Relevance of CYP2D6 Gene Variants in Population Genetic Differentiation
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Larrey, D.; Distlerath, L.M.; Dannan, G.A.; Wilkinson, G.R.; Guengerich, F.P. Purification and characterization of the rat liver microsomal cytochrome P-450 involved in the 4-hydroxylation of debrisoquine, a prototype for genetic variation in oxidative drug metabolism. Biochemistry 1984, 23, 2787–2795. [Google Scholar] [CrossRef] [PubMed]
- Eichelbaum, M.; Baur, M.; Dengler, H.; Osikowska-Evers, B.; Tieves, G.; Zekorn, C.; Rittner, C. Chromosomal assignment of human cytochrome P-450 (debrisoquine/sparteine type) to chromosome 22. Br. J. Clin. Pharmacol. 1987, 23, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Umeno, M.; Skoda, R.C.; A Meyer, U.; Gonzalez, F.J. The human debrisoquine 4-hydroxylase (CYP2D) locus: Sequence and identification of the polymorphic CYP2D6 gene, a related gene, and a pseudogene. Am. J. Hum. Genet. 1989, 45, 889–904. [Google Scholar] [PubMed]
- Daly, A.K.; Brockmoller, J.; Broly, F.; Eichelbaum, M.; E Evans, W.; Gonzalez, F.J.; Huang, J.D.; Idle, J.R.; Ingelman-Sundberg, M.; Ishizaki, T.; et al. Nomenclature for human CYP2D6 alleles. Pharmacogenetics 1996, 6, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Gresham, D.; Morar, B.; Underhill, P.A.; Passarino, G.; Lin, A.A.; Wise, C.; Angelicheva, D.; Calafell, F.; Oefner, P.J.; Shen, P.; et al. Origins and Divergence of the Roma (Gypsies). Am. J. Hum. Genet. 2001, 69, 1314–1331. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.; Crosby, I.; Yip, V.; Maguire, P.; Pirmohamed, M.; Turner, R.M. A Review of the Important Role of CYP2D6 in Pharmacogenomics. Genes 2020, 11, 1295. [Google Scholar] [CrossRef] [PubMed]
- Caudle, K.E.; Sangkuhl, K.; Whirl-Carrillo, M.; Swen, J.J.; Haidar, C.E.; Klein, T.E.; Gammal, R.S.; Relling, M.V.; Scott, S.A.; Hertz, D.L.; et al. Standardizing CYP 2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 2020, 13, 116–124. [Google Scholar] [CrossRef]
- The Human Cythchrome P450 (CYP) Allele Nomenclature Database. Available online: http://cypalleles.ki.se/ (accessed on 15 April 2022).
- PHARMGKB. Stanford University. Website. Available online: https://www.pharmgkb.org (accessed on 13 December 2021).
- Hicks, J.K.; Swen, J.J.; Gaedigk, A. Challenges in CYP2D6 Phenotype Assignment from Genotype Data: A Critical Assessment and Call for Standardization. Curr. Drug Metab. 2014, 15, 218–232. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Williams, I.S.; Gatchie, L.; Bharate, S.B.; Chaudhuri, B. Biotransformation, Using Recombinant CYP450-Expressing Baker’s Yeast Cells, Identifies a Novel CYP2D6.10A122V Variant Which Is a Superior Metabolizer of Codeine to Morphine Than the Wild-Type Enzyme. ACS Omega 2018, 3, 8903–8912. [Google Scholar] [CrossRef] [PubMed]
