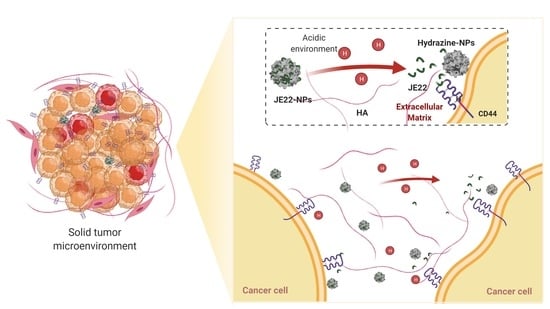
Selective Anticancer Therapy Based on a HA-CD44 Interaction Inhibitor Loaded on Polymeric Nanoparticles
Abstract
1. Introduction
2. Materials and Methods
2.1. Computational
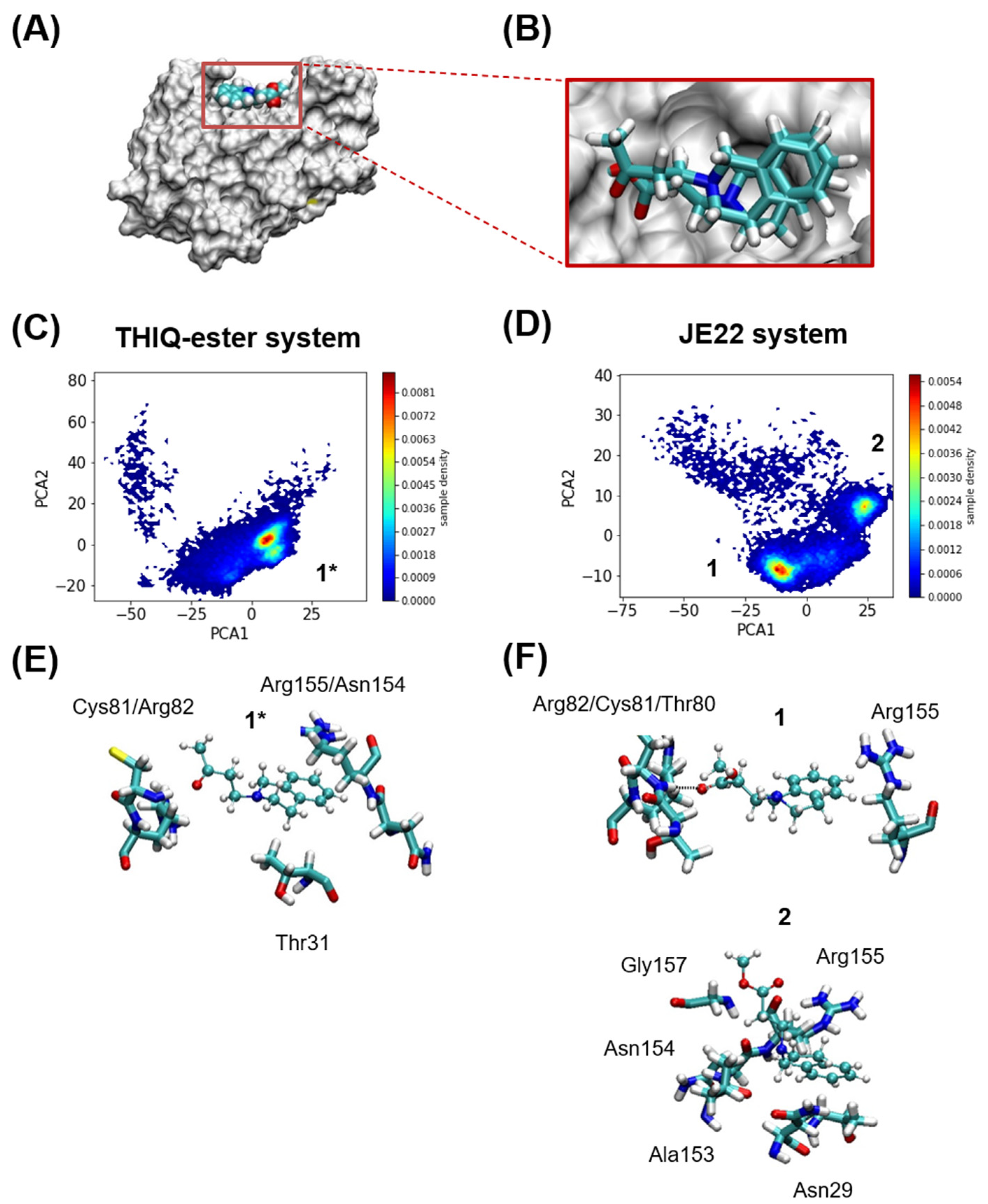
2.1.1. Systems Set Up
2.1.2. Molecular Dynamics Simulations
2.2. Chemistry and Characterization
2.2.1. General
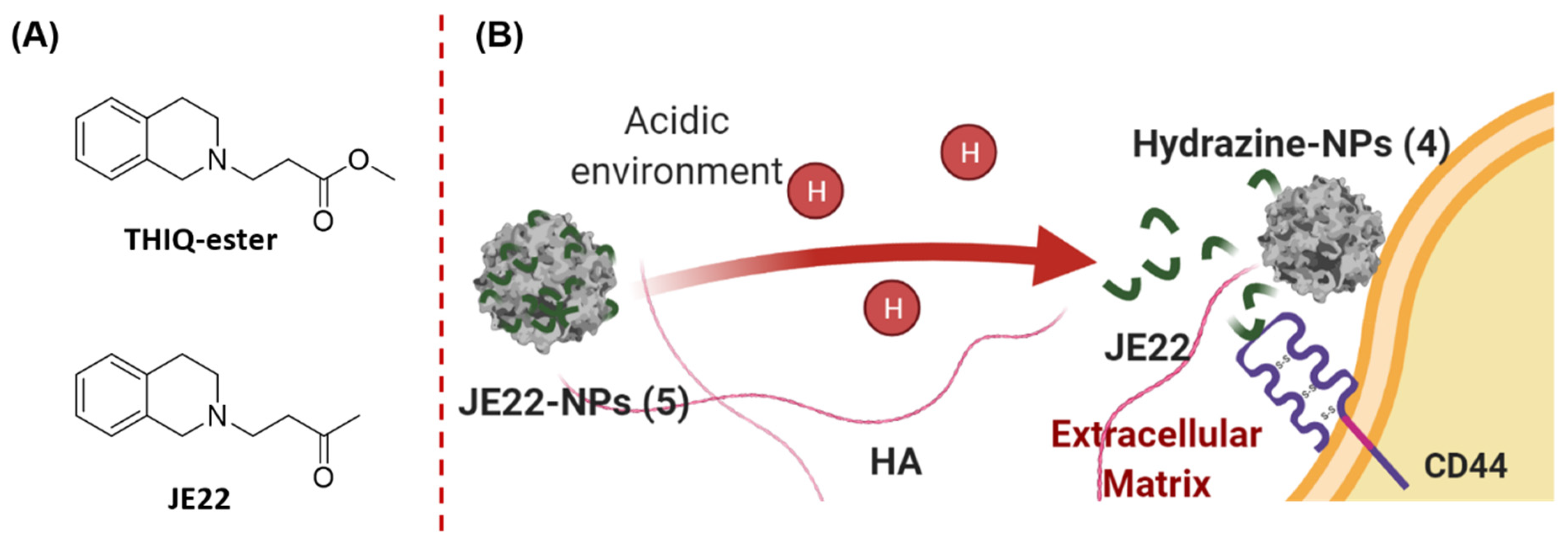
2.2.2. Synthesis of 4-(3,4-Dihydroisoquinolin-2(1H)-yl)butan-2-one (JE22)
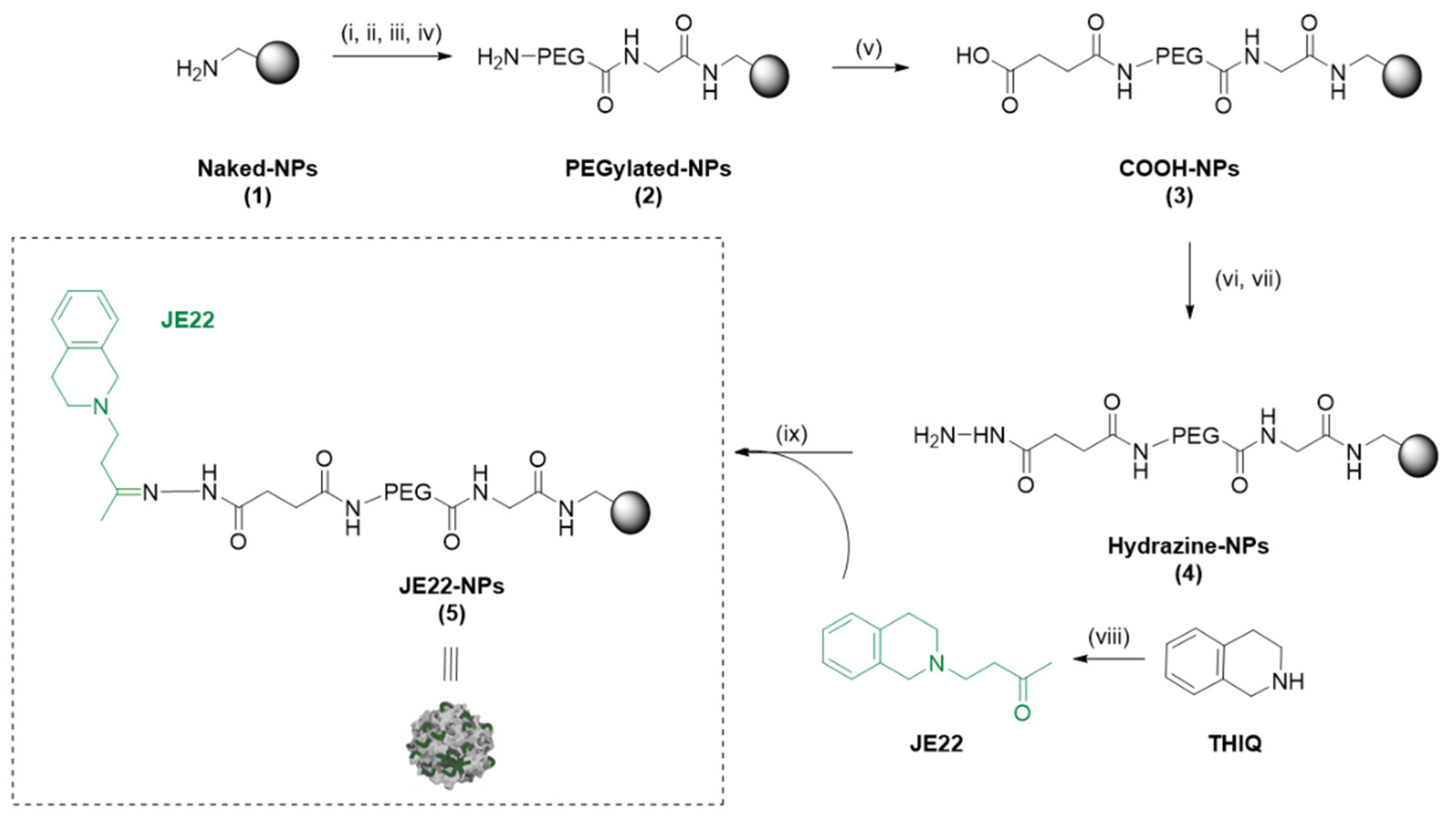
2.2.3. Preparation of Therapeutic Polymeric Nanoparticles JE22-NPs (5)
2.2.4. Characterization of JE22-NPs (5)
2.2.5. Stability Study of JE22-NPs (5)
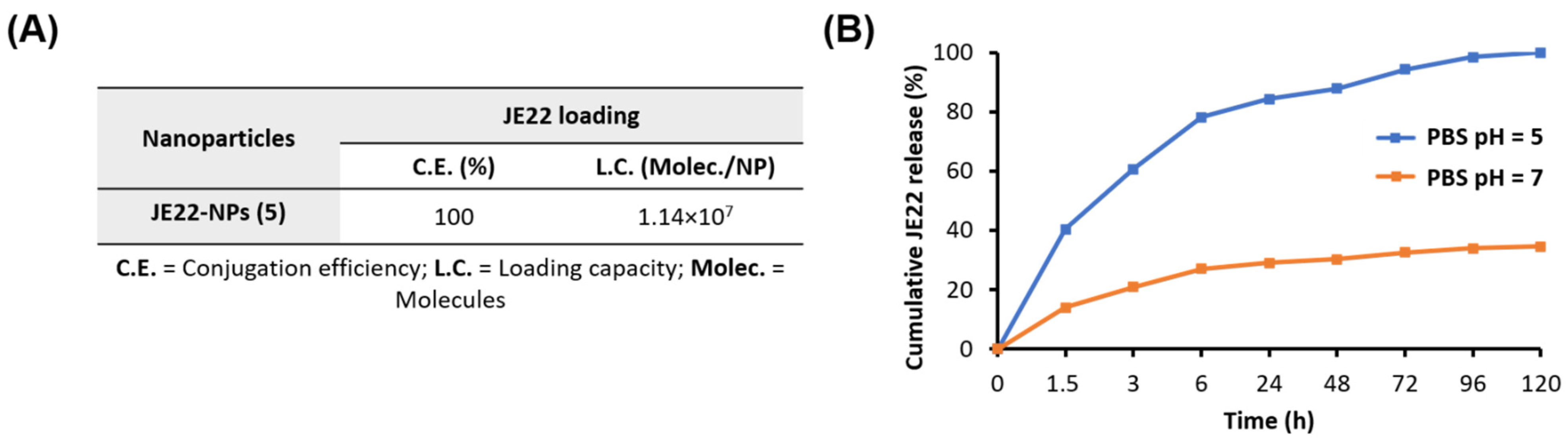
2.2.6. Determination of Conjugation Efficiency of JE22-NPs (5)
2.2.7. Evaluation of Drug Release Profile of JE22-NPs (5)
2.3. Biology
2.3.1. General
2.3.2. Cell Culture
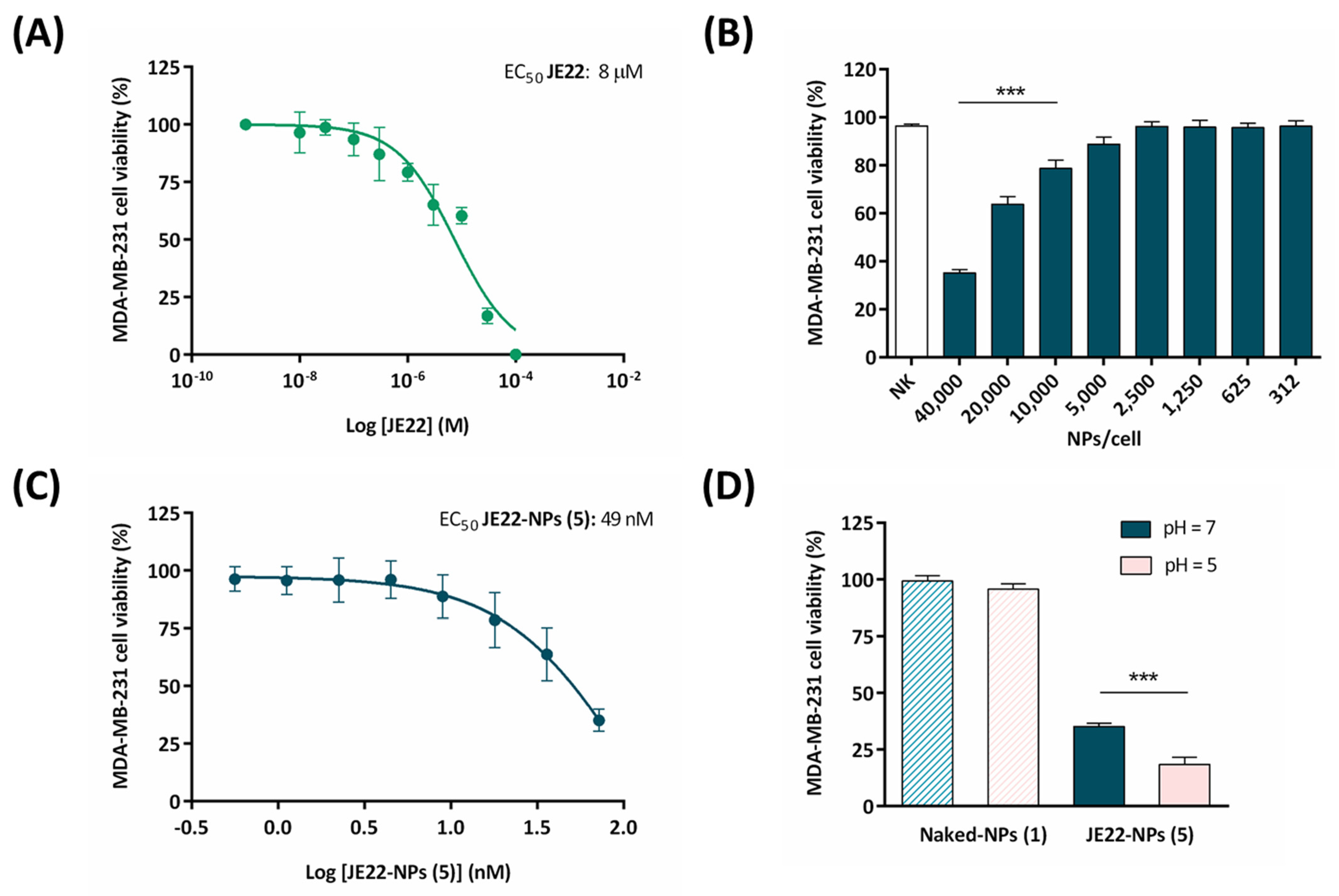
2.3.3. Cell Viability Assays
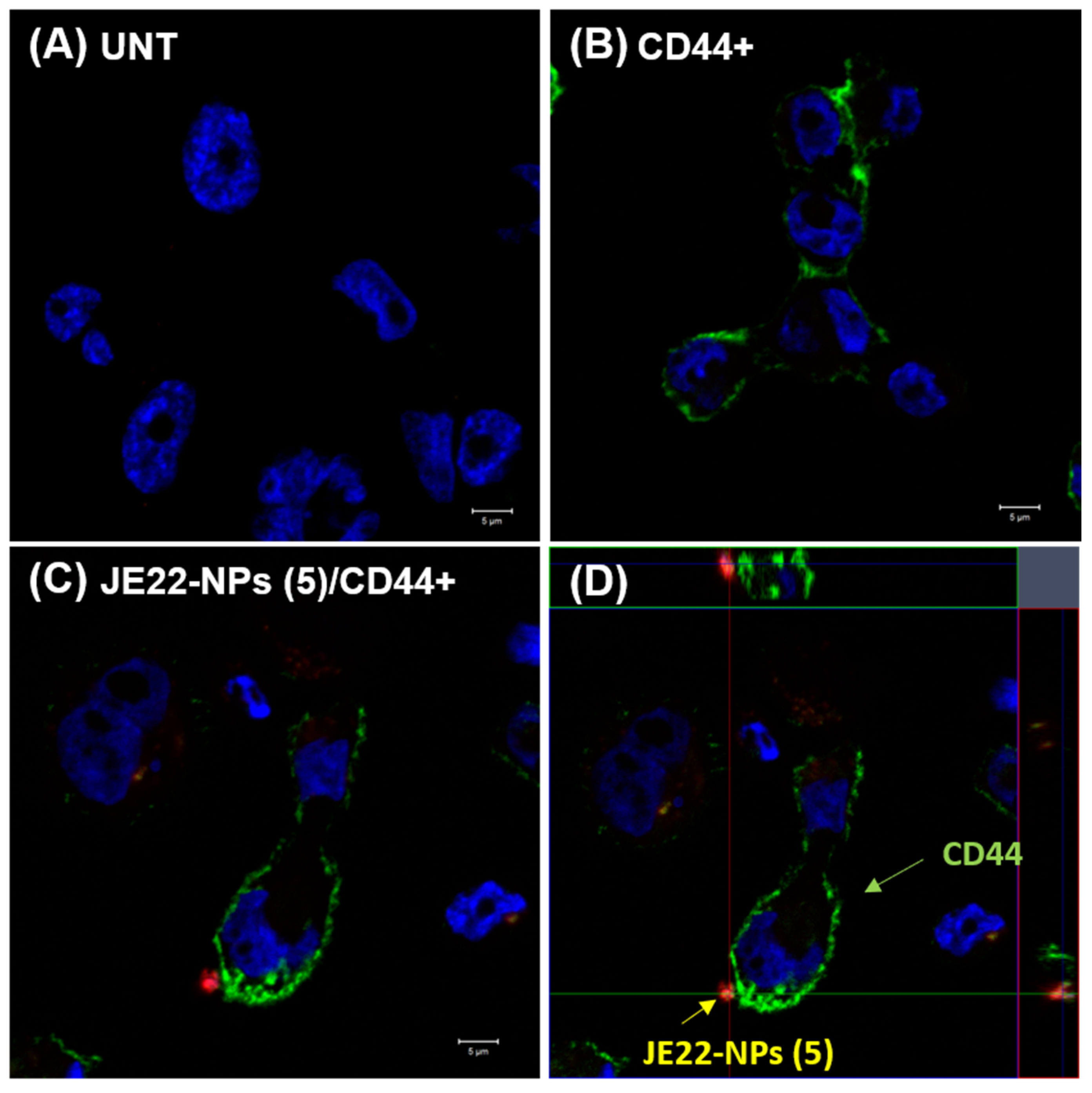
2.3.4. Confocal Microscopy Analysis
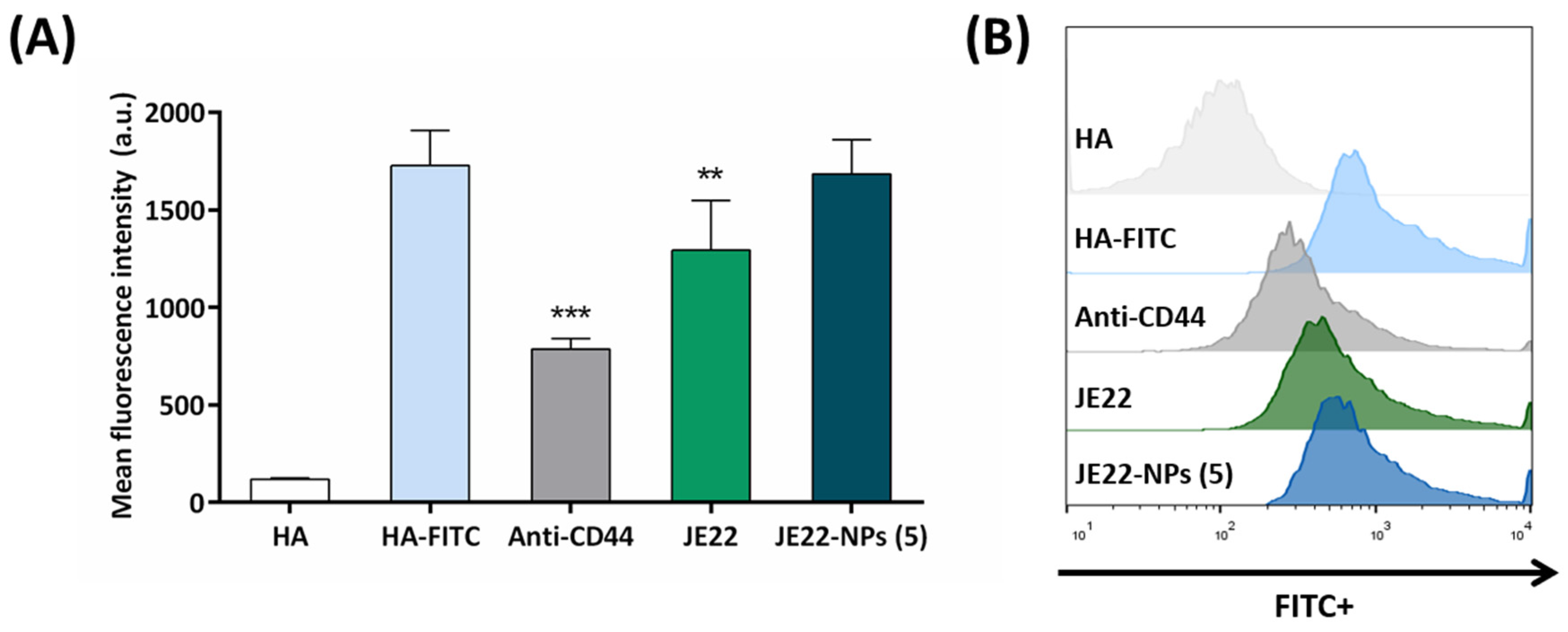
2.3.5. HA-FITC Binding Assay
2.3.6. Wound Healing Assay
2.3.7. Apoptosis Assay
2.3.8. Statistical Analysis
3. Results and Discussion
3.1. Study of the Effect of Structural Modification of THIQ on CD44 Interaction by Computational Studies
3.2. Synthesis and Physicochemical Characterization of the Nanodevice to Target CD44
3.2.1. Preparation of JE22-NPs (5)
3.2.2. Physicochemical Characterization of JE22-NPs (5)
3.2.3. Efficiency of Conjugation and Drug Release of JE22-NPs (5)
3.3. Evaluation of Efficiency of the Designed Nanodevice JE22-NP (5) for Antitumor Activity
Assessment of Biological Activity of JE22-NPs (5)
