AAV-CRISPR/Cas9 Gene Editing Preserves Long-Term Vision in the P23H Rat Model of Autosomal Dominant Retinitis Pigmentosa
Abstract
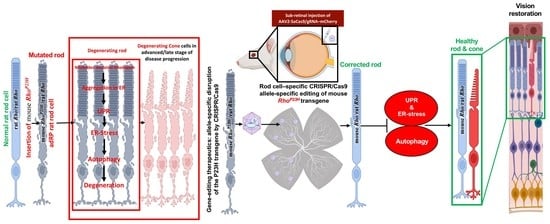
:1. Introduction
2. Materials and Methods
2.1. AAV-SaCas9/gRNA Vector Design
2.2. Animals and Injection Procedures
2.3. Visual Function Assessment
2.4. Retinal Cell Isolation, FACS-Based Sorting and Targeted Deep Sequencing
2.5. RNA Extraction and qPCR Analyses
2.6. Histologic Assessment, Immunofluorescent Staining, and Confocal Microscopy
2.7. Image Analysis
2.8. Statistical Significance
3. Results
3.1. Guide RNA Vector Design Strategy
3.2. SaCas9/gRNA Distribution in Retinas of P23H Rats following Subretinal Injection
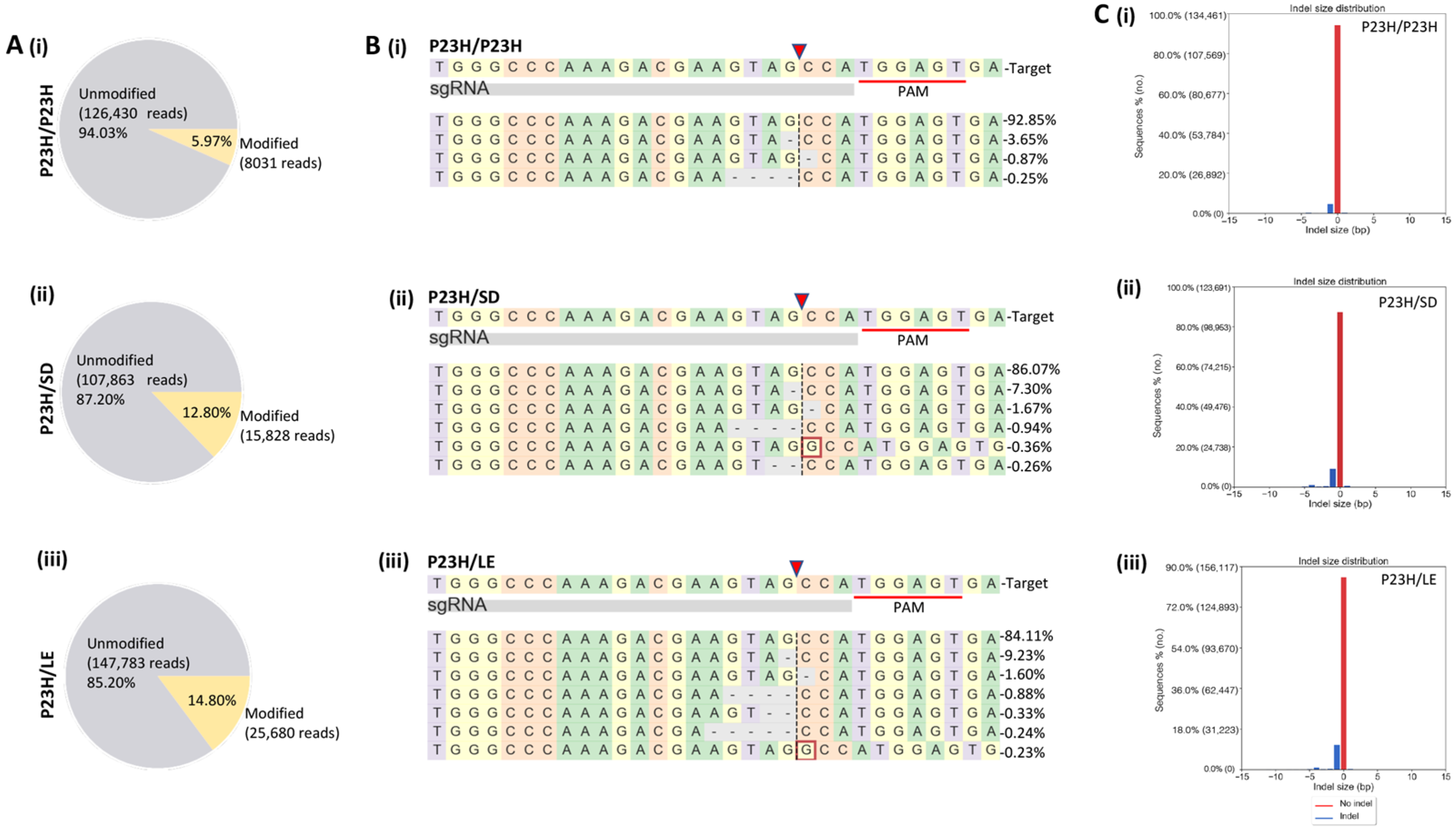
3.3. On- and Off-Target Genome Editing Analysis following In Vivo m-RhoP23H Disruption
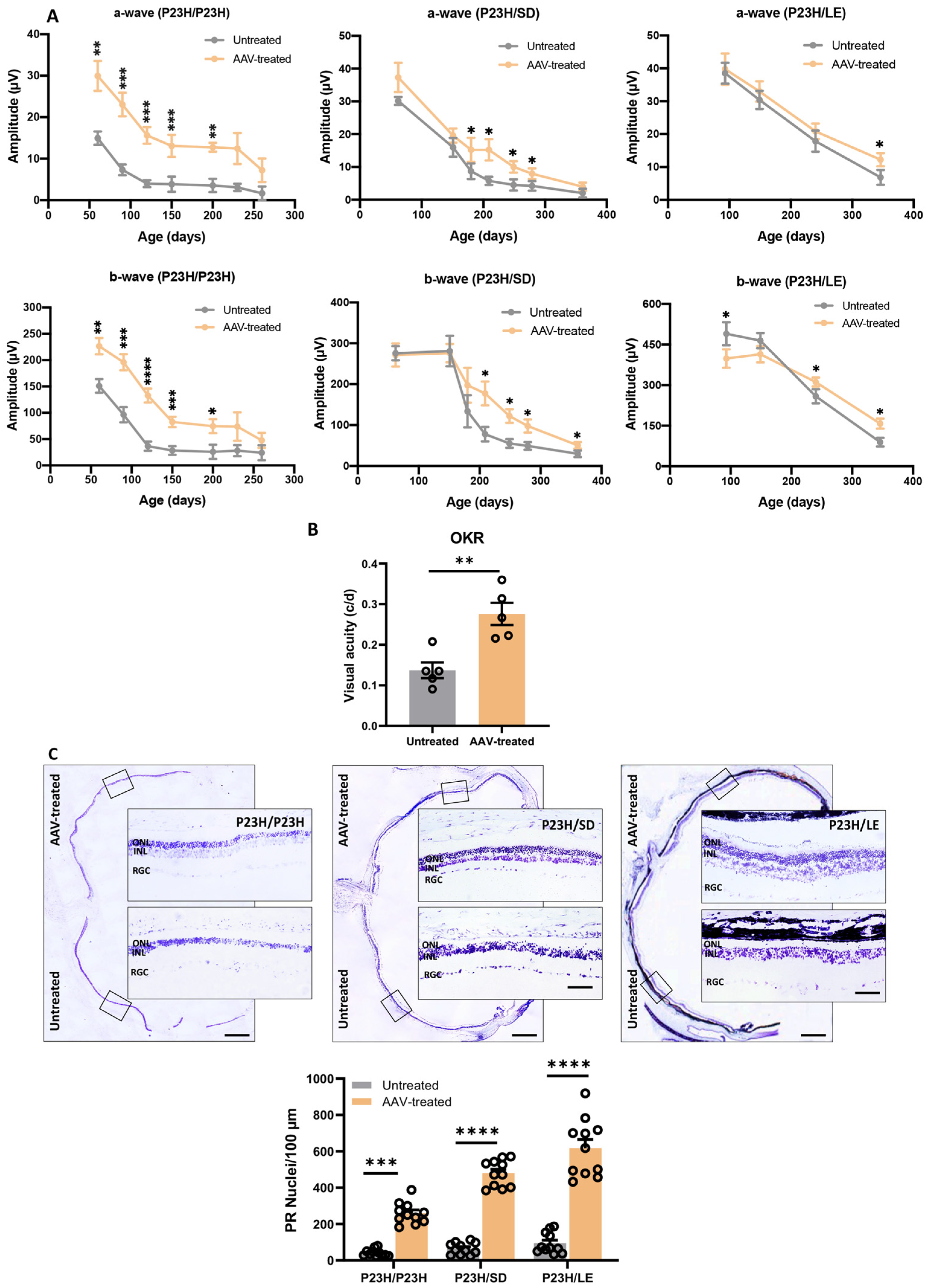
3.4. Long-Term Vision Rescue following m-RhoP23H Ablation
3.5. Histologic Evaluation of Retina following m-RhoP23H Ablation
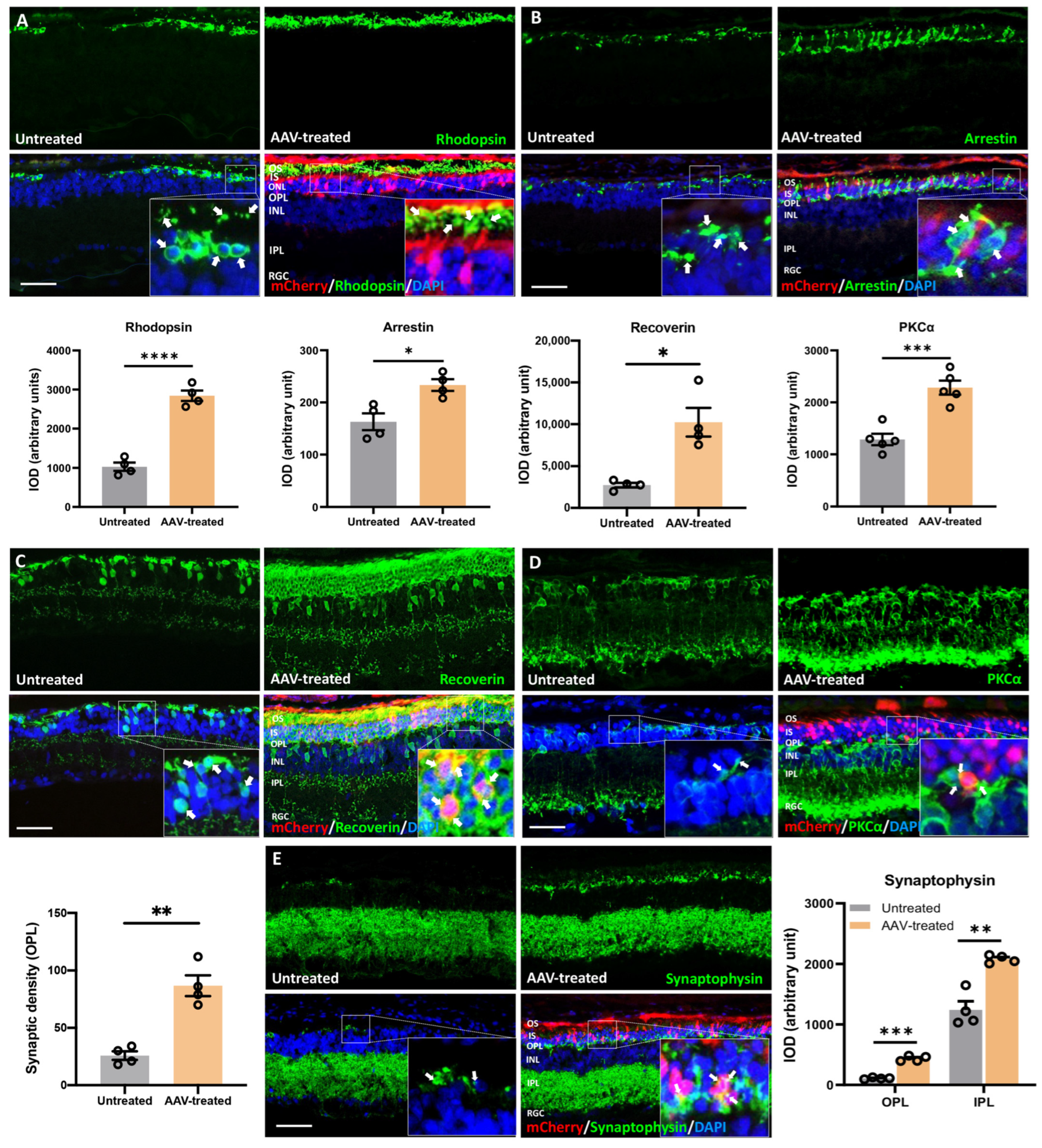
