Novel Ex Vivo Zymography Approach for Assessment of Protease Activity in Tissues with Activatable Antibodies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Materials
2.2. Pb-Tx Labeling
2.3. QZ Assay Protocol
2.4. In Vivo Efficacy Study
2.5. QZ Assay of Human Plasma Samples to Assess the Stability of CX-188 in Plasma In Vitro
2.6. Statistical Analysis
3. Results
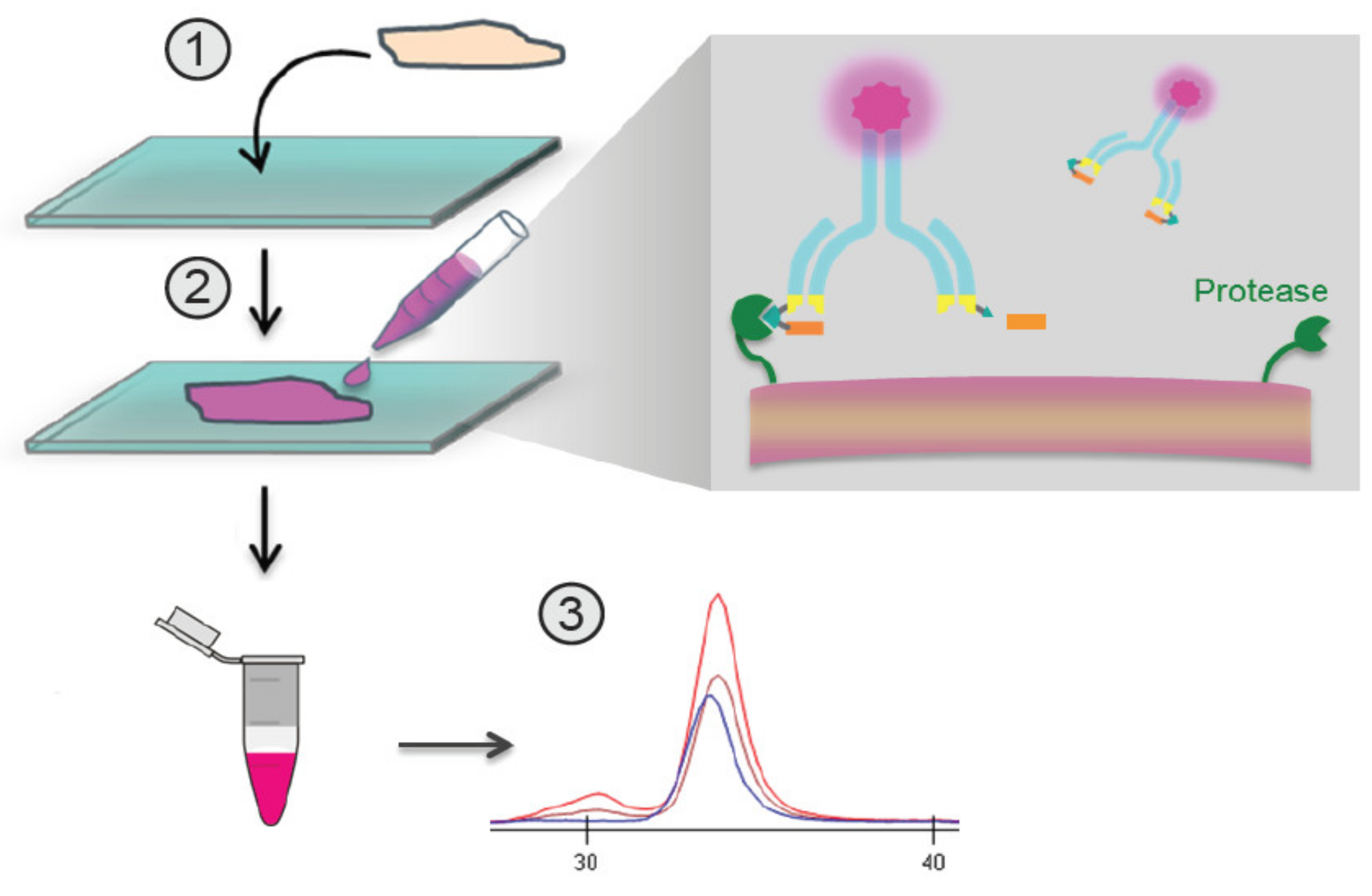
3.1. Quantitative Zymography (QZ) Technology for Assessment of Protease Activity Ex Vivo
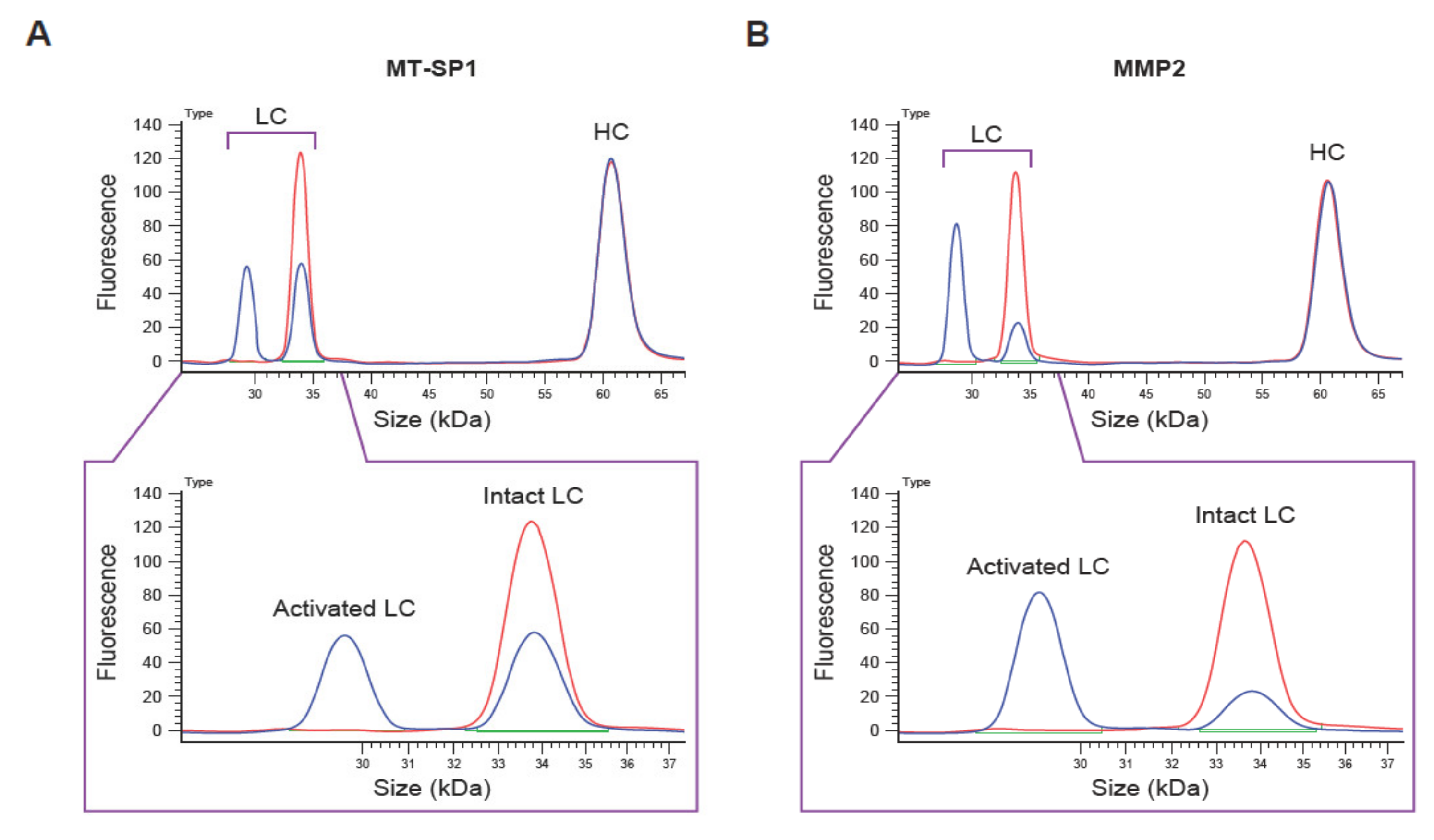
3.2. Optimization of QZ Assay Conditions
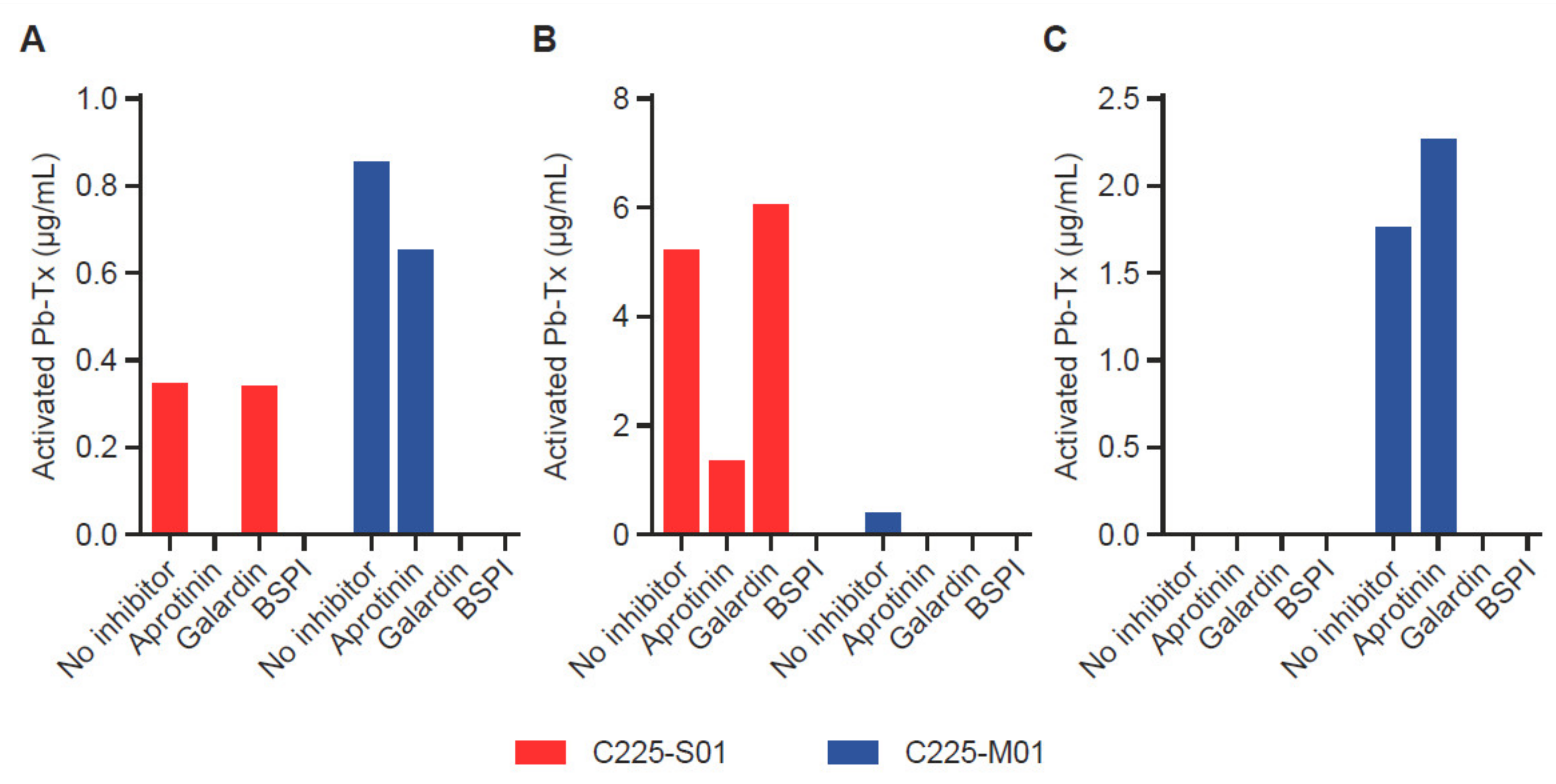
3.3. Assessment of Protease Activity of Different Specificities
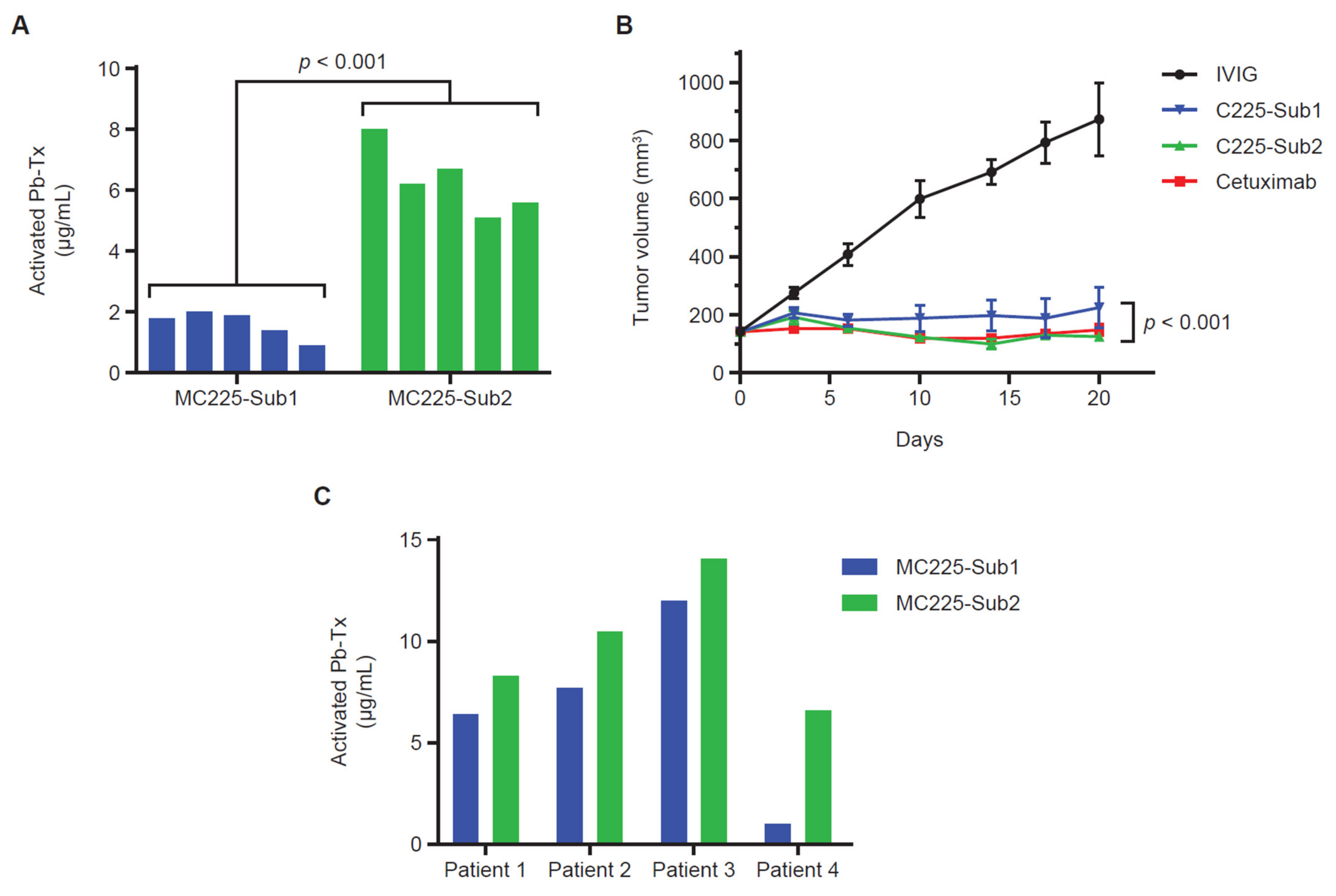
3.4. QZ Technology for Protease Characterization in H292 Xenograft Tumor Sections
3.5. QZ Technology for Assessment of Protease Activity in Patient-Derived Plasma Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Puente, X.S.; Sánchez, L.M.; Overall, C.M.; López-Otín, C. Human and mouse proteases: A comparative genomic approach. Nat. Rev. Genet. 2003, 4, 544–558. [Google Scholar] [CrossRef]
- Turk, B. Targeting proteases: Successes, failures and future prospects. Nat. Rev. Drug Discov. 2006, 5, 785–799. [Google Scholar] [CrossRef]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional enzymes in life and disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef] [Green Version]
- Narayanan, S. Multifunctional roles of thrombin. Ann. Clin. Lab. Sci. 1999, 29, 275–280. [Google Scholar]
- Heutinck, K.M.; ten Berge, I.J.; Hack, C.E.; Hamann, J.; Rowshani, A.T. Serine proteases of the human immune system in health and disease. Mol. Immunol. 2010, 47, 1943–1955. [Google Scholar] [CrossRef]