- Ingelman-Sundberg, M.; Sim, S.C.; Gomez, A.; Rodriguez-Antona, C. Influence of cytochrome P450 polymor-phisms on drug therapies: Pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 2007, 116, 496–526. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Turpeinen, M.; Klein, K.; Schwab, M. Functional pharmacogenetics/genomics of human cyto-chromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 2008, 392, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Fleeman, N.; Dundar, Y.; Dickson, R.; Jorgensen, A.; Pushpakom, S.; McLeod, C.; Pirmohamed, M.; Walley, T. Cytochrome P450 testing for prescribing antipsychotics in adults with schizophrenia: Systematic review and meta-analyses. Pharm. J. 2010, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Stingl, J.C.; Brockmöller, J.; Viviani, R. Genetic variability of drug-metabolizing enzymes: The dual impact on psychiatric therapy and regulation of brain function. Mol. Psychiatry 2012, 18, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Gaedigk, A. Complexities of CYP2D6 gene analysis and interpretation. Int. Rev. Psychiatry 2013, 25, 534–553. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.K.; Bishop, J.R.; Sangkuhl, K.; Müller, D.J.; Ji, Y.; Leckband, S.G.; Leeder, J.S.; Graham, R.L.; Chiulli, D.L.; Llerena, A.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther. 2015, 98, 127–134. [Google Scholar] [CrossRef]
- Beoris, M.; Wilson, J.A.; Garces, J.A.; Lukowiak, A.A. CYP2D6 copy number distribution in the US population. Pharm. Genom. 2016, 26, 96–99. [Google Scholar] [CrossRef][Green Version]
- Christensen, P.M.; Gotzsche, P.C.; Brosen, K. The sparteine/debrisoquine (CYP2D6) oxidation polymorphism and the risk of Parkinson’s disease: A meta-analysis. Pharmacogenetics 1998, 8, 473–479. [Google Scholar] [CrossRef]
- Lu, Y.; Peng, Q.; Zeng, Z.; Wang, J.; Deng, Y.; Xie, L.; Mo, C.; Zeng, J.; Qin, X.; Li, S. CYP2D6 phenotypes and Parkinson’s disease risk: A meta-analysis. J. Neurol. Sci. 2014, 336, 161–168. [Google Scholar] [CrossRef]
- Mishra, A.K.; Singh, M.P. Cytochrome P450 2D6 and Parkinson’s Disease: Polymorphism, Metabolic Role, Risk and Protection. Neurochem. Res. 2017, 42, 3353–3361. [Google Scholar] [CrossRef]
- Patsopoulos, N.; Ntzani, E.E.; Zintzaras, E.; Ioannidis, J.P. CYP2D6 polymorphisms and the risk of tardive dyskinesia in schizophrenia: A meta-analysis. Pharm. Genom. 2005, 15, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Scordo, M.G.; Dahl, M.-L.; Spina, E.; Cordici, F.; Arena, M.G. No association between CYP2D6 polymorphism and Alzheimer’s disease in an Italian population. Pharmacol. Res. 2006, 53, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.L.; Tang, N.L.S.; Wat, K.H.Y.; Tang, J.H.Y.; Lau, K.H.; Law, C.B.; Chiu, J.; Tam, C.C.W.; Poon, T.K.; Lin, K.L.; et al. Effect of CYP2D6 and CYP3A4 Genotypes on the Efficacy of Cholinesterase Inhibitors in Southern Chinese Patients With Alzheimer’s Disease. Am. J. Alzheimer’s Dis. Other Dement. 2019, 34, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Agundez, J.A. Cytochrome P450 Gene Polymorphism and Cancer. Curr. Drug Metab. 2004, 5, 211–224. [Google Scholar] [CrossRef]
- Rodriguez-Antona, C.; Gomez, A.; Karlgren, M.; Sim, S.C.; Ingelman-Sundberg, M. Molecular genetics and epigenetics of the cytochrome P450 gene family and its relevance for cancer risk and treatment. Qual. Life Res. 2009, 127, 1–17. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functional diversity. Pharm. J. 2004, 5, 6–13. [Google Scholar] [CrossRef]