3.6. Long-Term Preservation of Photoreceptors, Rod/Cone-Bipolar Dendritic Arborization, and Retinal Synaptic Connections following m-RhoP23H-Specific Ablation
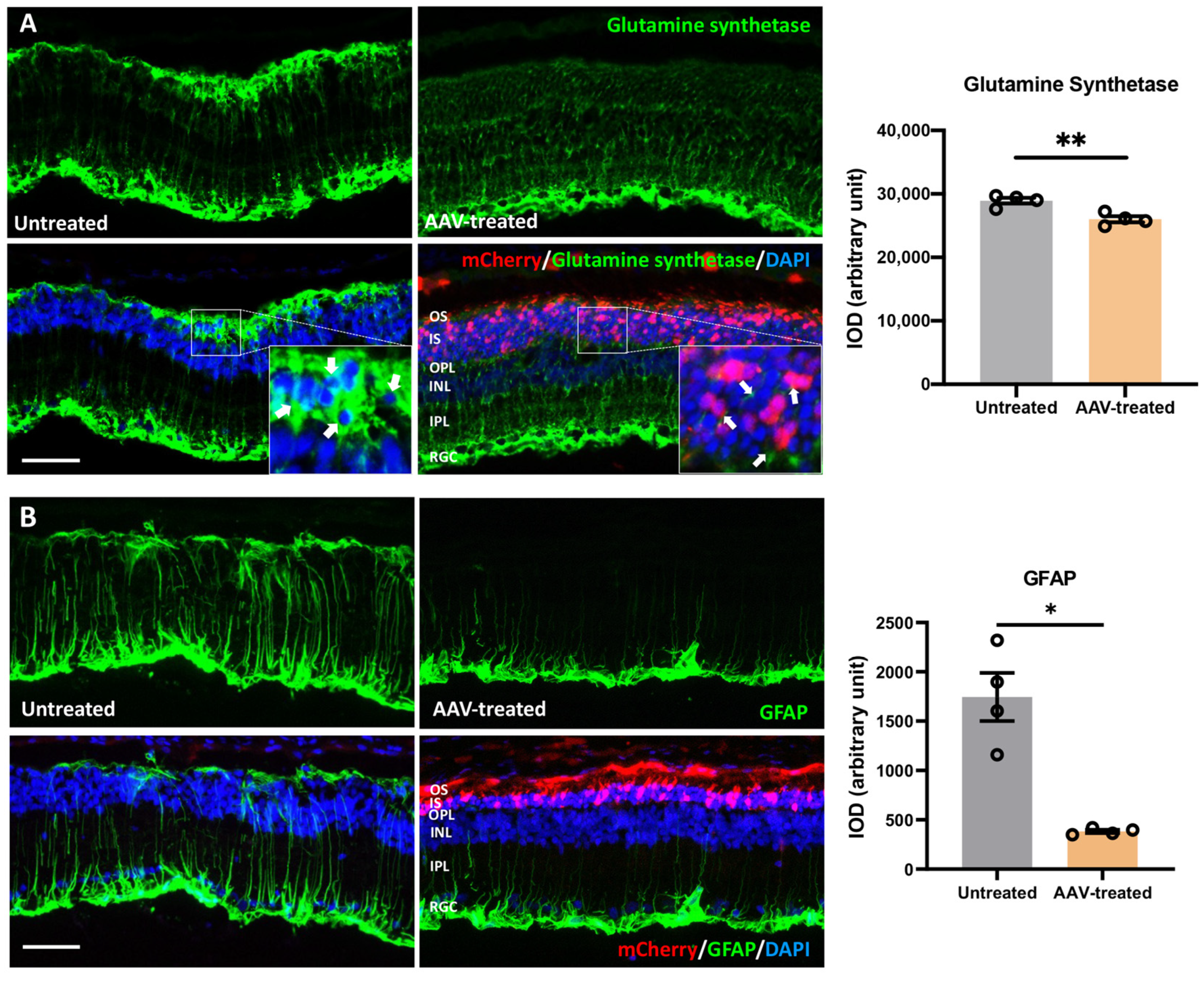
3.7. Suppression of Gliosis and Phagocytic Activity in Müller Glia following m-RhoP23H Ablation
3.8. m-RhoP23H -Specific Ablation Rescues PRs and Preserves Long-Term Vision by Suppressing UPR/ERS-Mediated Autophagy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parmeggiani, F.; Sorrentino, F.S.; Ponzin, D.; Barbaro, V.; Ferrari, S.; Di Iorio, E. Retinitis Pigmentosa: Genes and Disease Mechanisms. CG 2011, 12, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Hamel, C. Retinitis Pigmentosa. Orphanet J. Rare Dis. 2006, 1, 40. [Google Scholar] [CrossRef] [PubMed]
- O’Neal, T.B.; Luther, E.E. Retinitis Pigmentosa. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Miraldi Utz, V.; Coussa, R.G.; Antaki, F.; Traboulsi, E.I. Gene Therapy for RPE65 -Related Retinal Disease. Ophthalmic Genet. 