- Haley, S.A.; Wessel, G.M. Regulated proteolysis by cortical granule serine protease 1 at fertilization. Mol. Biol. Cell 2004, 15, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- Lamkanfi, M.; Festjens, N.; Declercq, W.; Vanden Berghe, T.; Vandenabeele, P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007, 14, 44–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Yuan, J. Caspases in apoptosis and beyond. Oncogene 2008, 27, 6194–6206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Sevenich, L.; Joyce, J.A. Pericellular proteolysis in cancer. Genes Dev. 2014, 28, 2331–2347. [Google Scholar] [CrossRef] [Green Version]
- Vandooren, J.; Geurts, N.; Martens, E.; Van den Steen, P.E.; Opdenakker, G. Zymography methods for visualizing hydrolytic enzymes. Nat. Methods 2013, 10, 211–220. [Google Scholar] [CrossRef]
- Hrabec, E.; Strek, M.; Nowak, D.; Greger, J.; Suwalski, M.; Hrabec, Z. Activity of type IV collagenases (MMP-2 and MMP-9) in primary pulmonary carcinomas: A quantitative analysis. J. Cancer Res. Clin. Oncol. 2002, 128, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Snoek-van Beurden, P.A.; Von den Hoff, J.W. Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. Biotechniques 2005, 38, 73–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.J.; Blomme, E.A.G. In situ zymography: A molecular pathology technique to localize endogenous protease activity in tissue sections. Vet. Pathol. 2003, 40, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Jedeszko, C.; Sameni, M.; Olive, M.B.; Moin, K.; Sloane, B.F. Visualizing protease activity in living cells: From two dimensions to four dimensions. Curr. Protoc. Cell Biol. 2008, 4–20. [Google Scholar] [CrossRef] [Green Version]