- Aklillu, E.; Herrlin, K.; Gustafsson, L.L.; Bertilsson, L.; Ingelman-Sundberg, M. Evidence for environmental influence on CYP2D6-catalysed debrisoquine hydroxylation as demonstrated by phenotyping and genotyping of Ethiopians living in Ethiopia or in Sweden. Pharmacogenetics 2002, 12, 375–383. [Google Scholar] [CrossRef]
- Podgorná, E.; Diallo, I.; Vangenot, C.; Sanchez-Mazas, A.; Sabbagh, A.; Černý, V.; Poloni, E.S. Variation in NAT2 acetylation phenotypes is associated with differences in food-producing subsistence modes and ecoregions in Africa. BMC Evol. Biol. 2015, 15, 263. [Google Scholar] [CrossRef]
- Fuselli, S.; de Filippo, C.; Mona, S.; Sistonen, J.; Fariselli, P.; Destro-Bisol, G.; Barbujani, G.; Bertorelle, G.; Sajantila, A. Evolution of detoxifying systems: The role of environment and population history in shaping genetic diversity at human CYP2D6 locus. Pharm. Genom. 2010, 20, 485–499. [Google Scholar] [CrossRef]
- Thomas, J.H. Rapid Birth–Death Evolution Specific to Xenobiotic Cytochrome P450 Genes in Vertebrates. PLoS Genet. 2007, 3, e67. [Google Scholar] [CrossRef]
- Delser, P.M.; Fuselli, S. Human loci involved in drug biotransformation: Worldwide genetic variation, population structure, and pharmacogenetic implications. Qual. Life Res. 2013, 132, 563–577. [Google Scholar] [CrossRef]
- Fuselli, S. Beyond drugs: The evolution of genes involved in human response to medications. Proc. R. Soc. B Boil. Sci. 2019, 286, 20191716. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.; E Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The Structure of Haplotype Blocks in the Human Genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef]
- Stephens, M.; Donnelly, P. A Comparison of Bayesian Methods for Haplotype Reconstruction from Population Genotype Data. Am. J. Hum. Genet. 2003, 73, 1162–1169. [Google Scholar] [CrossRef]
- Stephens, M.; Smith, N.J.; Donnelly, P. A New Statistical Method for Haplotype Reconstruction from Population Data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- van der Weide, J.; Steijns, L.S.W. Cytochrome P450 Enzyme System: Genetic Polymorphisms and Impact on Clinical Pharmacology. Ann. Clin. Biochem. Int. J. Lab. Med. 1999, 36, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.C.G.; Fernandes, M.R.; Ribeiro-Dos-Santos, A.M.; de Araújo, G.S.; de Souza, S.J.; Guerreiro, J.F.; Ribeiro-Dos-Santos, Â.; de Assumpção, P.P.; dos Santos, N.P.C.; Santos, S. Pharmacogenomic Profile of Amazonian Amerindians. J. Pers. Med. 2022, 12, 952. [Google Scholar] [CrossRef] [PubMed]
- Idda, M.L.; Zoledziewska, M.; Urru, S.A.M.; McInnes, G.; Bilotta, A.; Nuvoli, V.; Lodde, V.; Orrù, S.; Schlessinger, D.; Cucca, F.; et al. Genetic Variation among Pharmacogenes in the Sardinian Population. Int. J. Mol. Sci. 2022, 23, 10058. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.; Twesigomwe, D.; Nofziger, C.; Turner, A.J.; Helmecke, L.-S.; Broeckel, U.; Derezinski, A.D.; Hazelhurst, S.; Gaedigk, A. Characterization of Novel CYP2D6 Alleles across Sub-Saharan African Populations. J. Pers. Med. 2022, 12, 1575. [Google Scholar] [CrossRef] [PubMed]
