A pH- and Bioreducible Cationic Copolymer with Amino Acids and Piperazines for Adenovirus Delivery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cancer Cells and Ad Preparations
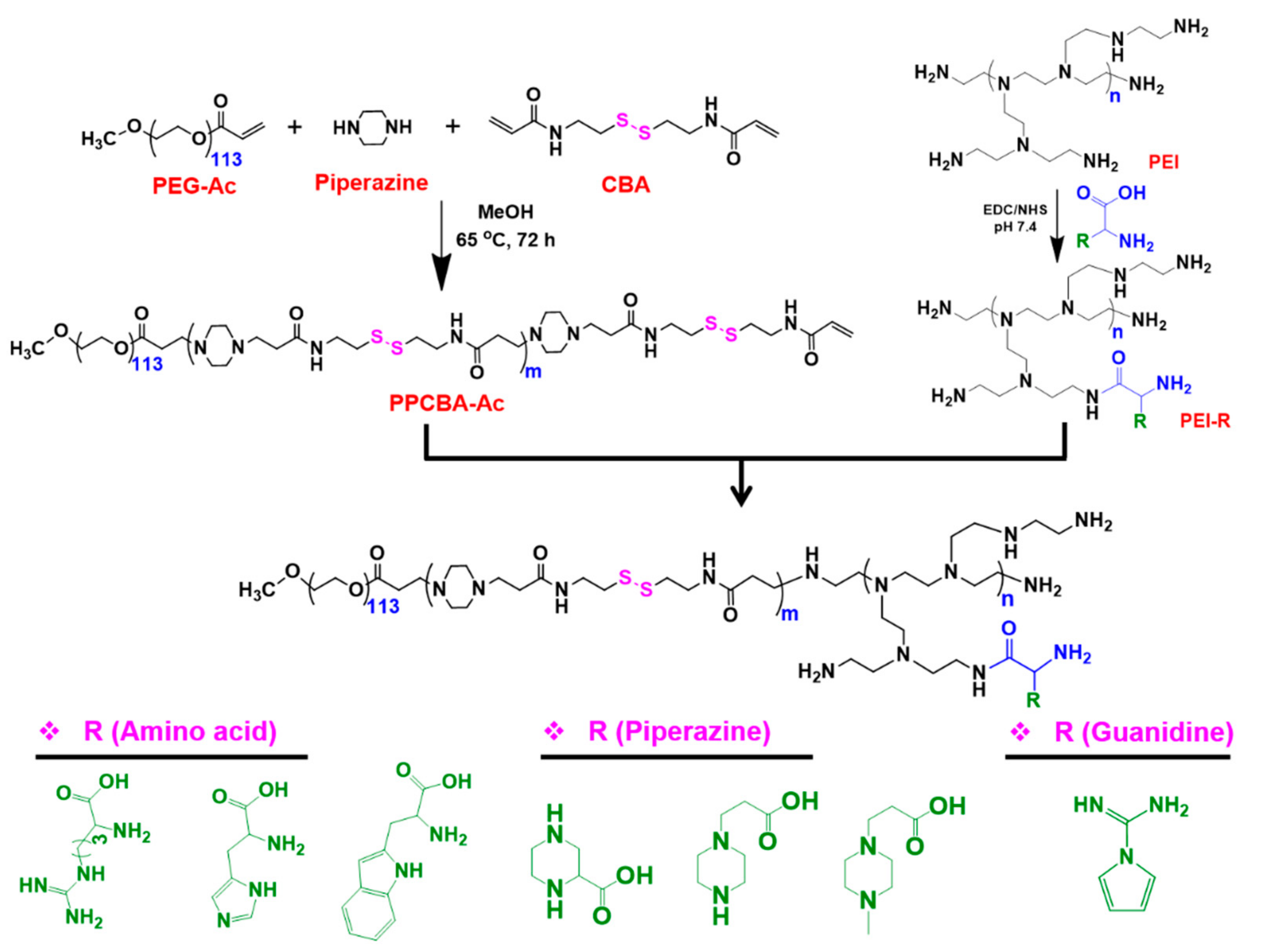
2.3. Synthesis of PPA Copolymers
2.4. Synthesis of Acrylate-Terminated Copolymer (PPCBA)
2.5. Synthesis of PEI Conjugates
2.6. Synthesis of PPA Copolymers
2.7. Synthesis of PPA-SPDP Copolymers
2.8. Preparation of PPA-SPDP Copolymer Conjugated Ad (PPA-Ad)
2.9. Characterization
2.9.1. 1H NMR Spectra
2.9.2. Size and Zeta Potential
2.9.3. Transmission Electron Microscopy (TEM)
2.9.4. Acid–Base Titration
2.10. Gel Retardation Assay
2.11. MTT Assay
2.12. Transduction of PPA-Ad Conjugate in Cancer Cells
2.13. Statistical Analysis
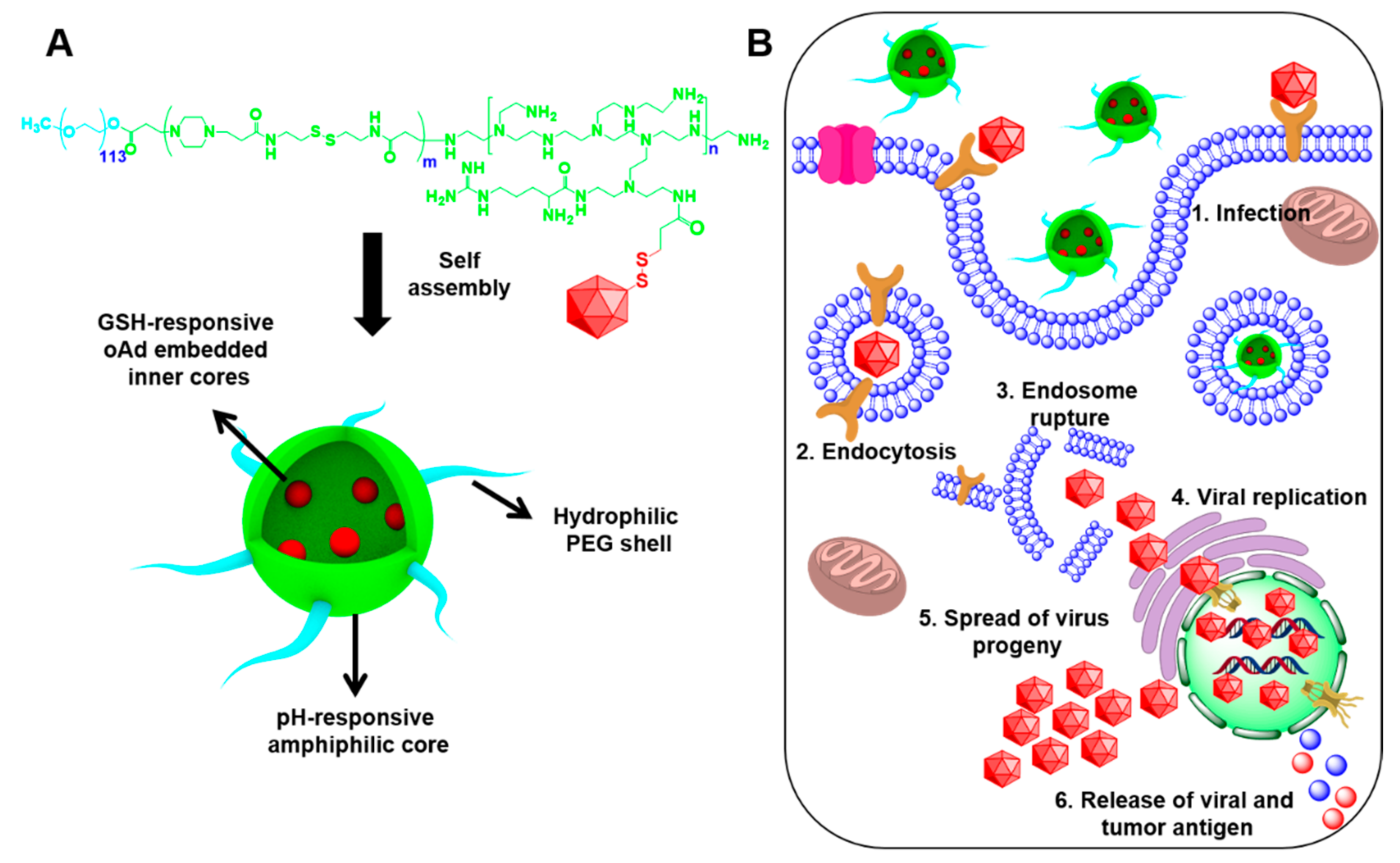
3. Results and Discussion
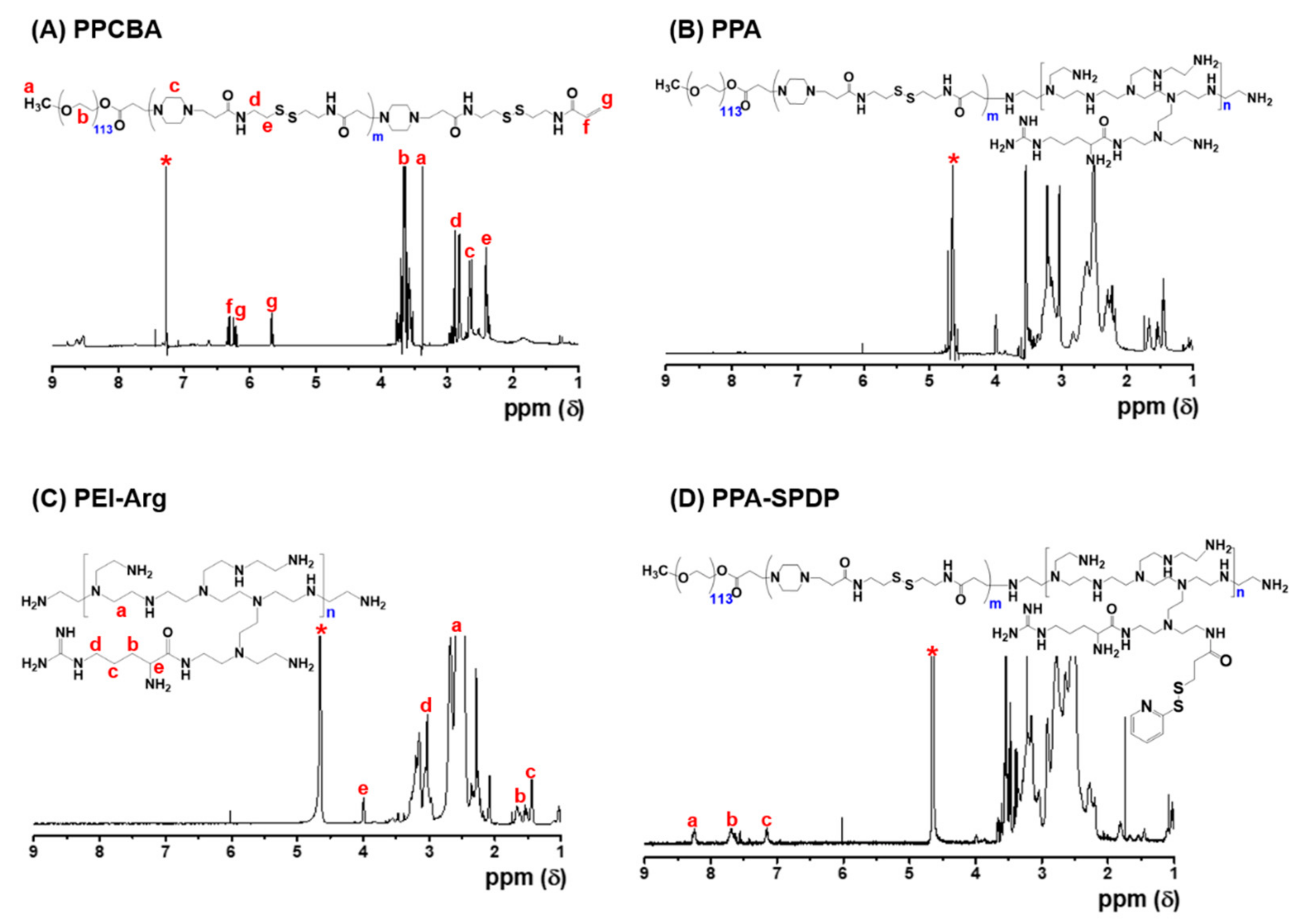
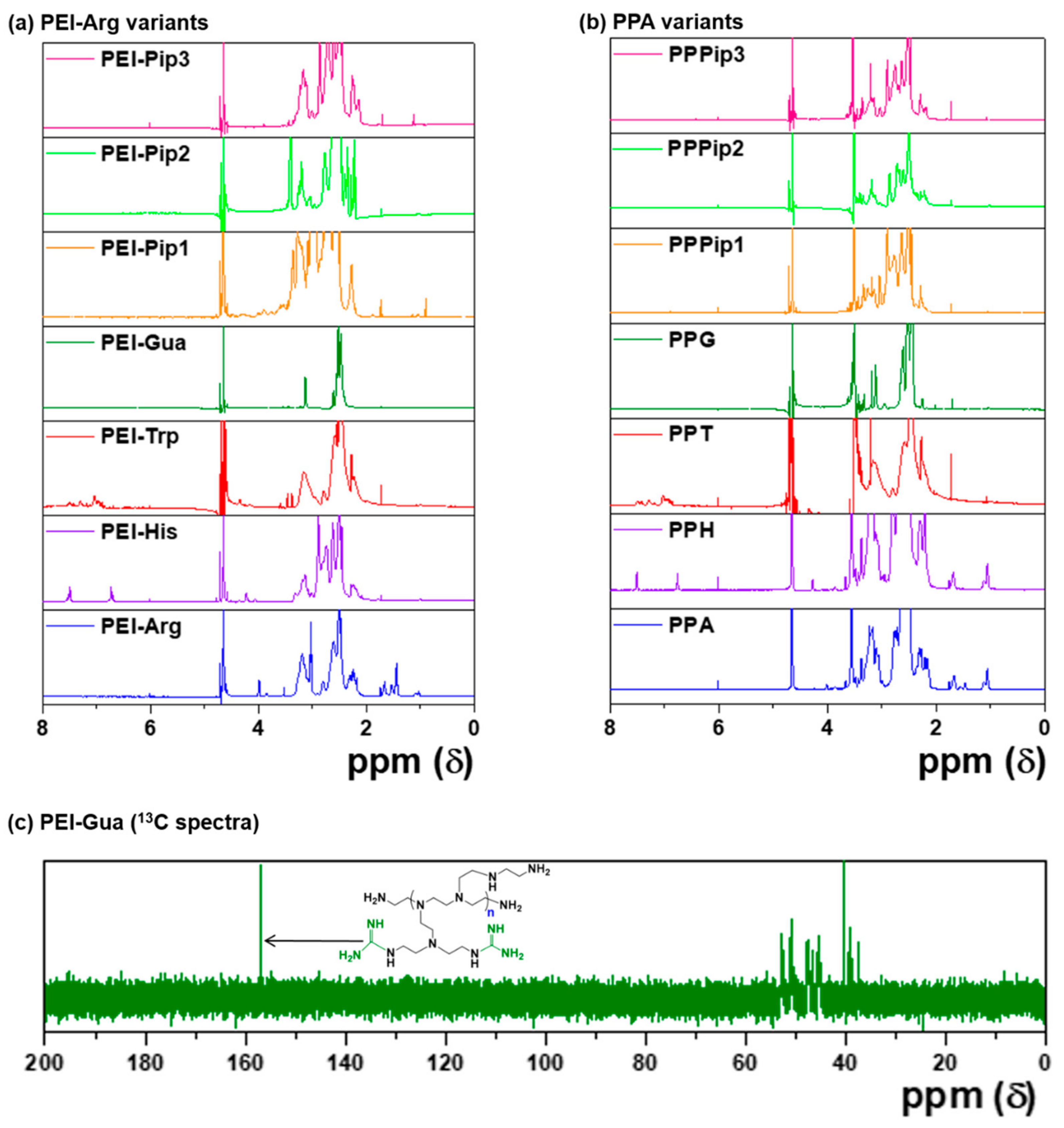
3.1. Synthesis and Characterization of PPA Copolymers
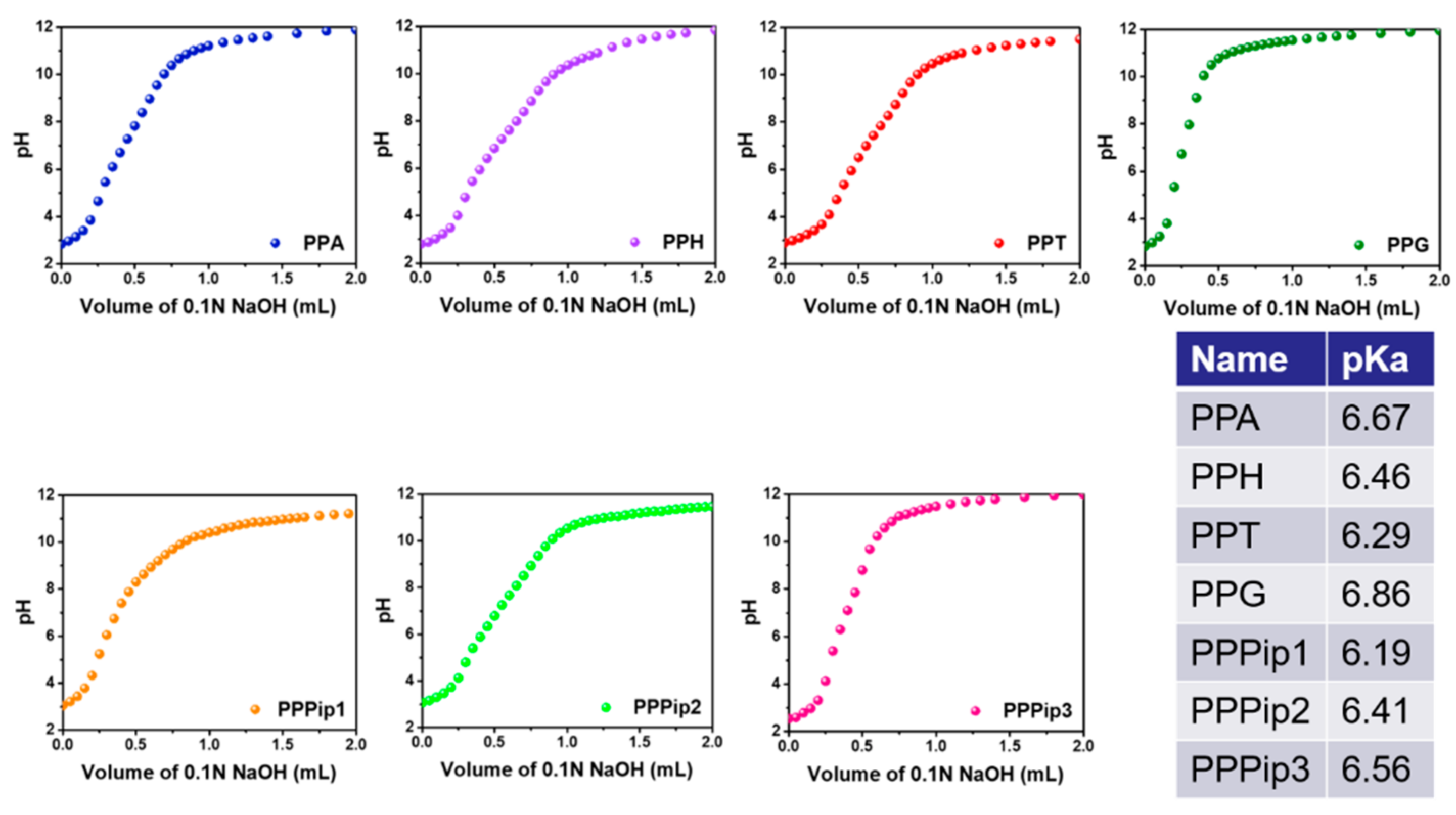
3.2. pH-Buffering Capacity of PPA Copolymers
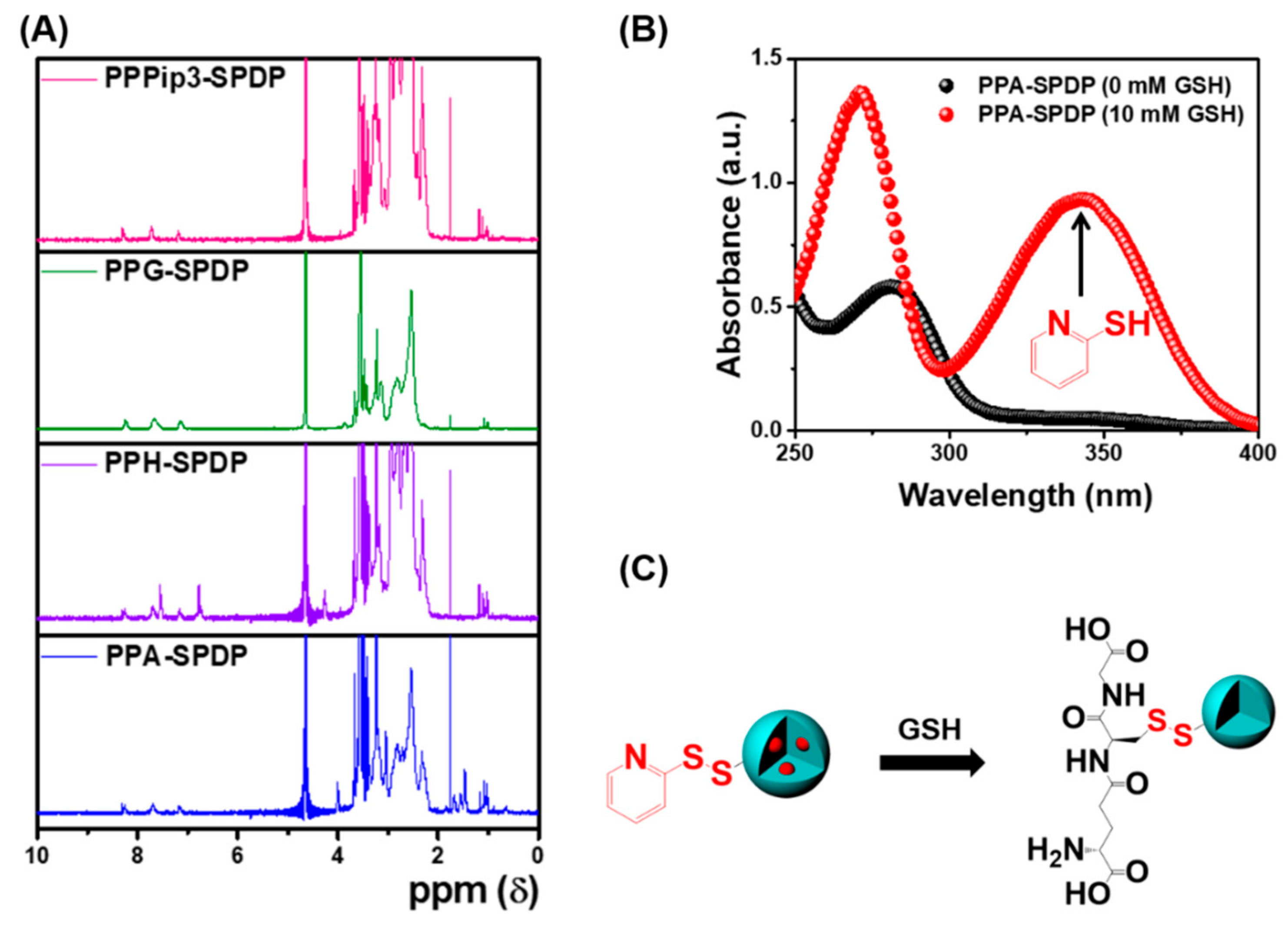
3.3. Characterization of PPA-SPDP Copolymers
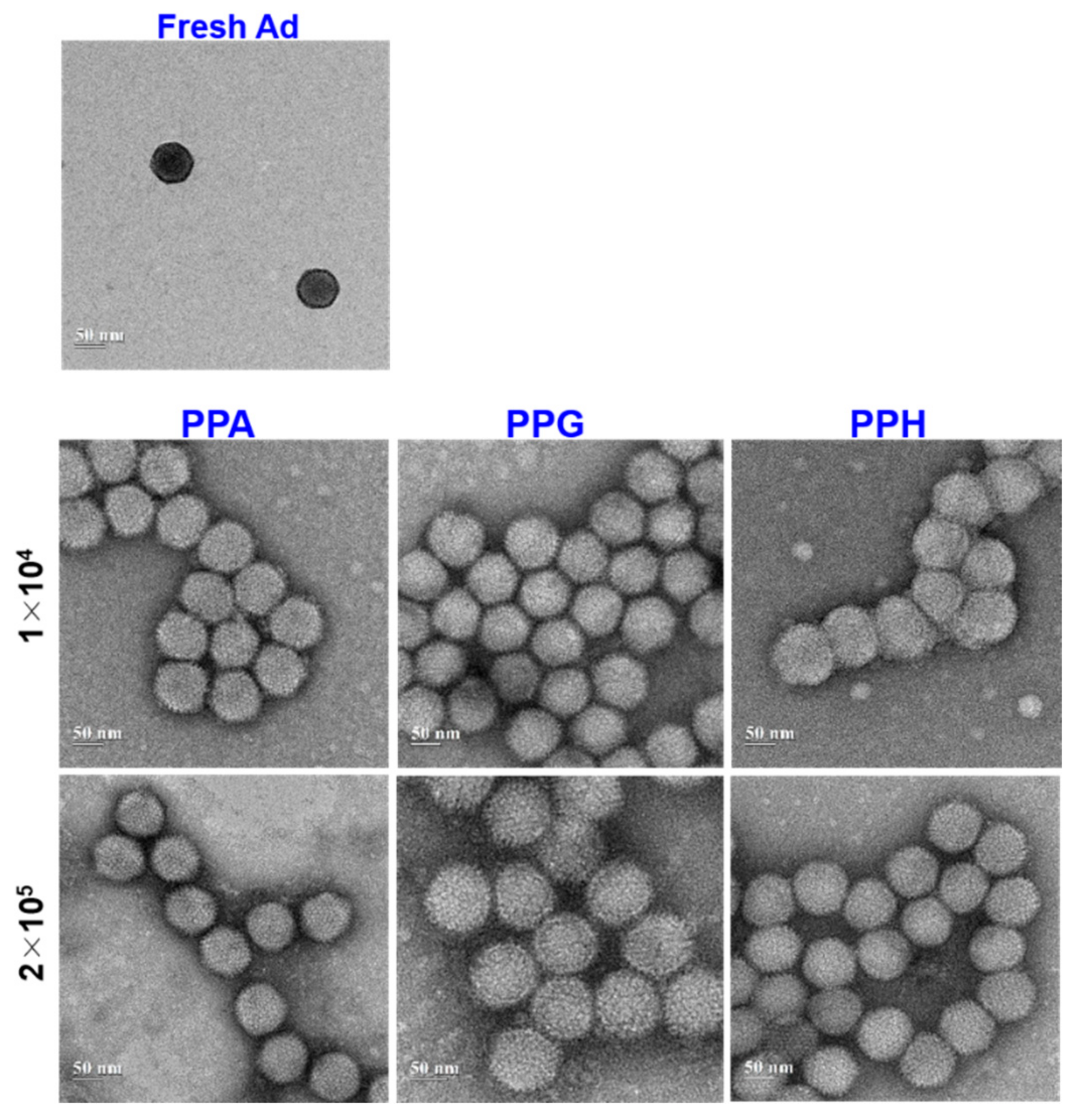
3.4. Physiochemical Characterization of PPA-Ad Conjugates
3.5. Gel Retardation Assay
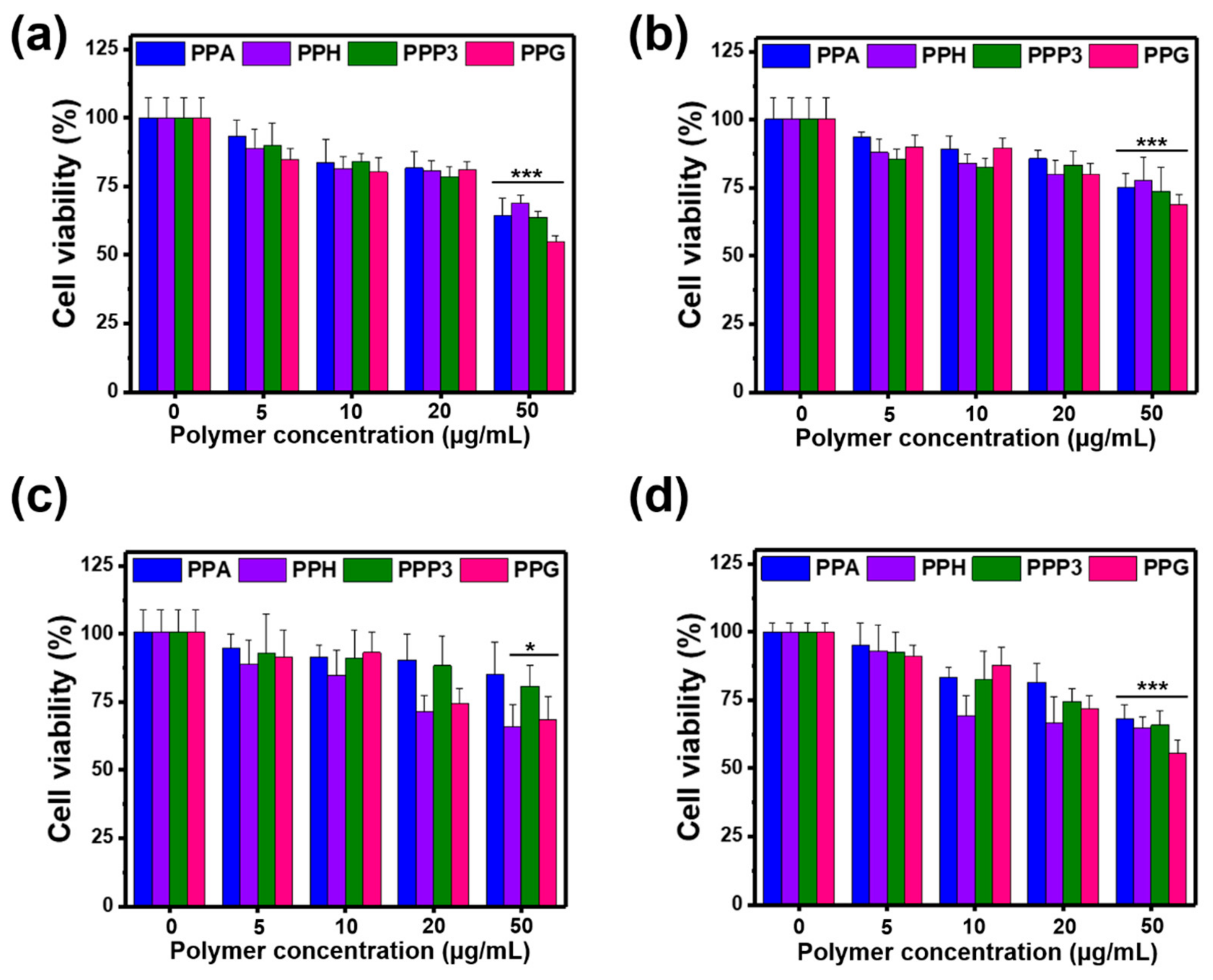
3.6. Cytotoxicity Test of PPA Copolymers
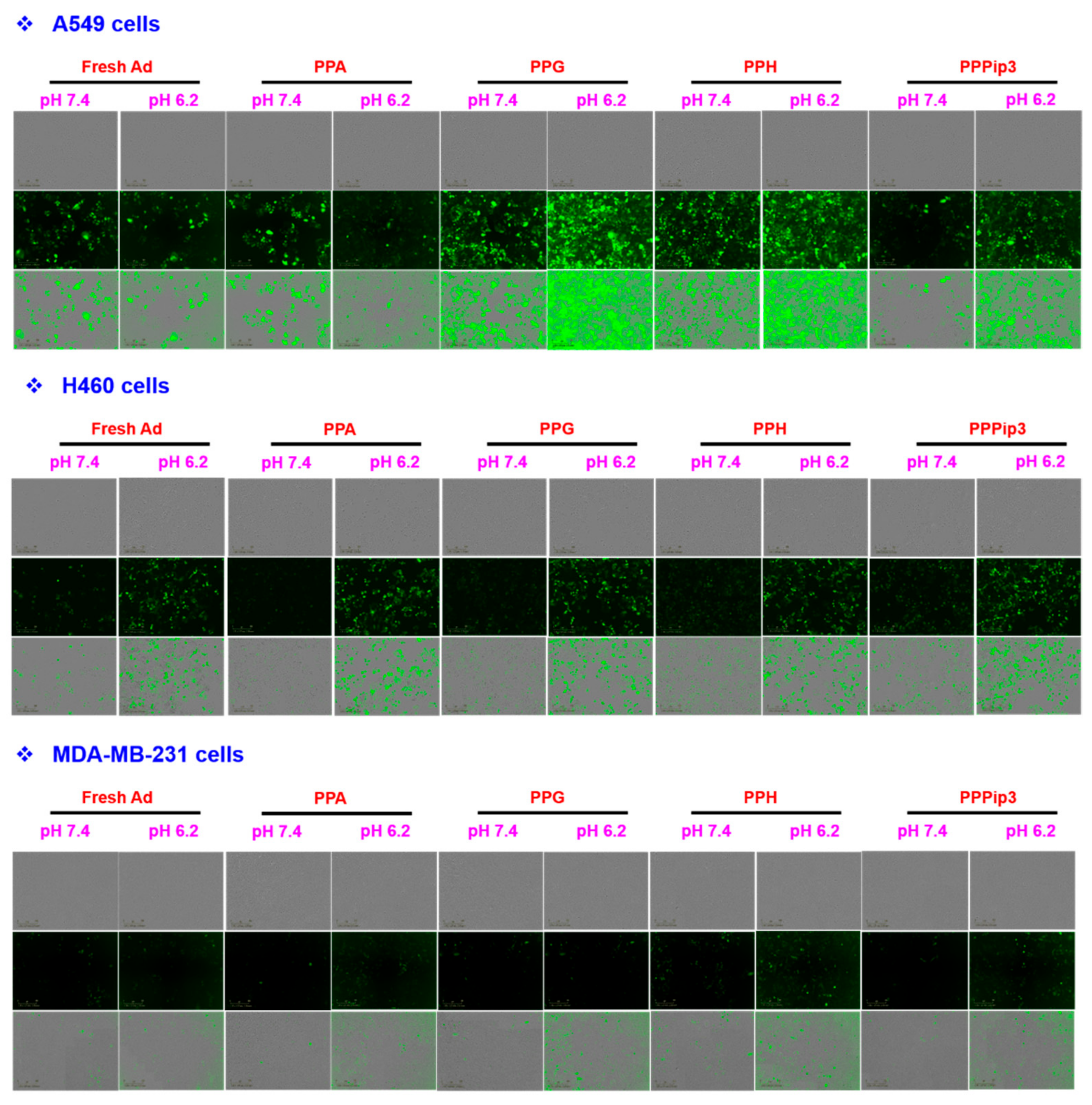
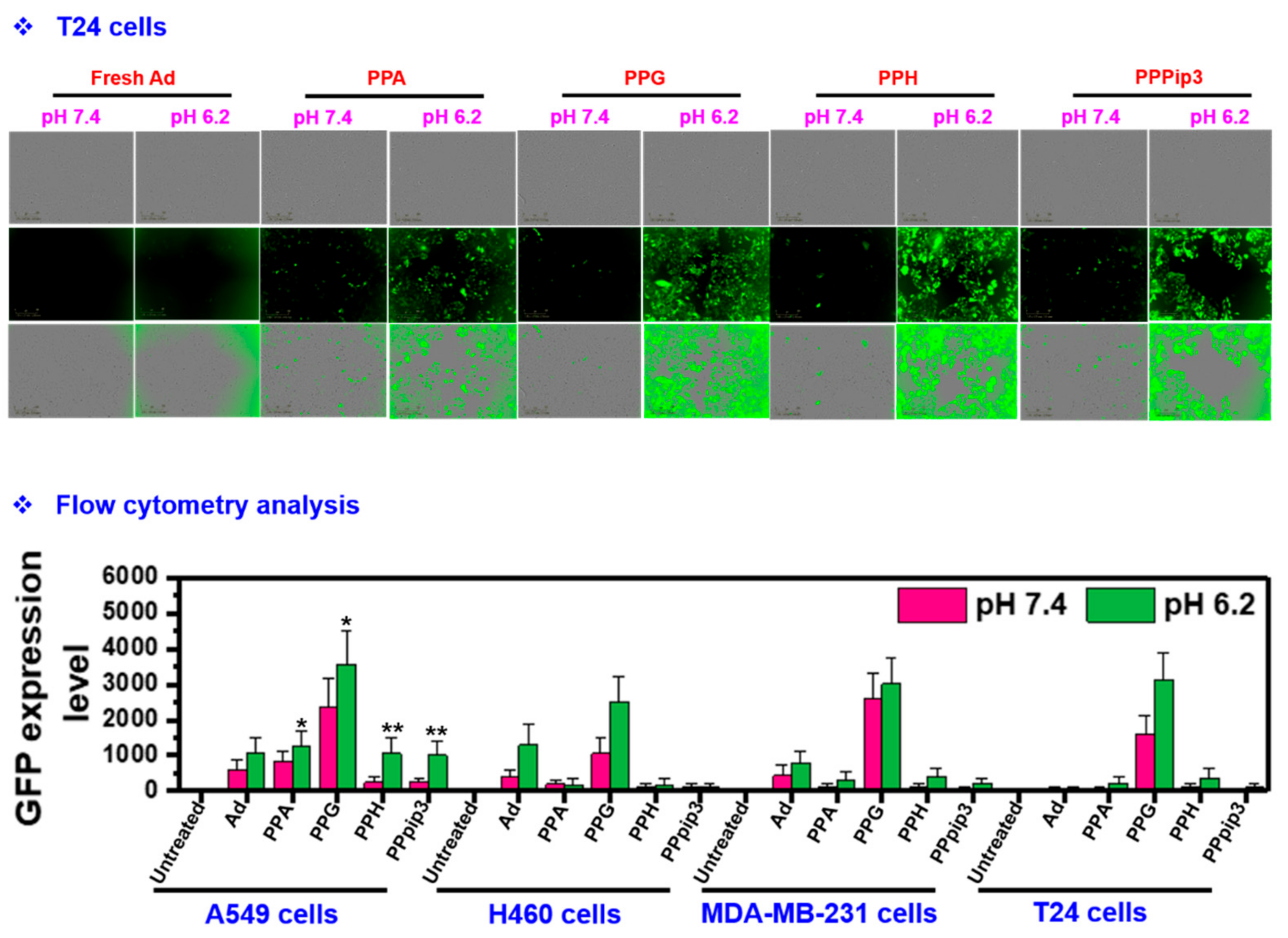
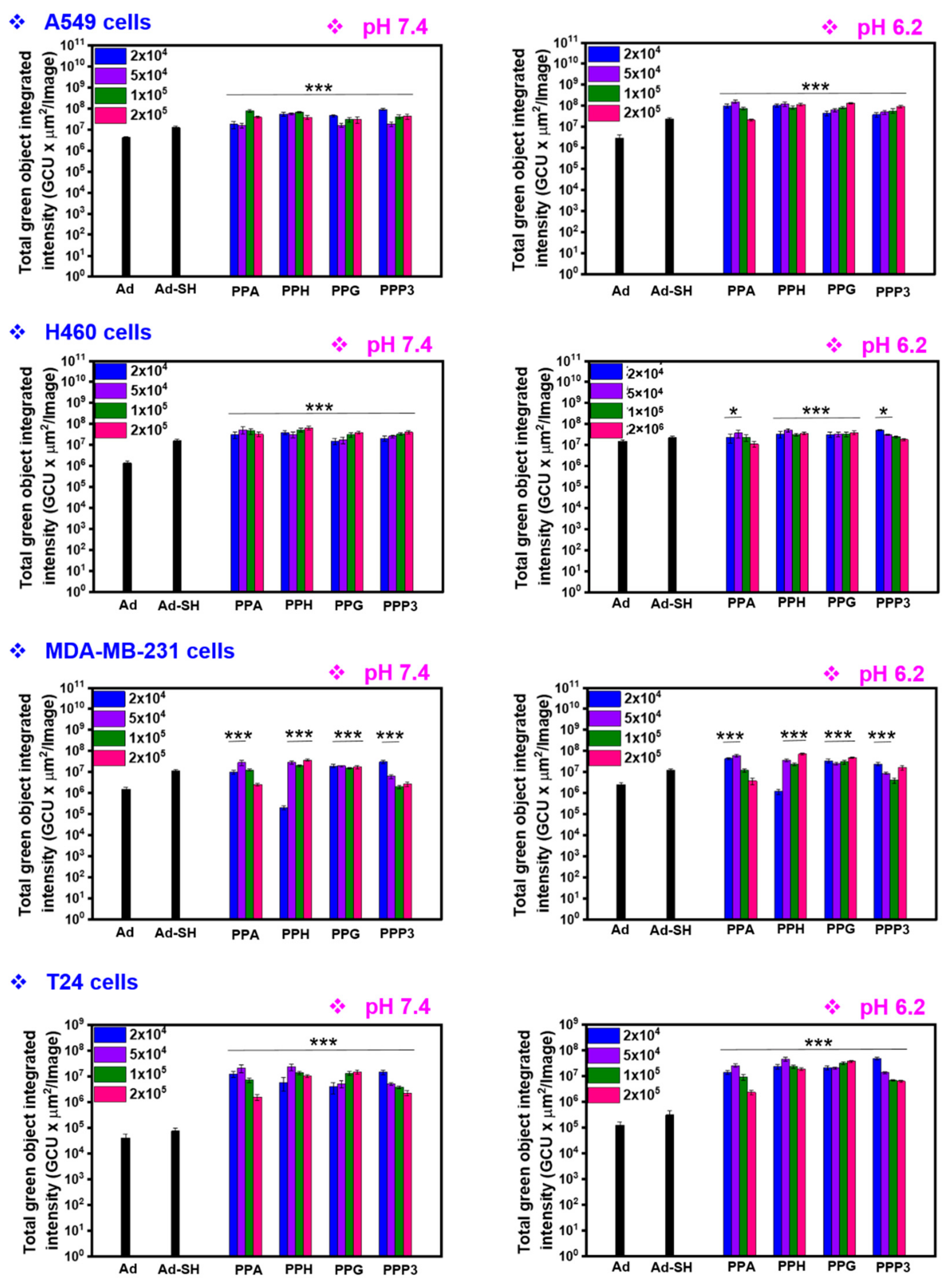
3.7. Transduction Efficiency of PPA-Ad Complex
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bulaklak, K.; Gersbach, C.A. The once and future gene therapy. Nat. Commun. 2020, 11, 5820. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorth, M.; Narvekar, A. Non viral vectors in gene therapy- an overview. J. Clin. Diagn. Res. 2015, 9, GE01–GE06. [Google Scholar] [CrossRef] [PubMed]
- Santana-Armas, M.L.; Tros de Ilarduya, C. Strategies for cancer gene-delivery improvement by non-viral vectors. Int. J. Pharm. 2021, 596, 120291. [Google Scholar] [CrossRef] [PubMed]