3.4. Evaluation of Efficiency of the Designed Nanodevice for the Inhibition of CD44 Receptor Binding
3.4.1. Analysis of the Interaction of the Designed Nanodevice JE22-NPs (5) with CD44+ Cells by Confocal Microscopy
3.4.2. Assessment of CD44-Binding Capacity
3.4.3. Influence of JE22-NPs (5) in Migration of CD44+ Cells
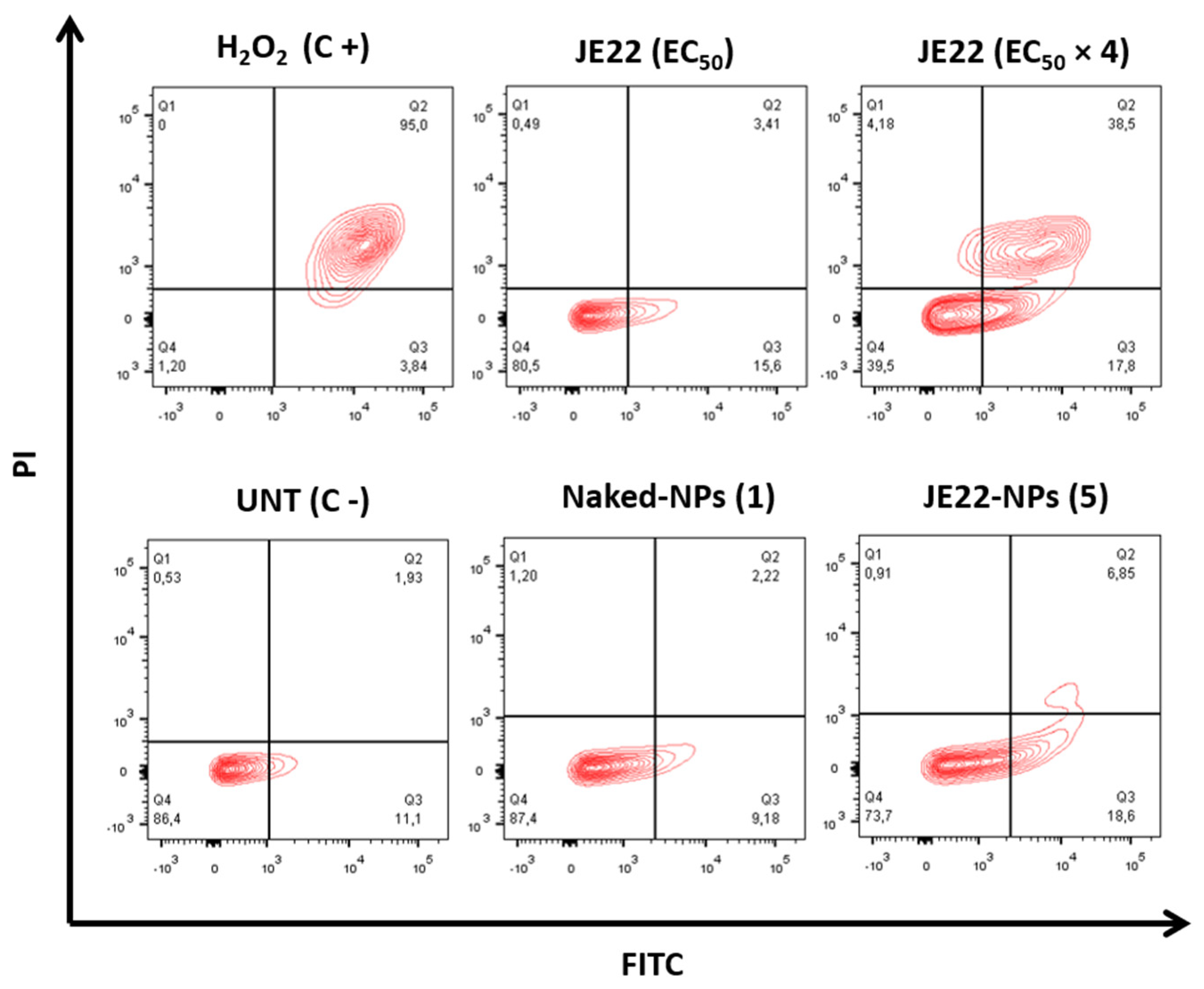
3.4.4. Apoptotic Activity of JE22-NPs (5)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fraser, J.R.; Laurent, T.C.; Laurent, U.B. Hyaluronan: Its nature, distribution, functions and turnover. J. Intern. Med. 1997, 242, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Toole, B.P. Hyaluronan: From extracellular glue to pericellular cue. Nat. Rev. Cancer 2004, 4, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Fallacara, A.; Baldini, E.; Manfredini, S.; Vertuani, S. Hyaluronic Acid in the Third Millennium. Polymers 2018, 10, 701. [Google Scholar] [CrossRef]
- Karousou, E.; Misra, S.; Ghatak, S.; Dobra, K.; Götte, M.; Vigetti, D.; Passi, A.; Karamanos, N.K.; Skandalis, S.S. Roles and targeting of the HAS/hyaluronan/CD44 molecular system in cancer. Matrix Biol. 2017, 59, 3–22. [Google Scholar]
- Chanmee, T.; Ontong, P.; Itano, N. Hyaluronan: A modulator of the tumor microenvironment. Cancer Lett. 2016, 375, 20–30. [Google Scholar] [CrossRef]
- Misra, S.; Heldin, P.; Hascall, V.C.; Karamanos, N.K.; Skandalis, S.S.; Markwald, R.R.; Ghatak, S. Hyaluronan-CD44 interactions as potential targets for cancer therapy. FEBS J. 2011, 278, 1429–1443. [Google Scholar]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef]
