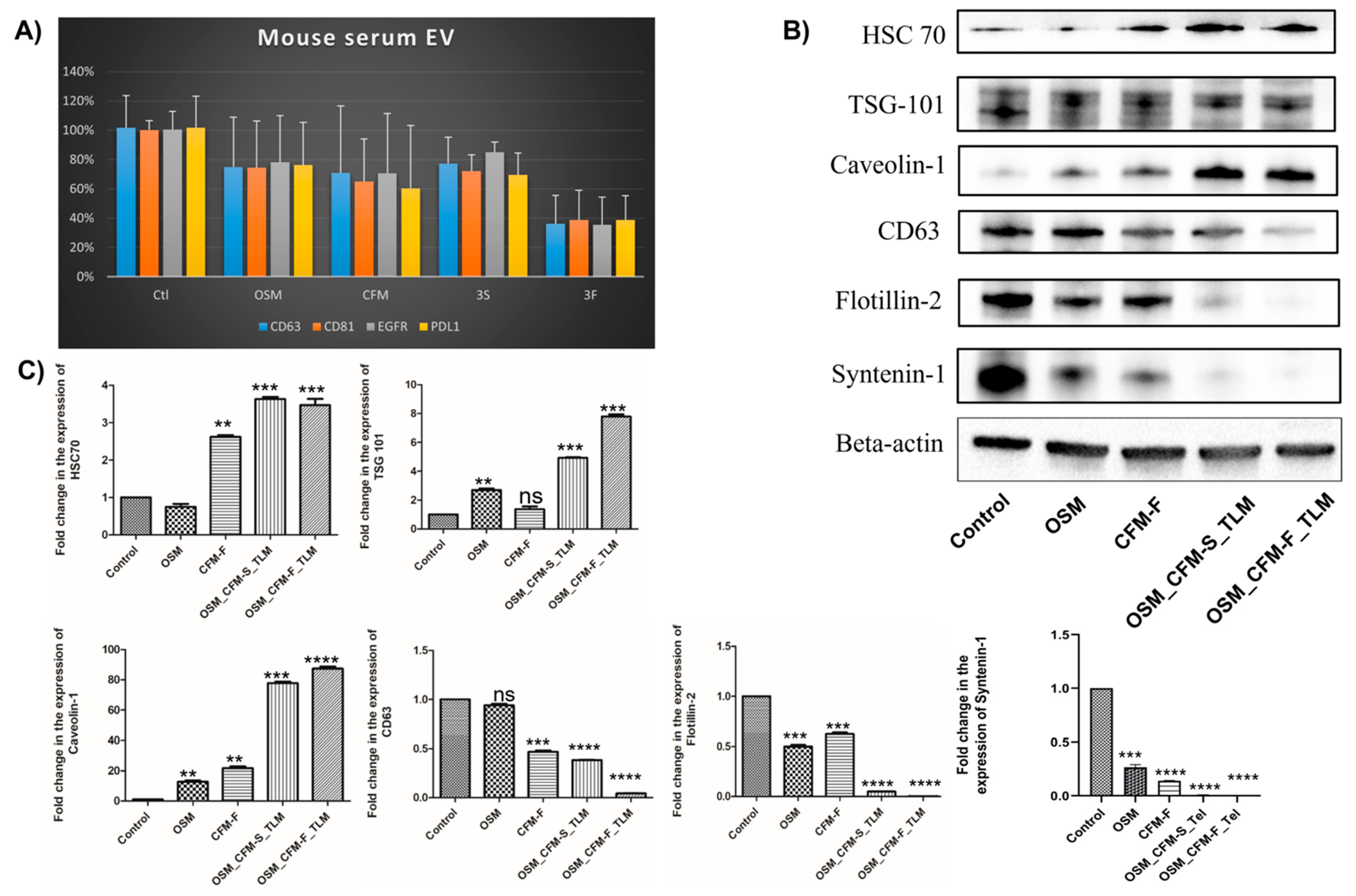
3.8. The Effects of TLM_CFM-F_OSM Combination on Exosomal Markers Expression in H1975 Lung Cancer
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.S.D.; Tanoue, L.T.; Matthay, R.A. Lung cancer: Epidemiology, etiology, and prevention. Clin. Chest Med. 2011, 32, 605–644. [Google Scholar] [CrossRef] [PubMed]
- Mitsudomi, T.; Morita, S.; Yatabe, Y.; Negoro, S.; Okamoto, I.; Tsurutani, J.; Seto, T.; Satouchi, M.; Tada, H.; Hirashima, T.; et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2009, 11, 121–128. [Google Scholar] [CrossRef]
- Rosell, R.; Moran, T.; Queralt, C.; Porta, R.; Cardenal, F.; Camps, C.; Majem, M.; Lopez-Vivanco, G.; Isla, D.; Provencio, M.; et al. Screening for Epidermal Growth Factor Receptor Mutations in Lung Cancer. N. Engl. J. Med. 2009, 361, 958–967. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, S.P.; Pietanza, M.C.; Johnson, M.L.; Riely, G.J.; Miller, V.A.; Sima, C.S.; Zakowski, M.F.; Rusch, V.W.; Ladanyi, M.; Kris, M.G. Incidence of EGFR Exon 19 Deletions and L858R in Tumor Specimens From Men and Cigarette Smokers with Lung Adenocarcinomas. J. Clin. Oncol. 2011, 29, 2066–2070. [Google Scholar] [CrossRef]
- Kommineni, N.; Nottingham, E.; Bagde, A.; Patel, N.; Rishi, A.K.; Dev, S.R.; Singh, M. Role of nano-lipid formulation of CARP-1 mimetic, CFM-4.17 to improve systemic exposure and response in OSM resistant non-small cell lung cancer. Eur. J. Pharm. Biopharm. 2021, 158, 172–184. [Google Scholar] [CrossRef]
- Tan, C.-S.; Gilligan, D.; Pacey, S.C. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015, 16, e447–e459. [Google Scholar] [CrossRef]
- Thress, K.S.; Paweletz, C.P.; Felip, E.; Cho, B.C.; Stetson, D.; Dougherty, B.; Lai, Z.; Markovets, A.; Vivancos, A.; Kuang, Y.; et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non–small cell lung cancer harboring EGFR T790M. Nat. Med. 2015, 21, 560–562. [Google Scholar] [CrossRef]
- Cross, D.; D’Cruz, C.; Eberlein, C.; Spitzler, P.; Ichihara, E.; Meador, C.; Ashton, S.; Mellor, M.; Stewart, R.; Smith, P.; et al. Targeting Resistance in Egfr-Mutant Non-Small Cell Lung Cancer (Nsclc): Preclinical Evidence Supporting the Combination of Egfr Tyrosine Kinase Inhibitors (Tkis) Azd9291 and Gefitinib with Molecularly Targeted Agents and Immunotherapeutics. Ann. Oncol. 2014, 25, iv155. [Google Scholar] [CrossRef]
- Patel, A.R.; Chougule, M.; Singh, M. EphA2 targeting pegylated nanocarrier drug delivery system for treatment of lung cancer. Pharm. Res. 2014, 31, 2796–2809. [Google Scholar] [CrossRef]
- Ichite, N.; Chougule, M.; Patel, A.R.; Jackson, T.; Safe, S.; Singh, M. Inhalation delivery of a novel diindolylmethane derivative for the treatment of lung cancer. Mol. Cancer Ther. 2010, 9, 3003–3014. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chougule, M.B.; Patel, A.; Sachdeva, P.; Jackson, T.; Singh, M. Enhanced anticancer activity of gemcitabine in combination with noscapine via antiangiogenic and apoptotic pathway against non-small cell lung cancer. PLoS ONE 2011, 6, e27394. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Ferdous, A.J.; Jackson, T.L. Stealth monensin liposomes as a potentiator of adriamycin in cancer treatment. J. Control. Release 1999, 59, 43–53. [Google Scholar] [CrossRef]