- Kwon, E.J.; Dudani, J.S.; Bhatia, S.N. Ultrasensitive tumour-penetrating nanosensors of protease activity. Nat. Biomed. Eng. 2017, 1, 54. [Google Scholar] [CrossRef] [Green Version]
- Soleimany, A.P.; Bhatia, S.N. Activity-based diagnostics: An emerging paradigm for disease detection and monitoring. Trends Mol. Med. 2020, 26, 450–468. [Google Scholar] [CrossRef]
- Poreba, M.; Groborz, K.M.; Rut, W.; Pore, M.; Snipas, S.J.; Vizovisek, M.; Turk, B.; Kuhn, P.; Drag, M.; Salvesen, G.S. Multiplexed probing of proteolytic enzymes using mass cytometry-compatible activity-based probes. J. Am. Chem. Soc. 2020, 142, 16704–16715. [Google Scholar] [CrossRef] [PubMed]
- Sela-Passwell, N.; Kikkeri, R.; Dym, O.; Rozenberg, H.; Margalit, R.; Arad-Yellin, R.; Eisenstein, M.; Brenner, O.; Shoham, T.; Danon, T.; et al. Antibodies targeting the catalytic zinc complex of activated matrix metalloproteinases show therapeutic potential. Nat. Med. 2011, 18, 143–147. [Google Scholar] [CrossRef]
- Schneider, E.L.; Lee, M.S.; Baharuddin, A.; Goetz, D.H.; Farady, C.J.; Ward, M.; Wang, C.I.; Craik, C.S. A reverse binding motif that contributes to specific protease inhibition by antibodies. J. Mol. Biol. 2012, 415, 699–715. [Google Scholar] [CrossRef] [Green Version]
- LeBeau, A.M.; Lee, M.; Murphy, S.T.; Hann, B.C.; Warren, R.S.; Delos Santos, R.; Kurhanewicz, J.; Hanash, S.M.; VanBrocklin, H.F.; Craik, C.S. Imaging a functional tumorigenic biomarker in the transformed epithelium. Proc. Natl. Acad. Sci. USA 2013, 110, 93–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeBeau, A.M.; Sevillano, N.; Markham, K.; Winter, M.B.; Murphy, S.T.; Hostetter, D.R.; West, J.; Lowman, H.; Craik, C.S.; VanBrocklin, H.F. Imaging