2018, 39, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Lewin, A.S.; Drenser, K.A.; Hauswirth, W.W.; Nishikawa, S.; Yasumura, D.; Flannery, J.G.; LaVail, M.M. Ribozyme Rescue of Photoreceptor Cells in a Transgenic Rat Model of Autosomal Dominant Retinitis Pigmentosa. Nat. Med. 1998, 4, 967–971. [Google Scholar] [CrossRef]
- LaVail, M.M.; Yasumura, D.; Matthes, M.T.; Drenser, K.A.; Flannery, J.G.; Lewin, A.S.; Hauswirth, W.W. Ribozyme Rescue of Photoreceptor Cells in P23H Transgenic Rats: Long-Term Survival and Late-Stage Therapy. Proc. Natl. Acad. Sci. USA 2000, 97, 11488–11493. [Google Scholar] [CrossRef] [Green Version]
- Mussolino, C.; Sanges, D.; Marrocco, E.; Bonetti, C.; Di Vicino, U.; Marigo, V.; Auricchio, A.; Meroni, G.; Surace, E.M. Zinc-Finger-Based Transcriptional Repression of Rhodopsin in a Model of Dominant Retinitis Pigmentosa. EMBO Mol. Med. 2011, 3, 118–128. [Google Scholar] [CrossRef] [Green Version]