- Fulzele, S.V.; Shaik, M.S.; Chatterjee, A.; Singh, M. Anti-cancer effect of celecoxib and aerosolized docetaxel against human non-small cell lung cancer cell line, A549. J. Pharm. Pharmacol. 2006, 58, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.R.; Chougule, M.B.; Townley, I.; Patlolla, R.; Wang, G.; Singh, M. Efficacy of aerosolized celecoxib encapsulated nanostructured lipid carrier in non-small cell lung cancer in combination with docetaxel. Pharm. Res. 2013, 30, 1435–1446. [Google Scholar] [CrossRef] [PubMed]
- Haynes, A.; Shaik, M.S.; Chatterjee, A.; Singh, M. Evaluation of an aerosolized selective COX-2 inhibitor as a potentiator of doxorubicin in a non-small-cell lung cancer cell line. Pharm. Res. 2003, 20, 1485–1495. [Google Scholar] [CrossRef]
- Verma, V.; Simone, C.B.; Werner-Wasik, M. Acute and late toxicities of concurrent chemoradiotherapy for locally-advanced non-small cell lung cancer. Cancers 2017, 9, 120. [Google Scholar] [CrossRef]
- Sun, G.; Rong, D.; Li, Z.; Sun, G.; Wu, F.; Li, X.; Cao, H.; Cheng, Y.; Tang, W.; Sun, Y. Role of Small Molecule Targeted Compounds in Cancer: Progress, Opportunities, and Challenges. Front. Cell Dev. Biol. 2021, 9, 2043. [Google Scholar] [CrossRef]
- Zhang, H. OSM making a breakthrough in lung cancer targeted therapy. OncoTargets Ther. 2016, 9, 5489. [Google Scholar] [CrossRef]
- Puliyappadamba, V.T.; Wu, W.; Bevis, D.; Zhang, L.; Polin, L.; Kilkuskie, R.; Finley, R., Jr.; Larsen, S.D.; Levi, E.; Miller, F.R.; et al. Antagonists of Anaphase-promoting Complex (APC)-2-Cell Cycle and Apoptosis Regulatory Protein (CARP)-1 Interaction Are Novel Regulators of Cell Growth and Apoptosis. J. Biol. Chem. 2011, 286, 38000–38017. [Google Scholar] [CrossRef]
- Lehman, N.L.; Tibshirani, R.; Hsu, J.Y.; Natkunam, Y.; Harris, B.T.; West, R.B.; Masek, A.; Montgomery, K.; van de Rijn, M.; Jackson, P.K. Oncogenic Regulators and Substrates of the Anaphase Promoting Complex/Cyclosome Are Frequently Overexpressed in Malignant Tumors. Am. J. Pathol. 2007, 170, 1793–1805. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muthu, M.; Cheriyan, V.T.; Rishi, A.K. CARP-1/CCAR1: A biphasic regulator of cancer cell growth and apoptosis. Oncotarget 2015, 6, 6499. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.-W.; Deng, C.-L.; Li, D.-D.; Liu, D.-D.; Chai, S.-M.; Wang, W.; Zhang, Y.; Chen, K.; Li, X.; Wang, J.; et al. Design, synthesis and biological evaluation of novel 4-aminoquinazolines as dual target inhibitors of EGFR-PI3Kα. Eur. J. Med. Chem. 2018, 146, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Swick, A.D.; Prabakaran, P.J.; Miller, M.C.; Javaid, A.M.; Fisher, M.M.; Sampene, E.; Ong, I.M.; Hu, R.; Iida, M.; Nickel, K.P.; et al. Cotargeting mTORC and EGFR Signaling as a Therapeutic Strategy in HNSCC. Mol. Cancer Ther. 2017, 16, 1257–1268. [Google Scholar] [CrossRef]