active urokinase plasminogen activator in prostate cancer. Cancer Res. 2015, 75, 1225–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weissleder, R.; Tung, C.H.; Mahmood, U.; Bogdanov, A., Jr. In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat. Biotechnol. 1999, 17, 375–378. [Google Scholar] [CrossRef]
- Blum, G.; Von Degenfeld, G.; Merchant, M.J.; Blau, H.M.; Bogyo, M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat. Chem. Biol. 2007, 3, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Kwong, G.A.; von Maltzahn, G.; Murugappan, G.; Abudayyeh, O.; Mo, S.; Papayannopoulos, I.A.; Sverdlov, D.Y.; Liu, S.B.; Warren, A.D.; Popov, Y.; et al. Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease. Nat. Biotechnol. 2013, 31, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Sanman, L.E.; Bogyo, M. Activity-based profiling of proteases. Annu. Rev. Biochem. 2014, 83, 249–273. [Google Scholar] [CrossRef] [Green Version]
- Garland, M.; Yim, J.J.; Bogyo, M. A bright future for precision medicine: Advances in fluorescent chemical probe design and their clinical application. Cell Chem. Biol. 2016, 23, 122–136. [Google Scholar] [CrossRef] [Green Version]
- Loynachan, C.N.; Soleimany, A.P.; Dudani, J.S.; Lin, Y.; Najer, A.; Bekdemir, A.; Chen, Q.; Bhatia, S.N.; Stevens, M.M. Renal clearable catalytic gold nanoclusters for in vivo disease monitoring. Nat. Nanotechnol. 2019, 14, 883–890. [Google Scholar] [CrossRef] [Green Version]
- Kirkpatrick, J.D.; Warren, A.D.; Soleimany, A.P.; Westcott, P.M.K.; Voog, J.C.; Martin-Alonso, C.; Fleming, H.E.; Tammela, T.; Jacks, T.; Bhatia, S.N. Urinary detection of lung cancer in mice via noninvasive pulmonary protease profiling. Sci. Transl. Med. 2020, 12, eaaw0262. [Google Scholar] [CrossRef]