- Chira, S.; Jackson, C.S.; Oprea, I.; Ozturk, F.; Pepper, M.S.; Diaconu, I.; Braicu, C.; Raduly, L.-Z.; Calin, G.A.; Berindan-Neagoe, I. Progresses towards safe and efficient gene therapy vectors. Oncotarget 2015, 6, 30675–30703. [Google Scholar] [CrossRef] [Green Version]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Waehler, R.; Russell, S.J.; Curiel, D.T. Engineering targeted viral vectors for gene therapy. Nat. Rev. Genet. 2007, 8, 573–587. [Google Scholar] [CrossRef]
- Finer, M.; Glorioso, J. A brief account of viral vectors and their promise for gene therapy. Gene Ther. 2017, 24, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Goswami, R.; Subramanian, G.; Silayeva, L.; Newkirk, I.; Doctor, D.; Chawla, K.; Chattopadhyay, S.; Chandra, D.; Chilukuri, N.; Betapudi, V. Gene Therapy Leaves a Vicious Cycle. Front. Oncol. 2019, 9, 297. [Google Scholar] [CrossRef]
- Lugin, M.L.; Lee, R.T.; Kwon, Y.J. Synthetically Engineered Adeno-Associated Virus for Efficient, Safe, and Versatile Gene Therapy Applications. ACS Nano 2020, 14, 14262–14283. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Kohlhapp, F.J.; Zloza, A. Oncolytic viruses: A new class of immunotherapy drugs. Nat. Rev. Drug Discov. 2015, 14, 642–662. [Google Scholar] [CrossRef]
- Bommareddy, P.K.; Shettigar, M.; Kaufman, H.L. Integrating oncolytic viruses in combination cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 498–513. [Google Scholar] [CrossRef] [PubMed]
- Le, T.M.D.; Duong, H.T.T.; Thambi, T.; Giang Phan, V.H.; Jeong, J.H.; Lee, D.S. Bioinspired pH- and Temperature-Responsive Injectable Adhesive Hydrogels with Polyplexes Promotes Skin Wound Healing. Biomacromolecules 2018, 19, 3536–3548. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Hong, J.W.; Thambi, T.; Yoon, A.-R.; Choi, J.-W.; Li, Y.; Bui, Q.N.; Lee, D.S.; Yun, C.-O. Optimizing Active Tumor Targeting Biocompatible Polymers for Efficient Systemic Delivery of Adenovirus. Cells 2021, 10, 1896. [Google Scholar] [CrossRef]