- Marcucci, F.; Corti, A. How to improve exposure of tumor cells to drugs—Promoter drugs increase tumor uptake and penetration of effector drugs. Adv. Drug Deliv. Rev. 2012, 64, 53–68. [Google Scholar] [CrossRef]
- Chauhan, V.P.; Martin, J.D.; Liu, H.; Lacorre, D.A.; Jain, S.R.; Kozin, S.V.; Stylianopoulos, T.; Mousa, A.S.; Han, X.; Adstamongkonkul, P.; et al. Angiotensin inhibition enhances drug delivery and potentiates chemotherapy by decompressing tumour blood vessels. Nat. Commun. 2013, 4, 2516. [Google Scholar] [CrossRef] [PubMed]
- Curnis, F.; Sacchi, A.; Corti, A. Improving chemotherapeutic drug penetration in tumors by vascular targeting and barrier alteration. J. Clin. Investig. 2002, 110, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Godugu, C.; Patel, A.R.; Doddapaneni, R.; Marepally, S.; Jackson, T.; Singh, M. Inhalation delivery of Telmisartan enhances intratumoral distribution of nanoparticles in lung cancer models. J. Control. Release 2013, 172, 86–95. [Google Scholar] [CrossRef]
- Patel, K.; Doddapaneni, R.; Chowdhury, N.; Boakye, C.H.; Behl, G.; Singh, M. Tumor stromal disrupting agent enhances the anticancer efficacy of docetaxel loaded PEGylated liposomes in lung cancer. Nanomedicine 2016, 11, 1377–1392. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, G.; Liu, S.; Su, H.; Wang, Y.; Li, J.; Luo, C. Remodeling the Tumor Microenvironment with Emerging Nanotherapeutics. Trends Pharmacol. Sci. 2018, 39, 59–74. [Google Scholar] [CrossRef]
- Green, R.; Howell, M.; Khalil, R.; Nair, R.; Yan, J.; Foran, E.; Katiri, S.; Banerjee, J.; Singh, M.; Bharadwaj, S.; et al. Actinomycin D and Telmisartan Combination Targets Lung Cancer Stem Cells Through the Wnt/Beta Catenin Pathway. Sci. Rep. 2019, 9, 18177. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Doddapaneni, R.; Sekar, V.; Chowdhury, N.; Singh, M. Combination approach of YSA peptide anchored docetaxel stealth liposomes with oral antifibrotic agent for the treatment of lung cancer. Mol. Pharm. 2016, 13, 2049–2058. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, L.; Yu, P.; Liu, B.; Zhu, J.; Yang, Y. Telmisartan exerts anti-tumor effects by activating peroxisome proliferator-activated receptor-γ in human lung adenocarcinoma A549 cells. Molecules 2014, 19, 2862–2876. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Y. Telmisartan inhibits NSCLC A549 cell proliferation and migration by regulating the PI3K/AKT signaling pathway. Oncol. Lett. 2018, 15, 5859–5864. [Google Scholar] [CrossRef]
- Surapaneni, S.K.; Nottingham, E.; Mondal, A.; Patel, N.; Arthur, P.; Gebeyehu, A.; Kalvala, A.K.; Rishi, A.K.; Singh, M. Telmisartan Facilitates the Anticancer Effects of CARP-1 Functional Mimetic and Sorafenib in Rociletinib Resistant Non-small Cell Lung Cancer. Anticancer Res. 2021, 41, 4215–4228. [Google Scholar] [CrossRef]
- Collins, I.; Workman, P. New approaches to molecular cancer therapeutics. Nat. Chem. Biol. 2006, 2, 689–700. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to OSM in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]