- Germain, M.; Caputo, F.; Metcalfe, S.; Tosi, G.; Spring, K.; Åslund, A.; Pottier, A.; Schiffelers, R.; Ceccaldi, A.; Schmid, R. Delivering the power of nanomedicine to patients today. J. Control. Release 2020, 326, 164–171. [Google Scholar] [CrossRef]
- Chiesa, E.; Greco, A.; Riva, F.; Dorati, R.; Conti, B.; Modena, T.; Genta, I. Hyaluronic Acid-Based Nanoparticles for Protein Delivery: Systematic Examination of Microfluidic Production Conditions. Pharmaceutics 2021, 13, 1565. [Google Scholar]
- Liang, X.; Li, X.; Duan, J.; Chen, Y.; Wang, X.; Pang, L.; Kong, D.; Song, B.; Li, C.; Yang, J. Nanoparticles with CD44 Targeting and ROS Triggering Properties as Effective in Vivo Antigen Delivery System. Mol. Pharm. 2018, 15, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Gaio, E.; Conte, C.; Esposito, D.; Reddi, E.; Quaglia, F.; Moret, F. CD44 Targeting Mediated by Polymeric Nanoparticles and Combination of Chlorine TPCS2a-PDT and Docetaxel-Chemotherapy for Efficient Killing of Breast Differentiated and Stem Cancer Cells In Vitro. Cancers 2020, 12, 278. [Google Scholar] [CrossRef]
- Shi, J.; Ren, Y.; Ma, J.; Luo, X.; Li, J.; Wu, Y.; Gu, H.; Fu, C.; Cao, Z.; Zhang, J. Novel CD44-targeting and pH/redox-dual-stimuli-responsive core–shell nanoparticles loading triptolide combats breast cancer growth and lung metastasis. J. Nanobiotechnol. 2021, 19, 188. [Google Scholar] [CrossRef]
- Navarro-Marchal, S.A.; Griñán-Lisón, C.; Entrena, J.M.; Ruiz-Alcalá, G.; Tristán-Manzano, M.; Martin, F.; Pérez-Victoria, I.; Peula-García, J.M.; Marchal, J.A. Anti-CD44-Conjugated Olive Oil Liquid Nanocapsules for Targeting Pancreatic Cancer Stem Cells. Biomacromolecules 2021, 22, 1374–1388. [Google Scholar] [CrossRef] [PubMed]