- Millington-Ward, S.; Chadderton, N.; O’Reilly, M.; Palfi, A.; Goldmann, T.; Kilty, C.; Humphries, M.; Wolfrum, U.; Bennett, J.; Humphries, P.; et al. Suppression and Replacement Gene Therapy for Autosomal Dominant Disease in a Murine Model of Dominant Retinitis Pigmentosa. Mol. Ther. 2011, 19, 642–649. [Google Scholar] [CrossRef]
- Bakondi, B.; Lv, W.; Lu, B.; Jones, M.K.; Tsai, Y.; Kim, K.J.; Levy, R.; Akhtar, A.A.; Breunig, J.J.; Svendsen, C.N.; et al. In Vivo CRISPR/Cas9 Gene Editing Corrects Retinal Dystrophy in the S334ter-3 Rat Model of Autosomal Dominant Retinitis Pigmentosa. Mol. Ther. 2016, 24, 556–563. [Google Scholar] [CrossRef] [Green Version]
- Latella, M.C.; Di Salvo, M.T.; Cocchiarella, F.; Benati, D.; Grisendi, G.; Comitato, A.; Marigo, V.; Recchia, A. In Vivo Editing of the Human Mutant Rhodopsin Gene by Electroporation of Plasmid-Based CRISPR/Cas9 in the Mouse Retina. Mol. Ther. Nucleic Acids 2016, 5, e389. [Google Scholar] [CrossRef]