- Jay, F.; Sjödin, P.; Jakobsson, M.; Blum, M.G. Anisotropic Isolation by Distance: The Main Orientations of Human Genetic Differentiation. Mol. Biol. Evol. 2013, 30, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Bamshad, M.; Wooding, S.P. Signatures of natural selection in the human genome. Nat. Rev. Genet. 2003, 4, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Runcharoen, C.; Fukunaga, K.; Sensorn, I.; Iemwimangsa, N.; Klumsathian, S.; Tong, H.; Vo, N.S.; Le, L.; Hlaing, T.M.; Thant, M.; et al. Prevalence of pharmacogenomic variants in 100 pharmacogenes among Southeast Asian populations under the collaboration of the Southeast Asian Pharmacogenomics Research Network (SEAPharm). Hum. Genome Var. 2021, 8, 1–6. [Google Scholar] [CrossRef]
- Zhang, C.; Jiang, X.; Chen, W.; Li, Q.; Yun, F.; Yang, X.; Dai, R.; Cheng, Y. Population genetic difference of pharmacogenomic VIP gene variants in the Lisu population from Yunnan Province. Medicine 2018, 97, e13674. [Google Scholar] [CrossRef]
- Li, D.; Peng, L.; Xing, S.; He, C.; Jin, T. Genetic analysis of pharmacogenomic VIP variants in the Wa population from Yunnan Province of China. BMC Genom. Data 2021, 22, 1–20. [Google Scholar] [CrossRef]
- Lee, J.Y.; Vinayagamoorthy, N.; Han, K.; Kwok, S.K.; Ju, J.H.; Park, K.S.; Jung, S.-H.; Park, S.-W.; Chung, Y.-J. Association of Polymorphisms of Cytochrome P450 2D6 With Blood Hydroxychloroquine Levels in Patients with Systemic Lupus Erythematosus. Arthritis Rheumatol. 2016, 68, 184–190. [Google Scholar] [CrossRef]
- López-García, M.A.; Feria-Romero, I.A.; Serrano, H.; Rayo-Mares, D.; Fagiolino, P.; Vázquez, M.; Escamilla-Núñez, C.; Grijalva, I.; Escalante-Santiago, D.; Orozco-Suarez, S. Influence of genetic variants of CYP2D6, CYP2C9, CYP2C19 and CYP3A4 on antiepileptic drug metabolism in pediatric patients with refractory epilepsy. Pharmacol. Rep. 2017, 69, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Lymperopoulos, A.; McCrink, K.A.; Brill, A. Impact of CYP2D6 Genetic Variation on the Response of the Cardiovascular Patient to Carvedilol and Metoprolol. Curr. Drug Metab. 2015, 17, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Masimirembwa, C.; Persson, I.; Bertilsson, L.; Hasler, J.; Ingelman-Sundberg, M. A novel mutant variant of the CYP2D6 gene (CYP2D617) common in a black African population: Association with diminished debrisoquine hydroxylase activity. Br. J. Clin. Pharmacol. 1996, 42, 713–719. [Google Scholar] [CrossRef]
- Muyambo, S.; Ndadza, A.; Soko, N.D.; Kruger, B.; Kadzirange, G.; Chimusa, E.; Masimirembwa, C.M.; Ntsekhe, M.; Nhachi, C.F.; Dandara, C. Warfarin Pharmacogenomics for Precision Medicine in Real-Life Clinical Practice in Southern Africa: Harnessing 73 Variants in 29 Pharmacogenes. OMICS: A J. Integr. Biol. 2022, 26, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Prohaska, A.; Racimo, F.; Schork, A.J.; Sikora, M.; Stern, A.J.; Ilardo, M.; Allentoft, M.E.; Folkersen, L.; Buil, A.; Moreno-Mayar, J.V.; et al. Human Disease Variation in the Light of Population Genomics. Cell 2019, 177, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Mazas, A.; Poloni, E.S.; Genetic Diversity in Africa. Encyclopedia of Life Sciences 2008. Available online: https://www.els.net/ (accessed on 15 April 2022).