- Hensen, L.C.M.; Hoeben, R.C.; Bots, S.T.F. Adenovirus Receptor Expression in Cancer and Its Multifaceted Role in Oncolytic Adenovirus Therapy. Int. J. Mol. Sci. 2020, 21, 6828. [Google Scholar] [CrossRef] [PubMed]
- Bilbao, R.; Srinivasan, S.; Reay, D.; Goldberg, L.; Hughes, T.; Roelvink, P.W.; Einfeld, D.A.; Wickham, T.J.; Clemens, P.R. Binding of adenoviral fiber knob to the coxsackievirus-adenovirus receptor is crucial for transduction of fetal muscle. Hum. Gene Ther. 2003, 14, 645–649. [Google Scholar] [CrossRef]
- Lyle, C.; McCormick, F. Integrin alphavbeta5 is a primary receptor for adenovirus in CAR-negative cells. Virol. J. 2010, 7, 148. [Google Scholar] [CrossRef] [Green Version]
- Mizuguchi, H.; Hayakawa, T. Targeted Adenovirus Vectors. Hum. Gene Ther. 2004, 15, 1034–1044. [Google Scholar] [CrossRef]
- Grove, J.; Marsh, M. The cell biology of receptor-mediated virus entry. J. Cell Biol. 2011, 195, 1071–1082. [Google Scholar] [CrossRef] [Green Version]
- Stoiber, S.; Cadilha, B.L.; Benmebarek, M.-R.; Lesch, S.; Endres, S.; Kobold, S. Limitations in the Design of Chimeric Antigen Receptors for Cancer Therapy. Cells 2019, 8, 472. [Google Scholar] [CrossRef] [Green Version]
- Reeh, M.; Bockhorn, M.; Görgens, D.; Vieth, M.; Hoffmann, T.; Simon, R.; Izbicki, J.R.; Sauter, G.; Schumacher, U.; Anders, M. Presence of the coxsackievirus and adenovirus receptor (CAR) in human neoplasms: A multitumour array analysis. Br. J. Cancer 2013, 109, 1848–1858. [Google Scholar] [CrossRef]
- Seidman, M.A.; Hogan, S.M.; Wendland, R.L.; Worgall, S.; Crystal, R.G.; Leopold, P.L. Variation in Adenovirus Receptor Expression and Adenovirus Vector-Mediated Transgene Expression at Defined Stages of the Cell Cycle. Mol. Ther. 2001, 4, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Gao, Y.-G.; Lin, X.; Li, Y.; Dang, K.; Tian, Y.; Zhang, W.-J.; Jiang, S.-F.; Qadir, A.; Qian, A.-R. The Development of Functional Non-Viral Vectors for Gene Delivery. Int. J. Mol. Sci. 2019, 20, 5491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, Y.K.; Kim, S.W. Recent advances in the development of gene delivery systems. Biomater. Res. 2019, 23, 8. [Google Scholar] [CrossRef] [PubMed]