- Desnoyers, L.R.; Vasiljeva, O.; Richardson, J.H.; Yang, A.; Menendez, E.E.; Liang, T.W.; Wong, C.; Bessette, P.H.; Kamath, K.; Moore, S.J.; et al. Tumor-specific activation of an EGFR-targeting probody enhances therapeutic index. Sci. Transl. Med. 2013, 5, 207ra144. [Google Scholar] [CrossRef]
- Trang, V.H.; Zhang, X.; Yumul, R.C.; Zeng, W.; Stone, I.J.; Wo, S.W.; Dominguez, M.M.; Cochran, J.H.; Simmons, J.K.; Ryan, M.C.; et al. A coiled-coil masking domain for selective activation of therapeutic antibodies. Nat. Biotechnol. 2019, 37, 761–765. [Google Scholar] [CrossRef]
- Wang, D.; Wang, T.; Yu, H.; Feng, B.; Zhou, L.; Zhou, F.; Hou, B.; Zhang, H.; Luo, M.; Li, Y. Engineering nanoparticles to locally activate T cells in the tumor microenvironment. Sci. Immunol. 2019, 4, eaau6584. [Google Scholar] [CrossRef]
- Millar, D.G.; Ramjiawan, R.R.; Kawaguchi, K.; Gupta, N.; Chen, J.; Zhang, S.; Nojiri, T.; Ho, W.W.; Aoki, S.; Jung, K.; et al. Antibody-mediated delivery of viral epitopes to tumors harnesses CMV-specific T cells for cancer therapy. Nat. Biotechnol. 2020, 38, 420–425. [Google Scholar] [CrossRef]
- Vasiljeva, O.; Hostetter, D.R.; Moore, S.J.; Winter, M.B. The multifaceted roles of tumor-associated proteases and harnessing their activity for prodrug activation. Biol. Chem. 2019, 400, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Kavanaugh, W.M. Antibody prodrugs for cancer. Expert Opin. Biol. Ther. 2020, 20, 163–171. [Google Scholar] [CrossRef]