- Li, P.; Kleinstiver, B.P.; Leon, M.Y.; Prew, M.S.; Navarro-Gomez, D.; Greenwald, S.H.; Pierce, E.A.; Joung, J.K.; Liu, Q. Allele-Specific CRISPR-Cas9 Genome Editing of the Single-Base P23H Mutation for Rhodopsin-Associated Dominant Retinitis Pigmentosa. CRISPR J. 2018, 1, 55–64. [Google Scholar] [CrossRef]
- Patrizi, C.; Llado, M.; Benati, D.; Iodice, C.; Marrocco, E.; Guarascio, R.; Surace, E.M.; Cheetham, M.E.; Auricchio, A.; Recchia, A. Allele-Specific Editing Ameliorates Dominant Retinitis Pigmentosa in a Transgenic Mouse Model. Am. J. Hum. Genet. 2021, 108, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Burnight, E.R.; Wiley, L.A.; DeLuca, A.P.; Oppedal, D.J.; Scheetz, T.E.; Mullins, R.F.; Stone, E.M.; Tucker, B.A. CRISPR/Cas9-Mediated Genome Editing for Correction of Inherited Retinal Disease Mutations. Investig. Ophthalmol. Vis. Sci. 2016, 57, 1157. [Google Scholar]
- Ledford, H. Quest to Use CRISPR against Disease Gains Ground. Nature 2020, 577, 156. [Google Scholar] [CrossRef] [PubMed]
- RetNet—Retinal Information Network. Available online: https://sph.uth.edu/retnet/ (accessed on 28 October 2021).