- Morille, M.; Passirani, C.; Vonarbourg, A.; Clavreul, A.; Benoit, J.-P. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials 2008, 29, 3477–3496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Bruggen, C.; Hexum, J.K.; Tan, Z.; Dalal, R.J.; Reineke, T.M. Nonviral Gene Delivery with Cationic Glycopolymers. Acc. Chem. Res. 2019, 52, 1347–1358. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Kim, J.; Kim, P.-H.; Kim, S.W.; Yun, C.-O. Enhancing the therapeutic efficacy of adenovirus in combination with biomaterials. Biomaterials 2012, 33, 1838–1850. [Google Scholar] [CrossRef] [Green Version]
- Duong, H.T.T.; Yin, Y.; Thambi, T.; Nguyen, T.L.; Giang Phan, V.H.; Lee, M.S.; Lee, J.E.; Kim, J.; Jeong, J.H.; Lee, D.S. Smart vaccine delivery based on microneedle arrays decorated with ultra-pH-responsive copolymers for cancer immunotherapy. Biomaterials 2018, 185, 13–24. [Google Scholar] [CrossRef]
- Lara-Velazquez, M.; Alkharboosh, R.; Norton, E.S.; Ramirez-Loera, C.; Freeman, W.D.; Guerrero-Cazares, H.; Forte, A.J.; Quiñones-Hinojosa, A.; Sarabia-Estrada, R. Chitosan-Based Non-viral Gene and Drug Delivery Systems for Brain Cancer. Front. Neurol. 2020, 11, 740. [Google Scholar] [CrossRef]
- Lee, C.-H.; Kasala, D.; Na, Y.; Lee, M.S.; Kim, S.W.; Jeong, J.H.; Yun, C.-O. Enhanced therapeutic efficacy of an adenovirus-PEI-bile-acid complex in tumors with low coxsackie and adenovirus receptor expression. Biomaterials 2014, 35, 5505–5516. [Google Scholar] [CrossRef]