- Li, J.; Zhang, L.; Zhou, H.; Stoneking, M.; Tang, K. Global patterns of genetic diversity and signals of natural selection for human ADME genes. Hum. Mol. Genet. 2011, 20, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Marković, A.S.; Petranović, M.Z.; Škobalj, M.; Poloni, E.S.; Oberški, L.P.; Škarić-Jurić, T.; Salihović, M.P. From dietary adaptation in the past to drug metabolism of today: An example of NAT genes in the Croatian Roma. Am. J. Biol. Anthr. 2022, 178, 140–153. [Google Scholar] [CrossRef]
- Marković, A.S.; Petranović, M.Z.; Tomas, Z.; Puljko, B.; Šetinc, M.; Škarić-Jurić, T.; Salihović, M.P. Untangling SNP Variations within CYP2D6 Gene in Croatian Roma. J. Pers. Med. 2022, 12, 374. [Google Scholar] [CrossRef]
- Petranovic, M.Z.; Tomas, Z.; Skaric-Juric, T.; Narancic, N.S.; Janicijevic, B.; Markovic, A.S.; Salihovic, M.P. The variability of multi-drug resistance ABCB1 gene in the Roma population from Croatia. Mol. Exp. Biol. Med. 2019, 2, 10–18. [Google Scholar] [CrossRef]
- Dlouhá, L.; Adámková, V.; Šedová, L.; Olišarová, V.; Hubáček, J.A.; Tóthová, V. Five genetic polymorphisms of cytochrome P450 enzymes in the Czech non-Roma and Czech Roma population samples. Drug Metab. Pers. Ther. 2020, 35, 20200103. [Google Scholar] [CrossRef]
- Weber, A.; Szalai, R.; Sipeky, C.; Magyari, L.; Melegh, M.; Jaromi, L.; Matyas, P.; Duga, B.; Kovesdi, E.; Hadzsiev, K.; et al. Increased prevalence of functional minor allele variants of drug metabolizing CYP2B6 and CYP2D6 genes in Roma population samples. Pharmacol. Rep. 2015, 67, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Moyà, G.; Dorado, P.; Ferreiro, V.; Naranjo, M.E.G.; Peñas-Lledó, E.M.; Llerena, A. High frequency of CYP2D6 ultrarapid metabolizer genotypes in an Ashkenazi Jewish population from Argentina. Pharm. J. 2016, 17, 378–381. [Google Scholar] [CrossRef]