- Lu, B.; Morgans, C.W.; Girman, S.; Luo, J.; Zhao, J.; Du, H.; Lim, S.; Ding, S.; Svendsen, C.; Zhang, K.; et al. Neural Stem Cells Derived by Small Molecules Preserve Vision. Transl. Vis. Sci. Technol. 2013, 2, 1. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.; Lu, B.; Bakondi, B.; Girman, S.; Sahabian, A.; Sareen, D.; Svendsen, C.N.; Wang, S. Human IPSC-Derived Neural Progenitors Preserve Vision in an AMD-Like Model. Stem Cells 2015, 33, 2537–2549. [Google Scholar] [CrossRef] [Green Version]
- Clement, K.; Rees, H.; Canver, M.C.; Gehrke, J.M.; Farouni, R.; Hsu, J.Y.; Cole, M.A.; Liu, D.R.; Joung, J.K.; Bauer, D.E.; et al. Accurate and Rapid Analysis of Genome Editing Data from Nucleases and Base Editors with CRISPResso2. Nat. BioTechnol. 2019, 37, 224–226. [Google Scholar] [CrossRef]
- Wang, S.; Lu, B.; Girman, S.; Duan, J.; McFarland, T.; Zhang, Q.; Grompe, M.; Adamus, G.; Appukuttan, B.; Lund, R. Non-Invasive Stem Cell Therapy in a Rat Model for Retinal Degeneration and Vascular Pathology. PLoS ONE 2010, 5, e9200. [Google Scholar] [CrossRef] [Green Version]
- Shahin, S.; Banerjee, S.; Swarup, V.; Singh, S.P.; Chaturvedi, C.M. From the Cover: 2.45-GHz Microwave Radiation Impairs Hippocampal Learning and Spatial Memory: Involvement of Local Stress Mechanism-Induced Suppression of IGluR/ERK/CREB Signaling. Toxicol. Sci. 2018, 161, 349–374. [Google Scholar] [CrossRef]
- Shihan, M.H.; Novo, S.G.; Le Marchand, S.J.; Wang, Y.; Duncan, M.K. A Simple Method for Quantitating Confocal Fluorescent Images. Biochem. Biophys. Rep. 2021, 25, 100916. [Google Scholar] [CrossRef]
- Flannery, J.G.; Zolotukhin, S.; Vaquero, M.I.; LaVail, M.M.; Muzyczka, N.; Hauswirth, W.W. Efficient Photoreceptor-Targeted Gene Expression in Vivo by Recombinant Adeno-Associated Virus. Proc. Natl. Acad. Sci. USA 1997, 94, 6916–6921. [Google Scholar] [CrossRef] [Green Version]
- Carrigan, M.; Duignan, E.; Humphries, P.; Palfi, A.; Kenna, P.F.; Farrar, G.J. A Novel Homozygous Truncating GNAT1 Mutation Implicated in Retinal Degeneration. Br. J. OphthalMol. 2016, 100, 495–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CRISPR RGEN Tools. Available online: http://www.rgenome.net/cas-offinder/ (accessed on 28 October 2021).
- Lu, B.; Morgans, C.W.; Girman, S.; Lund, R.; Wang, S. Retinal Morphological and Functional Changes in an Animal Model of Retinitis Pigmentosa. Vis. NeuroSci. 2013, 30, 77–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter-Dawson, L.D.; LaVail, M.M. Rods and Cones in the Mouse Retina. I. Structural Analysis Using Light and Electron Microscopy. J. Comp. Neurol. 1979, 188, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sánchez, L.; Lax, P.; Campello, L.; Pinilla, I.; Cuenca, N. Astrocytes and Müller Cell Alterations During Retinal Degeneration in a Transgenic Rat Model of Retinitis Pigmentosa. Front. Cell NeuroSci. 2015, 9, 484. [Google Scholar] [CrossRef] [Green Version]
- Sakami, S.; Imanishi, Y.; Palczewski, K. Müller Glia Phagocytose Dead Photoreceptor Cells in a Mouse Model of Retinal Degenerative Disease. FASEB J. 2019, 33, 3680–3692. [Google Scholar] [CrossRef]
- Nomura-Komoike, K.; Saitoh, F.; Fujieda, H. Phosphatidylserine Recognition and Rac1 Activation Are Required for Müller Glia Proliferation, Gliosis and Phagocytosis after Retinal Injury. Sci. Rep. 2020, 10, 1488. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.H.; Li, H.; Yasumura, D.; Cohen, H.R.; Zhang, C.; Panning, B.; Shokat, K.M.; Lavail, M.M.; Walter, P. IRE1 Signaling Affects Cell Fate during the Unfolded Protein Response. Science 2007, 318, 944–949. [Google Scholar] [CrossRef] [Green Version]