- Manouchehri, S.; Zarrintaj, P.; Saeb, M.R.; Ramsey, J.D. Advanced Delivery Systems Based on Lysine or Lysine Polymers. Mol. Pharm. 2021, 18, 3652–3670. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, H.; Liu, J.; Wang, Z. Poly (amidoamine) (PAMAM) dendrimer mediated delivery of drug and pDNA/siRNA for cancer therapy. Int. J. Pharm. 2018, 546, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Kamaly, N.; Yameen, B.; Wu, J.; Farokhzad, O.C. Degradable Controlled-Release Polymers and Polymeric Nanoparticles: Mechanisms of Controlling Drug Release. Chem. Rev. 2016, 116, 2602–2663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshayes, S.; Kasko, A.M. Polymeric biomaterials with engineered degradation. J. Polym. Sci. Part A Polym. Chem. 2013, 51, 3531–3566. [Google Scholar] [CrossRef]
- Elmowafy, E.M.; Tiboni, M.; Soliman, M.E. Biocompatibility, biodegradation and biomedical applications of poly(lactic acid)/poly(lactic-co-glycolic acid) micro and nanoparticles. J. Pharm. Investig. 2019, 49, 347–380. [Google Scholar] [CrossRef]
- Visan, A.I.; Popescu-Pelin, G.; Socol, G. Degradation Behavior of Polymers Used as Coating Materials for Drug Delivery-A Basic Review. Polymers 2021, 13, 1272. [Google Scholar] [CrossRef]
- Schmid, M.; Ernst, P.; Honegger, A.; Suomalainen, M.; Zimmermann, M.; Braun, L.; Stauffer, S.; Thom, C.; Dreier, B.; Eibauer, M.; et al. Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control. Nat. Commun. 2018, 9, 450. [Google Scholar] [CrossRef]
- Chen, C.-K.; Huang, P.-K.; Law, W.-C.; Chu, C.-H.; Chen, N.-T.; Lo, L.-W. Biodegradable Polymers for Gene-Delivery Applications. Int. J. Nanomed. 2020, 15, 2131–2150. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Lv, X.; Ding, P.; Wang, L.; Sun, Y.; Li, S.; Zhang, H.; Gao, Z. Exploring the functions of polymers in adenovirus-mediated gene delivery: Evading immune response and redirecting tropism. Acta Biomater. 2019, 97, 93–104. [Google Scholar] [CrossRef]
- Kasala, D.; Lee, S.-H.; Hong, J.W.; Choi, J.-W.; Nam, K.; Chung, Y.H.; Kim, S.W.; Yun, C.-O. Synergistic antitumor effect mediated by a paclitaxel-conjugated polymeric micelle-coated oncolytic adenovirus. Biomaterials 2017, 145, 207–222. [Google Scholar] [CrossRef]