- Cho, W.C. Proteomics technologies and challenges. Genom. Proteom. Bioinform. 2007, 5, 77–85. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Orsburn, B.C. Proteome Discoverer—A Community Enhanced Data Processing Suite for Protein Informatics. Proteomes 2021, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Simpson, H. The MASCOT method. Softw. Eng. J. 1986, 1, 103–120. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Meckes, D.G. Multiplex protein profiling method for extracellular vesicle protein detection. Sci. Rep. 2021, 11, 12477. [Google Scholar] [CrossRef]
- Di Liegro, C.M.; Schiera, G.; Di Liegro, I. H1. 0 linker histone as an epigenetic regulator of cell proliferation and differentiation. Genes 2018, 9, 310. [Google Scholar] [CrossRef]
- Zhang, M.-Y.; Han, Y.-C.; Han, Q.; Liang, Y.; Luo, Y.; Wei, L.; Yan, T.; Yang, Y.; Liu, S.-L.; Wang, E.-H. Lamin B2 promotes the malignant phenotype of non-small cell lung cancer cells by upregulating dimethylation of histone 3 lysine 9. Exp. Cell Res. 2020, 393, 112090. [Google Scholar] [CrossRef]
- Kielbassa, K.; Vegna, S.; Ramirez, C.; Akkari, L. Understanding the origin and diversity of macrophages to tailor their targeting in solid cancers. Front. Immunol. 2019, 10, 2215. [Google Scholar] [CrossRef]
- Talarico, M.; Nunes, R.; Silva, G.; Costa, L.; Cardoso, M.; Esteves, S.; Sarian, L.Z.; Zeferino, L.; Termini, L. High Expression of SOD2 Protein Is a Strong Prognostic Factor for Stage IIIB Squamous Cell Cervical Carcinoma. Antioxidants 2021, 10, 724. [Google Scholar] [CrossRef]
- Xia, Y.; Shen, S.; Verma, I.M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef]
- Syed, V. TGF-β Signaling in Cancer. J. Cell. Biochem. 2016, 117, 1279–1287. [Google Scholar] [CrossRef]
- Zou, S.; Tong, Q.; Liu, B.; Huang, W.; Tian, Y.; Fu, X. Targeting STAT3 in cancer immunotherapy. Mol. Cancer 2020, 19, 145. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, N.; Liu, P.; Xie, X. AMPK and Cancer. AMP-Act. Protein Kinase 2016, 203–226. [Google Scholar] [CrossRef]
- Collisson, E.; Campbell, J.; Brooks, A.; Berger, A.; Lee, W.; Chmielecki, J.; Beer, D.; Cope, L.; Creighton, C.; Ding, L. Comprehensive molecular profiling of lung adenocarcinoma: The cancer genome atlas research network. Nature 2014, 511, 543–550. [Google Scholar]
- Rosenzweig, S.A. Acquired resistance to drugs targeting tyrosine kinases. Adv. Cancer Res. 2018, 138, 71–98. [Google Scholar] [PubMed]
- Wang, S.; Song, Y.; Yan, F.; Liu, D. Mechanisms of resistance to third-generation EGFR tyrosine kinase inhibitors. Front. Med. 2016, 10, 383–388. [Google Scholar] [CrossRef]
- Botting, G.M.; Rastogi, I.; Chhabra, G.; Nlend, M.; Puri, N. Mechanism of resistance and novel targets mediating resistance to EGFR and c-Met tyrosine kinase inhibitors in non-small cell lung cancer. PLoS ONE 2015, 10, e0136155. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.O.; Sjin, R.T.T.; Haringsma, H.J.; Ohashi, K.; Sun, J.; Lee, K.; Dubrovskiy, A.; Labenski, M.; Zhu, Z.; Wang, Z.; et al. Discovery of a Mutant-Selective Covalent Inhibitor of EGFR that Overcomes T790M-Mediated Resistance in NSCLC. Cancer Discov. 2013, 3, 1404–1415. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.-C.; Hsiao, A.Y.; Allen, S.G.; Torisawa Y-s Ho, M.; Takayama, S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 2011, 136, 473–478. [Google Scholar] [CrossRef]