- Banerji, S.; Wright, A.J.; Noble, M.; Mahoney, D.J.; Campbell, I.D.; Day, A.J.; Jackson, D.G. Structures of the Cd44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat. Struct. Mol. Biol. 2007, 14, 234–239. [Google Scholar] [CrossRef]
- Liu, L.K.; Finzel, B.C. Fragment-based identification of an inducible binding site on cell surface receptor CD44 for the design of protein-carbohydrate interaction inhibitors. J. Med. Chem. 2014, 57, 2714–2725. [Google Scholar] [CrossRef] [PubMed]
- Unciti-Broceta, A.; Johansson, E.; Yusop, R.M.; Sánchez-Martín, R.M.; Bradley, M. Synthesis of polystyrene microspheres and functionalization with Pd0 nanoparticles to perform bioorthogonal organometallic chemistry in living cells. Nat. Protoc. 2012, 7, 1207–1218. [Google Scholar] [CrossRef]
- Altea-Manzano, P.; Unciti-Broceta, J.D.; Cano-Cortes, V.; Ruiz-Blas, M.P.; Valero-Griñan, T.; Diaz-Mochon, J.J.; Sanchez-Martin, R. Tracking cell proliferation using a nanotechnology-based approach. Nanomedicine 2017, 12, 1591–1605. [Google Scholar] [CrossRef] [PubMed]
- Valero, T.; Delgado-González, A.; Unciti-Broceta, J.D.; Cano-Cortés, V.; Pérez-López, A.M.; Unciti-Broceta, A.; Sánchez Martín, R.M. Drug “Clicking” on Cell-Penetrating Fluorescent Nanoparticles for In Cellulo Chemical Proteomics. Bioconjug. Chem. 2018, 29, 3154–3160. [Google Scholar]
- Cano-Cortes, M.V.; Navarro-Marchal, S.A.; Ruiz-Blas, M.P.; Diaz-Mochon, J.J.; Marchal, J.A.; Sanchez-Martin, R.M. A Versatile Theranostic Nanodevice Based on an Orthogonal Bioconjugation Strategy for Efficient Targeted Treatment and Monitoring of Triple Negative Breast Cancer. Nanomed. Nanotechnol. Biol. Med. 2020, 24, 102120. [Google Scholar] [CrossRef] [PubMed]