- Chomet, M.; Schreurs, M.; Nguyen, M.; Howng, B.; Villanueva, R.; Krimm, M.; Vasiljeva, O.; van Dongen, G.A.M.S.; Vugts, D.J. The tumor targeting performance of anti-CD166 Probody drug conjugate CX-2009 and its parental derivatives as monitored by 89Zr-immuno-PET in xenograft bearing mice. Theranostics 2020, 10, 5815–5828. [Google Scholar] [CrossRef] [PubMed]
- Giesen, D.; Broer, L.N.; Lub-de Hooge, M.N.; Popova, I.; Howng, B.; Nguyen, M.; Vasiljeva, O.; de Vries, E.G.E.; Pool, M. Probody therapeutic design of 89Zr-CX-072 promotes accumulation in PD-L1-expressing tumors compared to normal murine lymphoid tissue. Clin. Cancer Res. 2020, 26, 3999–4009. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.R.; Menendez, E.; Craik, C.S.; Kavanaugh, W.M.; Vasiljeva, O. In vivo imaging of protease activity by Probody therapeutic activation. Biochimie 2016, 122, 62–67. [Google Scholar] [CrossRef] [Green Version]
- Polu, K.R.; Lowman, H.B. Probody therapeutics for targeting antibodies to diseased tissue. Expert Opin. Biol. Ther. 2014, 14, 1049–1053. [Google Scholar] [CrossRef] [Green Version]
- Naing, A.; Thistlethwaite, F.C.; Spira, A.I.; Garcia-Corbacho, J.; Randhawa, M.; Eskens, F.; O’Neil, B.; Lavernia, J.; Uboha, N.V.; Hamid, O.; et al. CX-072, a PD-L1 Probody therapeutic, as monotherapy in patients with advanced solid tumors: Preliminary results of PROCLAIM-CX-072. J. Clin. Oncol. 2019, 37, 2513. [Google Scholar] [CrossRef]
- Autio, K.A.; Boni, V.; Humphrey, R.W.; Naing, A. Probody therapeutics: An emerging class of therapies designed to enhance on-target effects with reduced off-tumor toxicity for use in immuno-oncology. Clin. Cancer Res. 2020, 26, 984–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasiljeva, O.; Menendez, E.; Nguyen, M.; Craik, C.S.; Kavanaugh, W.M. Monitoring protease activity