- Gorbatyuk, M.S.; Knox, T.; LaVail, M.M.; Gorbatyuk, O.S.; Noorwez, S.M.; Hauswirth, W.W.; Lin, J.H.; Muzyczka, N.; Lewin, A.S. Restoration of Visual Function in P23H Rhodopsin Transgenic Rats by Gene Delivery of BiP/Grp78. Proc. Natl. Acad. Sci. USA 2010, 107, 5961–5966. [Google Scholar] [CrossRef] [Green Version]
- Aguilà, M.; Bellingham, J.; Athanasiou, D.; Bevilacqua, D.; Duran, Y.; Maswood, R.; Parfitt, D.A.; Iwawaki, T.; Spyrou, G.; Smith, A.J.; et al. AAV-Mediated ERdj5 Overexpression Protects against P23H Rhodopsin Toxicity. Hum. Mol. Genet. 2020, 29, 1310–1318. [Google Scholar] [CrossRef]
- Yao, J.; Qiu, Y.; Frontera, E.; Jia, L.; Khan, N.W.; Klionsky, D.J.; Ferguson, T.A.; Thompson, D.A.; Zacks, D.N. Inhibiting Autophagy Reduces Retinal Degeneration Caused by Protein Misfolding. Autophagy 2018, 14, 1226–1238. [Google Scholar] [CrossRef] [Green Version]
- Kakavand, K.; Jobling, A.I.; Greferath, U.; Vessey, K.A.; de Iongh, R.U.; Fletcher, E.L. Photoreceptor Degeneration in Pro23His Transgenic Rats (Line 3) Involves Autophagic and Necroptotic Mechanisms. Front. NeuroSci. 2020, 14, 581579. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, S.G.; Luoni, M.; Castoldi, V.; Massimino, L.; Cabassi, T.; Angeloni, D.; Demontis, G.C.; Leocani, L.; Andreazzoli, M.; Broccoli, V. Cas9/SgRNA Selective Targeting of the P23H Rhodopsin Mutant Allele for Treating Retinitis Pigmentosa by Intravitreal AAV9.PHP.B-Based Delivery. Hum. Mol. Genet. 2018, 27, 761–779. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-T.; Wu, W.-H.; Lee, T.-T.; Wu, W.-P.; Xu, C.L.; Park, K.S.; Cui, X.; Justus, S.; Lin, C.-S.; Jauregui, R.; et al. Clustered Regularly Interspaced Short Palindromic Repeats-Based Genome Surgery for the Treatment of Autosomal Dominant Retinitis Pigmentosa. Ophthalmology 2018, 125, 1421–1430. [Google Scholar] [CrossRef] [PubMed]
- LaVail, M.M. Rod Outer Segment Disk Shedding in Rat Retina: Relationship to Cyclic Lighting. Science 1976, 194, 1071–1074. [Google Scholar] [CrossRef] [PubMed]
- Young, R.W. The Renewal of Photoreceptor Cell Outer Segments. J. Cell Biol. 1967, 33, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Legros, J.; Hicks, D. Renewal of Photoreceptor Outer Segments and Their Phagocytosis by the Retinal Pigment Epithelium. Int. Rev. Cytol. 2000, 196, 245–313. [Google Scholar] [CrossRef] [PubMed]
- Dryja, T.P.; McGee, T.L.; Reichel, E.; Hahn, L.B.; Cowley, G.S.; Yandell, D.W.; Sandberg, M.A.; Berson, E.L. A Point Mutation of the Rhodopsin Gene in One Form of Retinitis Pigmentosa. Nature 1990, 343, 364–366. [Google Scholar] [CrossRef]
- Nguyen, A.T.H.; Campbell, M.; Kiang, A.-S.; Humphries, M.M.; Humphries, P. Current Therapeutic Strategies for P23H RHO-Linked RP. Adv. Exp. Med. Biol. 2014, 801, 471–476. [Google Scholar] [CrossRef]
- Athanasiou, D.; Aguila, M.; Bellingham, J.; Li, W.; McCulley, C.; Reeves, P.J.; Cheetham, M.E. The Molecular and Cellular Basis of Rhodopsin Retinitis Pigmentosa Reveals Potential Strategies for Therapy. Prog. Retin. Eye Res. 2018, 62, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Sakami, S.; Maeda, T.; Bereta, G.; Okano, K.; Golczak, M.; Sumaroka, A.; Roman, A.J.; Cideciyan, A.V.; Jacobson, S.G.; Palczewski, K. Probing Mechanisms of Photoreceptor Degeneration in a New Mouse Model of the Common Form of Autosomal Dominant Retinitis Pigmentosa Due to P23H Opsin Mutations. J. Biol. Chem. 2011, 286, 10551–10567. [Google Scholar] [CrossRef] [Green Version]