- Cano-Cortes, M.V.; Laz-Ruiz, J.A.; Diaz-Mochon, J.J.; Sanchez-Martin, R.M. Characterization and Therapeutic Effect of a pH Stimuli Responsive Polymeric Nanoformulation for Controlled Drug Release. Polymers 2020, 12, 1265. [Google Scholar] [CrossRef] [PubMed]
- Cano-Cortes, M.V.; Altea-Manzano, P.; Laz-Ruiz, J.A.; Unciti-Broceta, J.D.; Lopez-Delgado, F.J.; Espejo-Roman, J.M.; Diaz-Mochon, J.J.; Sanchez-Martin, R.M. An effective polymeric nanocarrier that allows for active targeting and selective drug delivery in cell coculture systems. Nanoscale 2021, 13, 3500–3511. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Klauda, J.B.; Im, W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J. 2009, 96, 41a. [Google Scholar] [CrossRef]
- Kumar, R.; Iyer, V.G.; Im, W. CHARMM-GUI: A graphical user interface for the CHARMM users. Abstr. Pap. Am. Chem. S. 2007, 233, 273. [Google Scholar]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.F.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Davila-Contreras, E.M.; Qi, Y.F.; Lee, J.M.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder Toward Realistic Biological Membrane Simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; MacKerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.; Jo, S.; Brooks, C.L.; Lee, H.S.; Im, W. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 2017, 38, 1879–1886. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.; Jo, S.; Im, W. CHARMM-GUI Ligand Reader & Modeler. Biophys. J. 2017, 112, 289a. [Google Scholar]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Jones, K.; Karier, P.; Klussmann, M. C1-substituted N-alkyl tetrahydroisoquinoline derivatives through V-catalyzed oxidative coupling. ChemCatChem 2012, 4, 51–54. [Google Scholar] [CrossRef]