- Wen, Y.F.; Gaedigk, A.; Boone, E.C.; Wang, W.Y.; Straka, R.J. The Identification of Novel CYP2D6 Variants in US Hmong: Results from Genome Sequencing and Clinical Genotyping. Front. Pharmacol. 2022, 13, 867331. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lou, H.; Yang, X.; Lu, D.; Li, S.; Jin, L.; Pan, X.; Yang, W.; Song, M.; Mamatyusupu, D.; et al. Genetic architectures of ADME genes in five Eurasian admixed populations and implications for drug safety and efficacy. J. Med Genet. 2014, 51, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Sistonen, J.; Sajantila, A.; Lao, O.; Corander, J.; Barbujani, G.; Fuselli, S. CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharm. Genom. 2007, 17, 93–101. [Google Scholar] [CrossRef]
- Wennerholm, A.; Dandara, C.; Sayi, J.; Svensson, J.; Abdi, Y.A.; Ingelman-Sundberg, M.; Bertilsson, L.; Hasler, J.; Gustafsson, L.L. The African-specific CYP2D6*17 allele encodes an enzyme with changed substrate specificity. Clin. Pharmacol. Ther. 2002, 71, 77–88. [Google Scholar] [CrossRef]
- Bogni, A.; Monshouwer, M.; Moscone, A.; Hidestrand, M.; Ingelman-Sundberg, M.; Hartung, T.; Coecke, S. Substrate specific metabolism by polymorphic cytochrome P450 2D6 alleles. Toxicol. Vitr. 2005, 19, 621–629. [Google Scholar] [CrossRef]
- Shen, H.; He, M.M.; Liu, H.; Wrighton, S.A.; Wang, L.; Guo, B.; Li, C. Comparative Metabolic Capabilities and Inhibitory Profiles of CYP2D6.1, CYP2D6.10, and CYP2D6. Drug Metab. Dispos. 2007, 35, 1292–1300. [Google Scholar] [CrossRef]