- Verbeke, C.S.; Mooney, D.J. Injectable, Pore-Forming Hydrogels for In Vivo Enrichment of Immature Dendritic Cells. Adv. Healthc. Mater. 2015, 4, 2677–2687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turabee, M.H.; Thambi, T.; Lym, J.S.; Lee, D.S. Bioresorbable polypeptide-based comb-polymers efficiently improves the stability and pharmacokinetics of proteins in vivo. Biomater. Sci. 2017, 5, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Kaygisiz, K.; Synatschke, C.V. Materials promoting viral gene delivery. Biomater. Sci. 2020, 8, 6113–6156. [Google Scholar] [CrossRef] [PubMed]
- Kasala, D.; Yoon, A.R.; Hong, J.; Kim, S.W.; Yun, C.-O. Evolving lessons on nanomaterial-coated viral vectors for local and systemic gene therapy. Nanomed. (Lond) 2016, 11, 1689–1713. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.-W.; Lee, J.-S.; Kim, S.W.; Yun, C.-O. Evolution of oncolytic adenovirus for cancer treatment. Adv. Drug Deliv. Rev. 2012, 64, 720–729. [Google Scholar] [CrossRef]
- Gil, M.S.; Cho, J.; Thambi, T.; Giang Phan, V.H.; Kwon, I.; Lee, D.S. Bioengineered robust hybrid hydrogels enrich the stability and efficacy of biological drugs. J. Control. Release 2017, 267, 119–132. [Google Scholar] [CrossRef]
- Thambi, T.; Deepagan, V.G.; Yoon, H.Y.; Han, H.S.; Kim, S.-H.; Son, S.; Jo, D.-G.; Ahn, C.-H.; Suh, Y.D.; Kim, K.; et al. Hypoxia-responsive polymeric nanoparticles for tumor-targeted drug delivery. Biomaterials 2014, 35, 1735–1743. [Google Scholar] [CrossRef]
- Gil, M.S.; Thambi, T.; Phan, V.H.G.; Kim, S.H.; Lee, D.S. Injectable hydrogel-incorporated cancer cell-specific cisplatin releasing nanogels for targeted drug delivery. J. Mater. Chem. B 2017, 5, 7140–7152. [Google Scholar] [CrossRef]
- Hu, J.; Zhao, W.; Liu, K.; Yu, Q.; Mao, Y.; Lu, Z.; Zhang, Y.; Zhu, M. Low-Molecular Weight Polyethylenimine Modified with Pluronic 123 and RGD- or Chimeric RGD-NLS Peptide: Characteristics and Transfection Efficacy of Their Complexes with Plasmid DNA. Molecules 2016, 21, 655. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, S.; Qu, Y.; Dong, Z.; Zhao, J.; Rauf Khan, A.; Rehman, S.; Zhao, Z. Poly (β-amino esters) based potential drug delivery and targeting polymer; an overview and perspectives (review). Eur. Polym. J. 2020, 141, 110097. [Google Scholar] [CrossRef]