- Manchun, S.; Dass, C.R.; Sriamornsak, P. Targeted Therapy for Cancer Using Ph-Responsive Nanocarrier Systems. Life Sci. 2012, 90, 381–387. [Google Scholar] [CrossRef]
- Unciti-Broceta, J.D.; Cano-Cortés, V.; Altea-Manzano, P.; Pernagallo, S.; Díaz-Mochón, J.J.; Sánchez-Martín, R.M. Number of nanoparticles per cell through a spectrophotometric method—A key parameter to assess nanoparticle-based cellular assays. Sci. Rep. 2015, 5, 10091. [Google Scholar] [CrossRef]
- Pietrovito, L.; Cano-Cortés, V.; Gamberi, T.; Magherini, F.; Bianchi, L.; Bini, L.; Sánchez-Martín, R.M.; Fasano, M.; Modesti, A. Cellular response to empty and palladium-conjugated amino-polystyrene nanospheres uptake: A proteomic study. Proteomics 2015, 15, 34–43. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef] [PubMed]
- Olsson, E.; Honeth, G.; Bendahl, P.; Saal, L.H.; Gruvberger-Saal, S.; Ringnér, M.; Vallon-Christersson, J.; Jönsson, G.; Holm, K.; Lövgren, K.; et al. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer 2011, 11, 418. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C.; Kishimoto, H.; Fuchs, R.K.; Fuchs, R.K.; Mehrotra, S.; Bhat-Nakshatri, P.; Turner, C.H.; Goulet, R.J.; Badve, S.; Nakshatri, H. CD44+/CD24-breast cancer cells exhibit enhanced invasive properties: An early step necessary for metastasis. Breast Cancer Res. 2006, 8, R59. [Google Scholar] [CrossRef] [PubMed]
- Michl, J.; Kyung, C.K.; Swietach, P. Evidence-based guidelines for controlling pH in mammalian live-cell culture systems. Commun. Biol. 2019, 2, 144. [Google Scholar] [CrossRef]