- Wang, B.; Yang, L.-P.; Zhang, X.-Z.; Huang, S.-Q.; Bartlam, M.; Zhou, S.-F. New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab. Rev. 2009, 41, 573–643. [Google Scholar] [CrossRef]
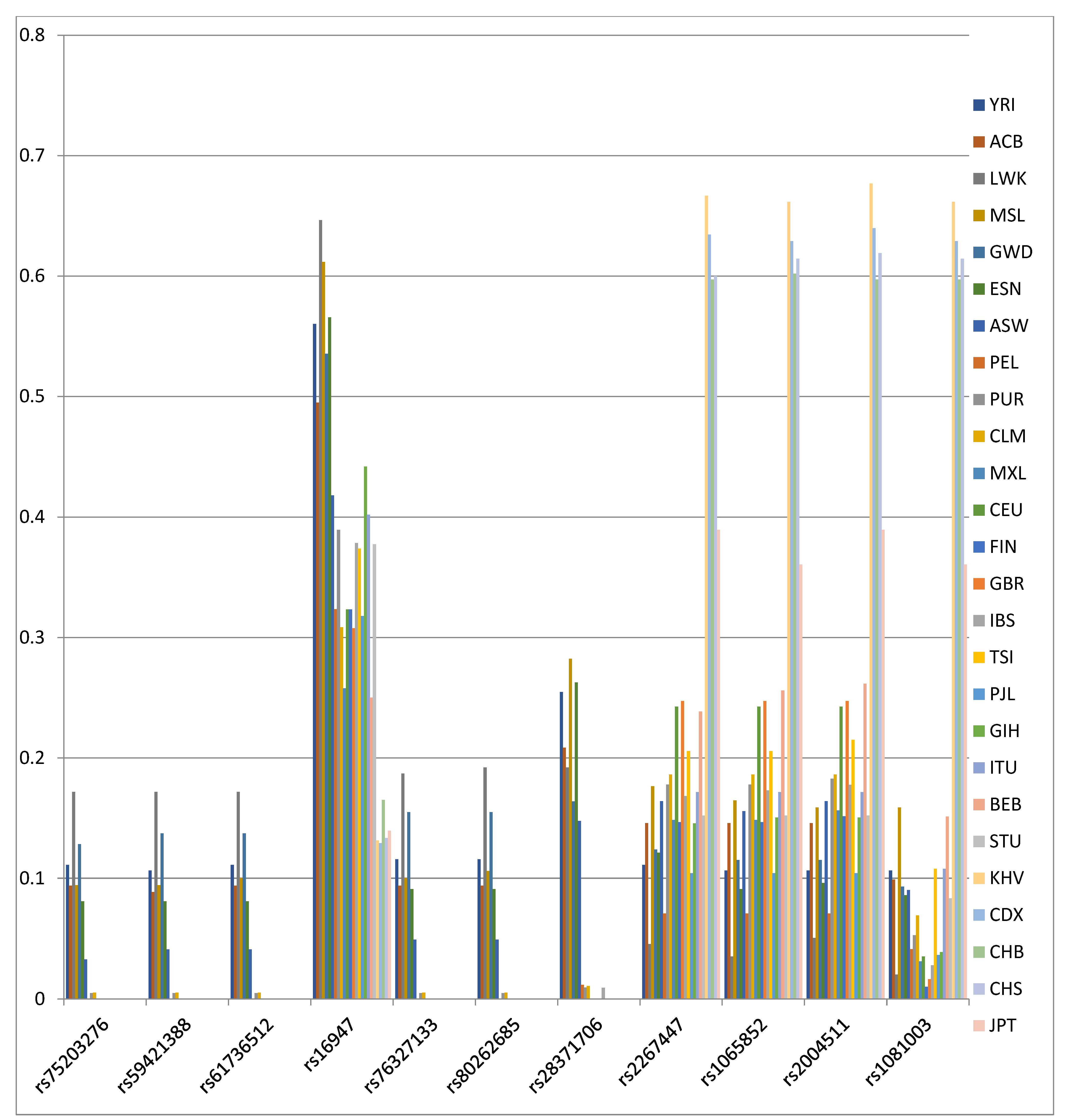

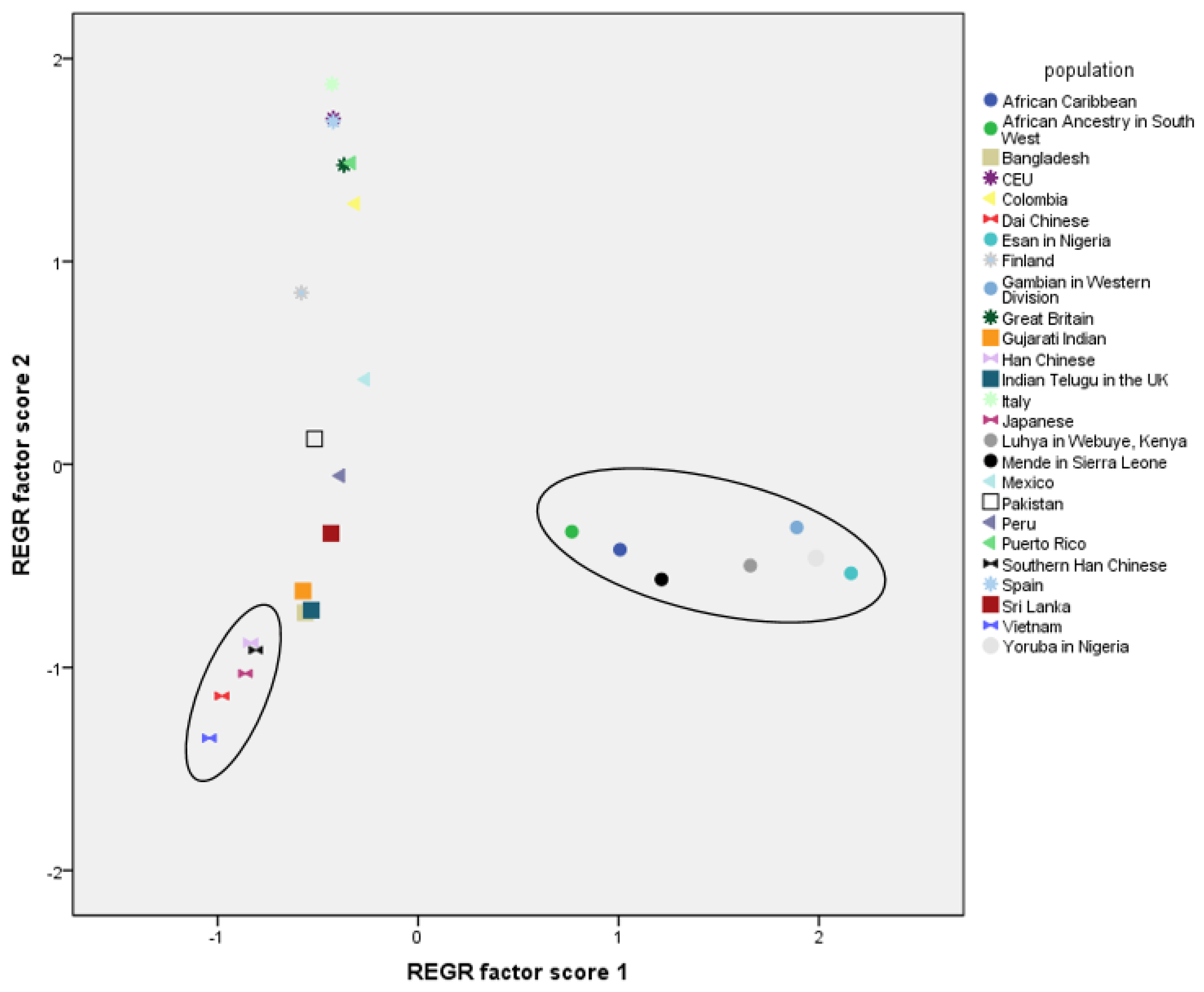
| Sample Size | No. of Haplotypes | No. of Polymorphic Sites | Haplotype Diversity | Nucleotide Diversity | ||
|---|---|---|---|---|---|---|
| African Ancestry | ESN | 198 | 36 | 50 | 0.935 | 0.113 |
| GWD | 226 | 37 | 49 | 0.918 | 0.114 | |
| MSL | 170 | 32 | 46 | 0.933 | 0.113 | |
| YRI | 216 | 31 | 48 | 0.926 | 0.109 | |
| LWK | 198 | 35 | 48 | 0.936 | 0.113 | |
| ACB | 192 | 36 | 48 | 0.933 | 0.115 | |
| ASW | 122 | 36 | 55 | 0.908 | 0.109 | |
| American Ancestry | CLM | 188 | 26 | 44 | 0.813 | 0.106 |
| MXL | 128 | 23 | 36 | 0.764 | 0.092 | |
| PEL | 170 | 16 | 34 | 0.602 | 0.086 | |
| PUR | 208 | 32 | 56 | 0.875 | 0.113 | |
| European Ancestry | CEU | 198 | 21 | 38 | 0.847 | 0.112 |
| FIN | 198 | 19 | 36 | 0.778 | 0.102 | |
| GBR | 182 | 17 | 35 | 0.834 | 0.112 | |
| IBS | 214 | 27 | 47 | 0.851 | 0.110 | |
| TSI | 214 | 22 | 41 | 0.845 | 0.113 | |
| South Asian Ancestry | BEB | 172 | 22 | 34 | 0.809 | 0.099 |
| GIH | 206 | 22 | 36 | 0.787 | 0.106 | |
| ITU | 204 | 21 | 34 | 0.794 | 0.105 | |
| PJL | 192 | 18 | 37 | 0.713 | 0.094 | |
| STU | 204 | 28 | 38 | 0.784 | 0.102 | |
| East Asian Ancestry | CDX | 186 | 18 | 26 | 0.620 | 0.069 |
| CHB | 206 | 19 | 30 | 0.637 | 0.075 | |
| CHS | 210 | 20 | 27 | 0.643 | 0.071 | |
| JPT | 208 | 11 | 23 | 0.717 | 0.074 | |
| KHV | 198 | 22 | 30 | 0.607 | 0.066 |
| SNP | Position | FST | p | FCT | p |
|---|---|---|---|---|---|
| rs1081003 | 42129754 | 0.366 | <0.00001 | 0.343 | 0.00001 |
| rs2004511 | 42127209 | 0.211 | <0.00001 | 0.193 | 0.00001 |
| rs1065852 | 42130692 | 0.209 | <0.00001 | 0.188 | <0.00001 |