in biological tissues using antibody prodrugs as sensing probes. Sci. Rep. 2020, 10, 5894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, J.J.; Schohn, A.; Bessette, P.H.; Boulware, K.T.; Daugherty, P.S. Bacterial display using circularly permuted outer membrane protein OmpX yields high affinity peptide ligands. Protein Sci. 2006, 15, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Boulware, K.T.; Daugherty, P.S. Protease specificity determination by using cellular libraries of peptide substrates (CliPS). Proc. Natl. Acad. Sci. USA 2006, 103, 7583–7588. [Google Scholar] [CrossRef] [Green Version]
- Meyerholz, D.K.; Beck, A.P. Principles and approaches for reproducible scoring of tissue stains in research. Lab. Investig. 2018, 98, 844–855. [Google Scholar] [CrossRef]
- Lechner, A.; Giorgetti, J.; Gahoual, R.; Beck, A.; Leize-Wagner, E.; François, Y.N. Insights from capillary electrophoresis approaches for characterization of monoclonal antibodies and antibody drug conjugates in the period 2016–2018. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1122–1123, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Gahoual, R.; Beck, A.; Leize-Wagner, E.; François, Y.N. Cutting-edge capillary electrophoresis characterization of monoclonal antibodies and related products. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016, 1032, 61–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soleimany, A.P.; Kirkpatrick, J.D.; Su, S.; Dudani, J.S.; Zhong, Q.; Bekdemir, A.; Bhatia, S.N. Activatable zymography probes enable in situ localization of protease dysregulation in cancer. Cancer Res. 2021, 81, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Lyman, S.; Naing, A.; Zein, I.A.; Ru, Y.; Howng, B.; Winter, M.B.; Vasiljeva, O.; Garner, W.; Zheng, B.; Stroh, M.; et al. Evidence of intratumoral localization, activation, and immunomodulatory effect of CX-072, a probody therapeutic targeting PD-L1, in a phase I/II trial. J. Clin. Oncol. 2020, 30, 3108. [Google Scholar] [CrossRef]
- Travis, J.; Salvesen, G.S. Human plasma proteinase inhibitors. Annu. Rev. Biochem. 1983, 52, 655–709. [Google Scholar] [CrossRef]
- Farady, C.J.; Craik, C.S. Mechanisms of macromolecular protease inhibitors. Chembiochem 2010, 11, 2341–2346. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Howng, B.; Winter, M.B.; LePage, C.; Popova, I.; Krimm, M.; Vasiljeva, O. Novel Ex Vivo Zymography Approach for Assessment of Protease Activity in Tissues with Activatable Antibodies. Pharmaceutics 2021, 13, 1390. https://doi.org/10.3390/pharmaceutics13091390
Howng B, Winter MB, LePage C, Popova I, Krimm M, Vasiljeva O. Novel Ex Vivo Zymography Approach for Assessment of Protease Activity in Tissues with Activatable Antibodies. Pharmaceutics. 2021; 13(9):1390. https://doi.org/10.3390/pharmaceutics13091390
Chicago/Turabian StyleHowng, Bruce, Michael B. Winter, Carol LePage, Irina Popova, Michael Krimm, and Olga Vasiljeva. 2021. "Novel Ex Vivo Zymography Approach for Assessment of Protease Activity in Tissues with Activatable Antibodies" Pharmaceutics 13, no. 9: 1390. https://doi.org/10.3390/pharmaceutics13091390
APA StyleHowng, B., Winter, M. B., LePage, C., Popova, I., Krimm, M., & Vasiljeva, O. (2021). Novel Ex Vivo Zymography Approach for Assessment of Protease Activity in Tissues with Activatable Antibodies. Pharmaceutics, 13(9), 1390. https://doi.org/10.3390/pharmaceutics13091390