- Ali, H.; Al-Yatama, M.K.; Abu-Farha, M.; Behbehani, K.; Al Madhoun, A. Multi-lineage differentiation of human umbilical cord Wharton’s Jelly Mesenchymal Stromal Cells mediates changes in the expression profile of stemness markers. PLoS ONE 2015, 10, e0122465. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, D.; Svechkarev, D.; Souchek, J.; Hill, T.; Taylor, M.; Natarajan, A.; Mohs, A. Impact of structurally modifying hyaluronic acid on CD44 interaction. J. Mater. Chem. B 2017, 5, 8183–8192. [Google Scholar] [CrossRef]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef]
- Aguirre-Alvarado, C.; Segura-Cabrera, A.; Velázquez-Quesada, I.; Hernández-Esquivel, M.A.; García-Pérez, C.A.; Guerrero-Rodríguez, S.L.; Ruiz-Moreno, A.G.; Rodríguez-Moreno, A.; Pérez-Tapia, S.-M.; Velasco-Velázquez, M.A. Virtual screening-driven repositioning of etoposide as CD44 antagonist in breast cancer cells. Oncotarget 2016, 7, 23772–23784. [Google Scholar] [CrossRef]
- Tsujimoto, Y.; Shimizu, S. Role of the mitochondrial membrane permeability transition in cell death. Apoptosis 2007, 5, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.F.; Tan, W.; Tian, K.; Yu, H.; Qiang, W.A.; Wang, Y.T. Combined effects of furanodiene and doxorubicin on the migration and invasion of MDA-MB-231 breast cancer cells in vitro. Oncol. Rep. 2017, 37, 2016–2024. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Verlet, L. Computer “Experiments” on Classical Fluids. I. Thermodynamical Properties of Lennard−Jones Molecules. Phys. Rev. 1967, 159, 98–103. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. SETTLE−an analytical version of the Shake and Rattle algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Tuckerman, M.; Berne, B.J.; Martyna, G.J. Reversible Multiple Time Scale Molecular−Dynamics. J. Chem. Phys. 1992, 97, 1990–2001. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Henin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.; Sánchez−Martín, R.; Bradley, M. Knocking (Anti)−Sense into Cells: The Microsphere Approach to Gene Silencing. Bioconjug. Chem. 2009, 20, 422–426. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espejo-Román, J.M.; Rubio-Ruiz, B.; Cano-Cortés, V.; Cruz-López, O.; Gonzalez-Resines, S.; Domene, C.; Conejo-García, A.; Sánchez-Martín, R.M. Selective Anticancer Therapy Based on a HA-CD44 Interaction Inhibitor Loaded on Polymeric Nanoparticles. Pharmaceutics 2022, 14, 788. https://doi.org/10.3390/pharmaceutics14040788
Espejo-Román JM, Rubio-Ruiz B, Cano-Cortés V, Cruz-López O, Gonzalez-Resines S, Domene C, Conejo-García A, Sánchez-Martín RM. Selective Anticancer Therapy Based on a HA-CD44 Interaction Inhibitor Loaded on Polymeric Nanoparticles. Pharmaceutics. 2022; 14(4):788. https://doi.org/10.3390/pharmaceutics14040788
Chicago/Turabian StyleEspejo-Román, José M., Belén Rubio-Ruiz, Victoria Cano-Cortés, Olga Cruz-López, Saúl Gonzalez-Resines, Carmen Domene, Ana Conejo-García, and Rosario M. Sánchez-Martín. 2022. "Selective Anticancer Therapy Based on a HA-CD44 Interaction Inhibitor Loaded on Polymeric Nanoparticles" Pharmaceutics 14, no. 4: 788. https://doi.org/10.3390/pharmaceutics14040788
APA StyleEspejo-Román, J. M., Rubio-Ruiz, B., Cano-Cortés, V., Cruz-López, O., Gonzalez-Resines, S., Domene, C., Conejo-García, A., & Sánchez-Martín, R. M. (2022). Selective Anticancer Therapy Based on a HA-CD44 Interaction Inhibitor Loaded on Polymeric Nanoparticles. Pharmaceutics, 14(4), 788. https://doi.org/10.3390/pharmaceutics14040788