- Cheriyan, V.T.; Alsaab, H.; Sekhar, S.; Venkatesh, J.; Mondal, A.; Vhora, I.; Sau, S.; Muthu, M.; Polin, L.A.; Levi, E.; et al. A CARP-1 functional mimetic compound is synergistic with BRAF-targeting in non-small cell lung cancers. Oncotarget 2018, 9, 29680–29697. [Google Scholar] [CrossRef]
- Sanders, M.J.; Grondin, P.O.; Hegarty, B.D.; Snowden, M.A.; Carling, D. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem. J. 2007, 403, 139–148. [Google Scholar] [CrossRef]
- Suter, M.; Riek, U.; Tuerk, R.; Schlattner, U.; Wallimann, T.; Neumann, D. Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. J. Biol. Chem. 2006, 281, 32207–32216. [Google Scholar] [CrossRef] [PubMed]
- Woods, A.; Johnstone, S.R.; Dickerson, K.; Leiper, F.C.; Fryer, L.G.; Neumann, D.; Schlattner, U.; Wallimann, T.; Carlson, M.; Carling, D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 2003, 13, 2004–2008. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.J.; Kosmatka, M.; Bardeesy, N.; Hurley, R.L.; Witters, L.A.; DePinho, R.A.; Cantley, L.C. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA 2004, 101, 3329–3335. [Google Scholar] [CrossRef] [PubMed]
- Hawley, S.A.; Boudeau, J.; Reid, J.L.; Mustard, K.J.; Udd, L.; Mäkelä, T.P.; Alessi, D.R.; Hardie, D.G. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2003, 2, 28. [Google Scholar] [CrossRef]
- Motoshima, H.; Goldstein, B.J.; Igata, M.; Araki, E. AMPK and cell proliferation–AMPK as a therapeutic target for atherosclerosis and cancer. J. Physiol. 2006, 574, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Jiang, J.; Xie, N.; Zhou, L.; Huang, Z.; Zhang, L.; Qin, S.; Fu, S.; Peng, L.; Gao, W.; et al. MCT1 relieves OSM-induced CRC suppression by promoting autophagy through the LKB1/AMPK signaling. Cell Death Dis. 2019, 10, 615. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.-J.; Park, J.-H.; Cho, D.-H. Activation of AMPK by Telmisartan Decreases Basal and PDGF-stimulated VSMC Proliferation via Inhibiting the mTOR/p70S6K Signaling Axis. J. Korean Med. Sci. 2020, 35, e289. [Google Scholar] [CrossRef]
- Oura, K.; Tadokoro, T.; Fujihara, S.; Morishita, A.; Chiyo, T.; Samukawa, E.; Yamana, Y.; Fujita, K.; Sakamoto, T.; Nomura, T.; et al. Telmisartan inhibits hepatocellular carcinoma cell proliferation in vitro by inducing cell cycle arrest. Oncol. Rep. 2017, 38, 2825–2835. [Google Scholar] [CrossRef]
- Jamal, S.; Cheriyan, V.T.; Muthu, M.; Munie, S.; Levi, E.; Ashour, A.; Pass, H.I.; Wali, A.; Singh, M.; Rishi, A.K. CARP-1 Functional Mimetics Are a Novel Class of Small Molecule Inhibitors of Malignant Pleural Mesothelioma Cells. PLoS ONE 2014, 9, e89146. [Google Scholar] [CrossRef]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef]