| rs2267447 | 42128694 | 0.204 | <0.00001 | 0.187 | <0.00001 |
| rs28371706 | 42129770 | 0.197 | <0.00001 | 0.163 | 0.0001 |
| rs80262685 | 42128576 | 0.117 | <0.00001 | 0.096 | 0.00015 |
| rs76327133 | 42128668 | 0.115 | <0.00001 | 0.094 | 0.00025 |
| rs16947 | 42127941 | 0.111 | <0.00001 | 0.099 | <0.00001 |
| rs61736512 | 42129132 | 0.105 | <0.00001 | 0.085 | 0.00026 |
| rs59421388 | 42127608 | 0.104 | <0.00001 | 0.084 | 0.0002 |
| rs75203276 | 42128499 | 0.103 | <0.00001 | 0.081 | 0.00014 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stojanović Marković, A.; Zajc Petranović, M.; Škarić-Jurić, T.; Celinšćak, Ž.; Šetinc, M.; Tomas, Ž.; Peričić Salihović, M. Relevance of CYP2D6 Gene Variants in Population Genetic Differentiation. Pharmaceutics 2022, 14, 2481. https://doi.org/10.3390/pharmaceutics14112481
Stojanović Marković A, Zajc Petranović M, Škarić-Jurić T, Celinšćak Ž, Šetinc M, Tomas Ž, Peričić Salihović M. Relevance of CYP2D6 Gene Variants in Population Genetic Differentiation. Pharmaceutics. 2022; 14(11):2481. https://doi.org/10.3390/pharmaceutics14112481
Chicago/Turabian StyleStojanović Marković, Anita, Matea Zajc Petranović, Tatjana Škarić-Jurić, Željka Celinšćak, Maja Šetinc, Željka Tomas, and Marijana Peričić Salihović. 2022. "Relevance of CYP2D6 Gene Variants in Population Genetic Differentiation" Pharmaceutics 14, no. 11: 2481. https://doi.org/10.3390/pharmaceutics14112481
APA StyleStojanović Marković, A., Zajc Petranović, M., Škarić-Jurić, T., Celinšćak, Ž., Šetinc, M., Tomas, Ž., & Peričić Salihović, M. (2022). Relevance of CYP2D6 Gene Variants in Population Genetic Differentiation. Pharmaceutics, 14(11), 2481. https://doi.org/10.3390/pharmaceutics14112481