- Thambi, T.; Li, Y.; Lee, D.S. Injectable hydrogels for sustained release of therapeutic agents. J. Control. Release 2017, 267, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Wang, K.; Zhao, Z.; Zhang, Y.; Sun, T.; Zhang, F.; Wu, J.; Fu, Y.; Du, Y.; Zhang, L.; et al. Brain-Targeted Delivery of Trans-Activating Transcriptor-Conjugated Magnetic PLGA/Lipid Nanoparticles. PLoS ONE 2014, 9, e106652. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.-J.; Kasala, D.; Choi, J.-W.; Lee, S.-H.; Hwang, J.K.; Kim, S.W.; Yun, C.-O. Safety Profiles and Antitumor Efficacy of Oncolytic Adenovirus Coated with Bioreducible Polymer in the Treatment of a CAR Negative Tumor Model. Biomacromolecules 2015, 16, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Suk, J.S.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutlar, L.; Zhou, D.; Gao, Y.; Zhao, T.; Greiser, U.; Wang, W.; Wang, W. Highly Branched Poly(β-Amino Esters): Synthesis and Application in Gene Delivery. Biomacromolecules 2015, 16, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-W.; Nam, J.-P.; Nam, K.; Lee, Y.S.; Yun, C.-O.; Kim, S.W. Oncolytic Adenovirus Coated with Multidegradable Bioreducible Core-Cross-Linked Polyethylenimine for Cancer Gene Therapy. Biomacromolecules 2015, 16, 2132–2143. [Google Scholar] [CrossRef]
- Agarwal, P.; Gammon, E.A.; Sajib, A.M.; Sandey, M.; Smith, B.F. Cell-Surface Integrins and CAR Are Both Essential for Adenovirus Type 5 Transduction of Canine Cells of Lymphocytic Origin. PLoS ONE 2017, 12, e0169532. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhang, L.; Zhu, W.; Guo, R.; Sun, H.; Chen, X.; Deng, N. Barriers and Strategies of Cationic Liposomes for Cancer Gene Therapy. Mol. Ther. Methods Clin. Dev. 2020, 18, 751–764. [Google Scholar] [CrossRef]