- Nader, N.; Ng, S.S.M.; Lambrou, G.I.; Pervanidou, P.; Wang, Y.; Chrousos, G.P.; Kino, T. AMPK regulates metabolic actions of glucocorticoids by phosphorylating the glucocorticoid receptor through p38 MAPK. Mol. Endocrinol. 2010, 24, 1748–1764. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-W.; Wong, L.L.-Y.; Tse, E.Y.-T.; Liu, H.-F.; Leong, V.Y.-L.; Lee, J.M.-F.; Hardie, D.G.; Ng, I.O.-L.; Ching, Y.-P. AMPK Promotes p53 Acetylation via Phosphorylation and Inactivation of SIRT1 in Liver Cancer Cells. Cancer Res. 2012, 72, 4394–4404. [Google Scholar] [CrossRef] [PubMed]
- Porras, A.; Zuluaga, S.; Black, E.; Valladares, A.; Álvarez, A.M.; Ambrosino, C.; Benito, M.; Nebreda, A.R. p38α Mitogen-activated Protein Kinase Sensitizes Cells to Apoptosis Induced by Different Stimuli. Mol. Biol. Cell 2004, 15, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.A.; Zheng, Q.; Liu, Z.; Thompson, J.E. Role of p38 and JNK MAPK signaling pathways and tumor suppressor p53 on induction of apoptosis in response to Ad-eIF5A1 in A549 lung cancer cells. Mol. Cancer 2013, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Naetar, N.; Ferraioli, S.; Foisner, R. Lamins in the nuclear interior− life outside the lamina. J. Cell Sci. 2017, 130, 2087–2096. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Fei, L.; Zhang, M.; Zhang, W.; Liu, X.; Wang, C.; Luo, Y.; Zhang, H.; Han, Y. Lamin B2 binding to minichromosome maintenance complex component 7 promotes non-small cell lung carcinogenesis. Oncotarget 2017, 8, 104813–104830. [Google Scholar] [CrossRef] [PubMed]
- Chandran, S.; Suggs, J.A.; Wang, B.J.; Han, A.; Bhide, S.; Cryderman, D.E.; Moore, S.A.; Bernstein, S.I.; Wallrath, L.L.; Melkani, G.C. Suppression of myopathic lamin mutations by muscle-specific activation of AMPK and modulation of downstream signaling. Hum. Mol. Genet. 2019, 28, 351–371. [Google Scholar] [CrossRef]
- Baud, V.; Karin, M. Is NF-κB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 2009, 8, 33–40. [Google Scholar] [CrossRef]
- Basseres, D.; Baldwin, A. Nuclear factor-κ B and inhibitor of κ B kinase pathways in oncogenic initiation and progression. Oncogene 2006, 25, 6817–6830. [Google Scholar] [CrossRef]
- Jiang, X.-M.; Xu, Y.-L.; Yuan, L.-W.; Huang, M.-Y.; Ye, Z.-H.; Su, M.-X.; Chen, X.P.; Zhu, H.; Ye, R.D.; Lu, J.J. TGFβ2-mediated epithelial–mesenchymal transition and NF-κB pathway activation contribute to OSM resistance. Acta Pharmacol. Sin. 2021, 42, 451–459. [Google Scholar] [CrossRef]
- Abdel-Fattah, M.M.; Salama, A.A.; Shehata, B.A.; Ismaiel, I.E. The potential effect of the angiotensin II receptor blocker telmisartan in regulating OVA-induced airway remodeling in experimental rats. Pharmacol. Rep. 2015, 67, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Shi, F.; Zhai, R.; Wang, H.; Li, K.; Xu, C.; Yao, W.; Zhou, F. TGF-β promote epithelial-mesenchymal transition via NF-κB/NOX4/ROS signal pathway in lung cancer cells. Mol. Biol. Rep. 2021, 48, 2365–2375. [Google Scholar] [CrossRef] [PubMed]