- Cideciyan, A.V.; Jacobson, S.G.; Beltran, W.A.; Sumaroka, A.; Swider, M.; Iwabe, S.; Roman, A.J.; Olivares, M.B.; Schwartz, S.B.; Komáromy, A.M.; et al. Human Retinal Gene Therapy for Leber Congenital Amaurosis Shows Advancing Retinal Degeneration despite Enduring Visual Improvement. Proc. Natl. Acad. Sci. USA 2013, 110, E517–E525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojno, A.P.; Pierce, E.A.; Bennett, J. Seeing the Light. Sci. Transl. Med. 2013, 5, 175fs8. [Google Scholar] [CrossRef] [PubMed]
- Nishiguchi, K.M.; Fujita, K.; Tokashiki, N.; Komamura, H.; Takemoto-Kimura, S.; Okuno, H.; Bito, H.; Nakazawa, T. Retained Plasticity and Substantial Recovery of Rod-Mediated Visual Acuity at the Visual Cortex in Blind Adult Mice with Retinal Dystrophy. Mol. Ther. 2018, 26, 2397–2406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crudele, J.M.; Chamberlain, J.S. Cas9 Immunity Creates Challenges for CRISPR Gene Editing Therapies. Nat. Commun. 2018, 9, 3497. [Google Scholar] [CrossRef]
- Lyu, P.; Javidi-Parsijani, P.; Atala, A.; Lu, B. Delivering Cas9/SgRNA Ribonucleoprotein (RNP) by Lentiviral Capsid-Based Bionanoparticles for Efficient “hit-and-Run” Genome Editing. Nucleic Acids Res. 2019, 47, e99. [Google Scholar] [CrossRef] [Green Version]
- Petris, G.; Casini, A.; Montagna, C.; Lorenzin, F.; Prandi, D.; Romanel, A.; Zasso, J.; Conti, L.; Demichelis, F.; Cereseto, A. Hit and Go CAS9 Delivered through a Lentiviral Based Self-Limiting Circuit. Nat. Commun. 2017, 8, 15334. [Google Scholar] [CrossRef]
- Xia, E.; Duan, R.; Shi, F.; Seigel, K.E.; Grasemann, H.; Hu, J. Overcoming the Undesirable CRISPR-Cas9 Expression in Gene Correction. Mol. Ther. Nucleic Acids 2018, 13, 699–709. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, P. The Laboratory Rat: Relating Its Age With Human’s. Int. J. Prev. Med. 2013, 4, 624–630. [Google Scholar]
- Zhang, X.-H.; Tee, L.Y.; Wang, X.-G.; Huang, Q.-S.; Yang, S.-H. Off-Target Effects in CRISPR/Cas9-Mediated Genome Engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef]
- Pickar-Oliver, A.; Gersbach, C.A. The next Generation of CRISPR-Cas Technologies and Applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahin, S.; Xu, H.; Lu, B.; Mercado, A.; Jones, M.K.; Bakondi, B.; Wang, S. AAV-CRISPR/Cas9 Gene Editing Preserves Long-Term Vision in the P23H Rat Model of Autosomal Dominant Retinitis Pigmentosa. Pharmaceutics 2022, 14, 824. https://doi.org/10.3390/pharmaceutics14040824
Shahin S, Xu H, Lu B, Mercado A, Jones MK, Bakondi B, Wang S. AAV-CRISPR/Cas9 Gene Editing Preserves Long-Term Vision in the P23H Rat Model of Autosomal Dominant Retinitis Pigmentosa. Pharmaceutics. 2022; 14(4):824. https://doi.org/10.3390/pharmaceutics14040824
Chicago/Turabian StyleShahin, Saba, Hui Xu, Bin Lu, Augustus Mercado, Melissa K. Jones, Benjamin Bakondi, and Shaomei Wang. 2022. "AAV-CRISPR/Cas9 Gene Editing Preserves Long-Term Vision in the P23H Rat Model of Autosomal Dominant Retinitis Pigmentosa" Pharmaceutics 14, no. 4: 824. https://doi.org/10.3390/pharmaceutics14040824
APA StyleShahin, S., Xu, H., Lu, B., Mercado, A., Jones, M. K., Bakondi, B., & Wang, S. (2022). AAV-CRISPR/Cas9 Gene Editing Preserves Long-Term Vision in the P23H Rat Model of Autosomal Dominant Retinitis Pigmentosa. Pharmaceutics, 14(4), 824. https://doi.org/10.3390/pharmaceutics14040824