| Library | PPCBA (Degree of Polymerization) | Acrylate Consumption (%) | bPEI Substitution Moieties | Degree of Substitution (Each Moieties) a | Degree of Substitution of SPDP a |
|---|---|---|---|---|---|
| PPA | 8.08 | 100 | Arginine | 5.03 | 2.36 |
| PPH | 8.08 | 100 | Histidine | 4.93 | 1.98 |
| PPT | 8.08 | 100 | Tryptophan | 4.72 | 1.71 |
| PPG | 8.08 | 100 | Guanidine | 3.86 | 1.78 |
| PPPip1 | 8.08 | 100 | Piperazine | 4.66 | 2.61 |
| PPPip2 | 8.08 | 100 | Piperazine | 3.64 | 2.05 |
| PPPip3 | 8.08 | 100 | Piperazine | 4.54 | 2.72 |
| Ad | Ad-SH | 1 × 104 | 2 × 104 | 5 × 104 | 1 × 105 | 2 × 105 | |
|---|---|---|---|---|---|---|---|
| Ad | 5209.719 | 4537.426 | 4419.841 | 3289.477 | 2776.355 | 231.556 | 91.435 |
| Ad | 2851.991 | 2762.406 | 2694.941 | 1672.941 | 328.385 | * | * |
| Ad | 2762.820 | 2690.234 | * | * | * | * | * |
| Ad | 3706.184 | 3220.062 | 622.335 | 435.385 | 46.192 | * | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thambi, T.; Lee, J.; Yoon, A.-R.; Kasala, D.; Yun, C.-O. A pH- and Bioreducible Cationic Copolymer with Amino Acids and Piperazines for Adenovirus Delivery. Pharmaceutics 2022, 14, 597. https://doi.org/10.3390/pharmaceutics14030597
Thambi T, Lee J, Yoon A-R, Kasala D, Yun C-O. A pH- and Bioreducible Cationic Copolymer with Amino Acids and Piperazines for Adenovirus Delivery. Pharmaceutics. 2022; 14(3):597. https://doi.org/10.3390/pharmaceutics14030597
Chicago/Turabian StyleThambi, Thavasyappan, Jeongmin Lee, A-Rum Yoon, Dayananda Kasala, and Chae-Ok Yun. 2022. "A pH- and Bioreducible Cationic Copolymer with Amino Acids and Piperazines for Adenovirus Delivery" Pharmaceutics 14, no. 3: 597. https://doi.org/10.3390/pharmaceutics14030597
APA StyleThambi, T., Lee, J., Yoon, A.-R., Kasala, D., & Yun, C.-O. (2022). A pH- and Bioreducible Cationic Copolymer with Amino Acids and Piperazines for Adenovirus Delivery. Pharmaceutics, 14(3), 597. https://doi.org/10.3390/pharmaceutics14030597