- Merchant, N.; Nagaraju, G.P.; Rajitha, B.; Lammata, S.; Jella, K.K.; Buchwald, Z.S.; Lakka, S.S.; Ali, A.N. Matrix metalloproteinases: Their functional role in lung cancer. Carcinogenesis 2017, 38, 766–780. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Tanigaki, T.; Heimer, D.; Wang, W.; Ross, W.G.; Murphy, G.A.; Sakai, A.; Sussman, H.H.; Vu, T.H.; Raffin, T.A. TGF-beta 1 causes increased endothelial ICAM-1 expression and lung injury. J. Appl. Physiol. 1994, 77, 1281–1287. [Google Scholar] [CrossRef]
- Harada, D.; Takigawa, N.; Kiura, K. The role of STAT3 in non-small cell lung cancer. Cancers 2014, 6, 708–722. [Google Scholar] [CrossRef]
- Fan, Y.; Mao, R.; Yang, J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef]
- Silva, M.; AMelo, S. Non-coding RNAs in exosomes: New players in cancer biology. Curr. Genom. 2015, 16, 295–303. [Google Scholar] [CrossRef]
- Shimada, Y.; Matsubayashi, J.; Kudo, Y.; Maehara, S.; Takeuchi, S.; Hagiwara, M.; Kakihana, M.; Ohira, T.; Nagao, T.; Ikeda, N. Serum-derived exosomal PD-L1 expression to predict anti-PD-1 response and in patients with non-small cell lung cancer. Sci. Rep. 2021, 11, 7830. [Google Scholar] [CrossRef]
- Ye, L.; Zhu, Z.; Chen, X.; Zhang, H.; Huang, J.; Gu, S.; Zhao, X. The Importance of Exosomal PD-L1 in Cancer Progression and Its Potential as a Therapeutic Target. Cells 2021, 10, 3247. [Google Scholar] [CrossRef]
- Wu, S.; Luo, M.; To, K.K.; Zhang, J.; Su, C.; Zhang, H.; An, S.; Wang, F.; Chen, D.; Fu, L. Intercellular transfer of exosomal wild type EGFR triggers OSM resistance in non-small cell lung cancer. Mol. Cancer 2021, 20, 17. [Google Scholar] [CrossRef]


| Gene | Primer |
|---|---|
| Lamin B2 | F: CGGAGAGTCCTGGATGAGAC |
| R: TCTTCTTGGCGCTCTTGTTG | |
| EGFR | F: AACACCCTGGTCTGGAAGTACG |
| R: TCGTTGGACAGCCTTCAAGACC | |
| GAPDH | GTCTCCTCTGACTTCAACAGCG |
| ACCACCCTGTTGCTGTAGCCAA |
| S.No | Antibody Name | Company | Catalog No. |
|---|---|---|---|
| 1 | Lamin B2 | Cell Signaling Technology | 12255 |
| 2 | SOD | Cell Signaling Technology | 13141 |
| 3 | SOX2 | Cell Signaling Technology | 3579 |
| 4 | EGFR | Cell Signaling Technology | 54359 |
| 5 | AMPK | Cell Signaling Technology | 2532 |
| 6 | TGF-beta | Cell Signaling Technology | 3709 |
| 7 | Bcl2 | Cell Signaling Technology | 4223 |
| 8 | Bax | Cell Signaling Technology | 5023 |
| 9 | p38 | Cell Signaling Technology | 8690 |
| 10 | P53 | Cell Signaling Technology | 2527 |
| 11 | HSC 70 | Santa Cruz Biotechnology | sc-7298 |
| 12 | TSG 101 | Cell Signaling Technology | 28405 |
| 13 | CD 63 | Cell Signaling Technology | 28405 |
| 14 | Calnexin | Cell Signaling Technology | 2679 |
| 15 | Flotillin-2 | Cell Signaling Technology | 3436 |
| 16 | Caveolin-1 | Cell Signaling Technology | 3267 |
| Protein Name | Role in Cancer | Expression in Treatment Group |
|---|---|---|
| Histone H1.0 | Histone H1.0 overexpression in all cancer cells promotes differentiation during tumor development [45]. | Downregulated |
| Lamin-B2 | By upregulating demethylation of histone 3 lysine 9, Lamin B2 increases the malignant phenotype of non-small cell lung cancer cells [46]. | Downregulated |
| Macrophage mannose receptor 1 | Tumor-associated macrophages (TAMs) that express the multi-ligand endocytic receptor mannose receptor (CD206/MRC1) have a role in angiogenesis, metastasi, tumor immunosuppression, and recurrence [47]. | Downregulated |
| SOD2 | High superoxide dismutase 2 (SOD2) expression is associated with a poor prognosis at many cancer sites, the presence of metastases, and more advanced cancer [48]. | Downregulated |
| NFKB | The oncogenesis process is influenced by the pleiotropic transcription factor NFKB, which upregulates genes involved in cell proliferation, metastasis, apoptosis suppression and angiogenesis [49] | Downregulated |
| TGF beta | TGF- is the most potent inducer of epithelial-mesenchymal transition in non-small cell lung cancer cells, and it is essential for the establishment of a tumor-promoting microenvironment in lung cancer tissue [50]. | Downregulated |
| STAT3 | Many malignancies have constitutively active STAT3, which plays a key role in tumor development and metastasis [51]. | Downregulated |
| AMPK | In cancer, AMPK plays a tumor suppressor role. Activation of AMPK reduces tumor growth by targeting several tumorigenesis-related signaling pathways at various phases of tumor formation [52]. | Upregulated |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nimma, R.; Kalvala, A.K.; Patel, N.; Surapaneni, S.K.; Sun, L.; Singh, R.; Nottingham, E.; Bagde, A.; Kommineni, N.; Arthur, P.; et al. Combined Transcriptomic and Proteomic Profiling to Unravel Osimertinib, CARP-1 Functional Mimetic (CFM 4.17) Formulation and Telmisartan Combo Treatment in NSCLC Tumor Xenografts. Pharmaceutics 2022, 14, 1156. https://doi.org/10.3390/pharmaceutics14061156
Nimma R, Kalvala AK, Patel N, Surapaneni SK, Sun L, Singh R, Nottingham E, Bagde A, Kommineni N, Arthur P, et al. Combined Transcriptomic and Proteomic Profiling to Unravel Osimertinib, CARP-1 Functional Mimetic (CFM 4.17) Formulation and Telmisartan Combo Treatment in NSCLC Tumor Xenografts. Pharmaceutics. 2022; 14(6):1156. https://doi.org/10.3390/pharmaceutics14061156
Chicago/Turabian StyleNimma, Ramesh, Anil Kumar Kalvala, Nilkumar Patel, Sunil Kumar Surapaneni, Li Sun, Rakesh Singh, Ebony Nottingham, Arvind Bagde, Nagavendra Kommineni, Peggy Arthur, and et al. 2022. "Combined Transcriptomic and Proteomic Profiling to Unravel Osimertinib, CARP-1 Functional Mimetic (CFM 4.17) Formulation and Telmisartan Combo Treatment in NSCLC Tumor Xenografts" Pharmaceutics 14, no. 6: 1156. https://doi.org/10.3390/pharmaceutics14061156
APA StyleNimma, R., Kalvala, A. K., Patel, N., Surapaneni, S. K., Sun, L., Singh, R., Nottingham, E., Bagde, A., Kommineni, N., Arthur, P., Nathani, A., Meckes, D. G., Jr., & Singh, M. (2022). Combined Transcriptomic and Proteomic Profiling to Unravel Osimertinib, CARP-1 Functional Mimetic (CFM 4.17) Formulation and Telmisartan Combo Treatment in NSCLC Tumor Xenografts. Pharmaceutics, 14(6), 1156. https://doi.org/10.3390/pharmaceutics14061156







