The Epidemiology of African Swine Fever in “Nonendemic” Regions of Zambia (1989–2015): Implications for Disease Prevention and Control
Abstract
:1. Introduction
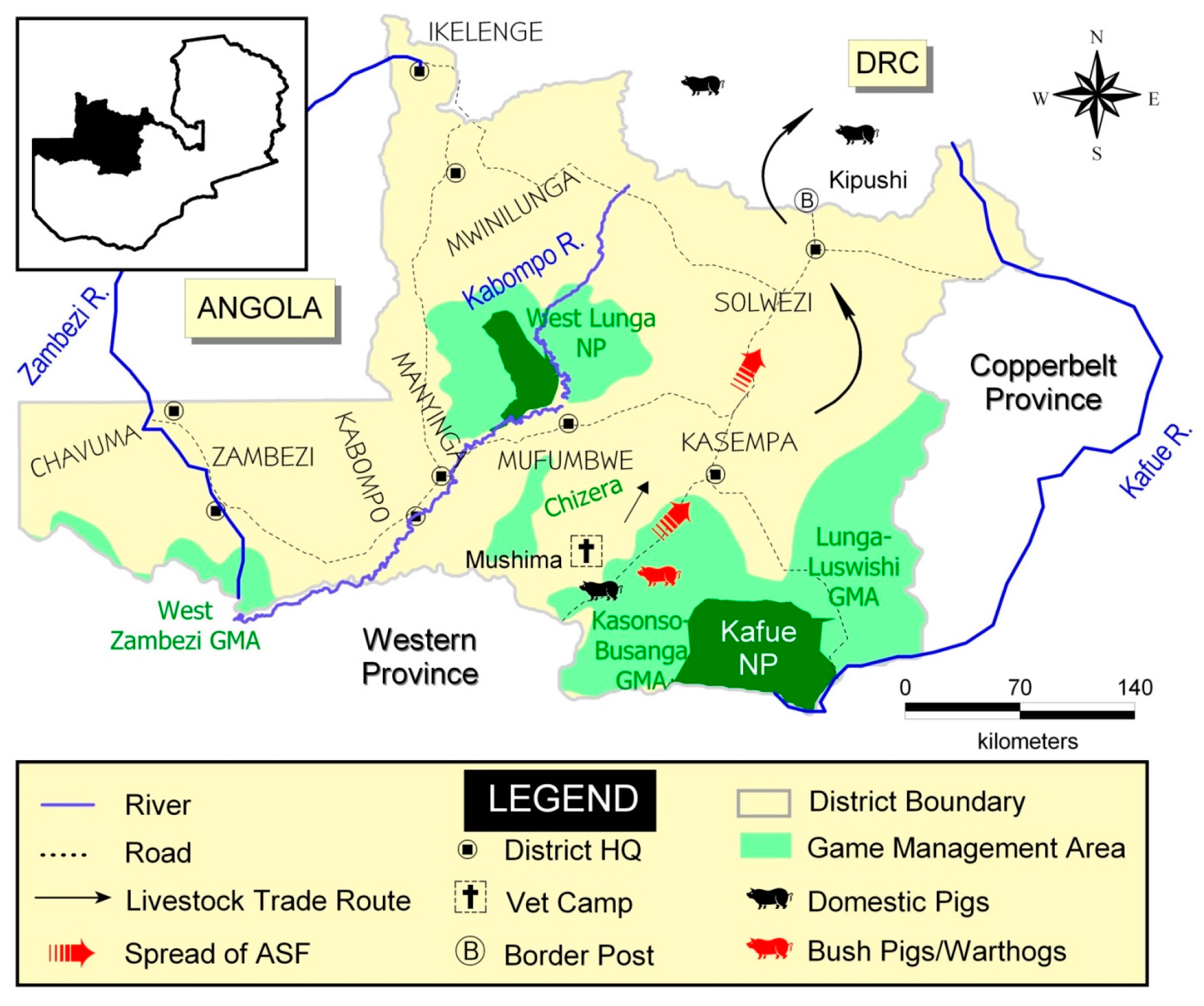
2. History of ASF Outbreaks in Nonendemic Areas outside Eastern Province
2.1. ASF Outbreak in Kabwe, Central Province (1989)
2.2. ASF Outbreaks in Lusaka Province (1993)
2.3. ASF Outbreaks in Lusaka and Southern Provinces (2001–2002)
2.4. ASF Outbreaks in Livingstone, Southern Province (2004 and 2006)
2.5. ASF Outbreaks in Northwestern Province (2006–2008)
2.6. ASF Outbreaks in Zambia during 2013–2015
3. Brief History and Status of ASF in Neighboring Countries of Zambia
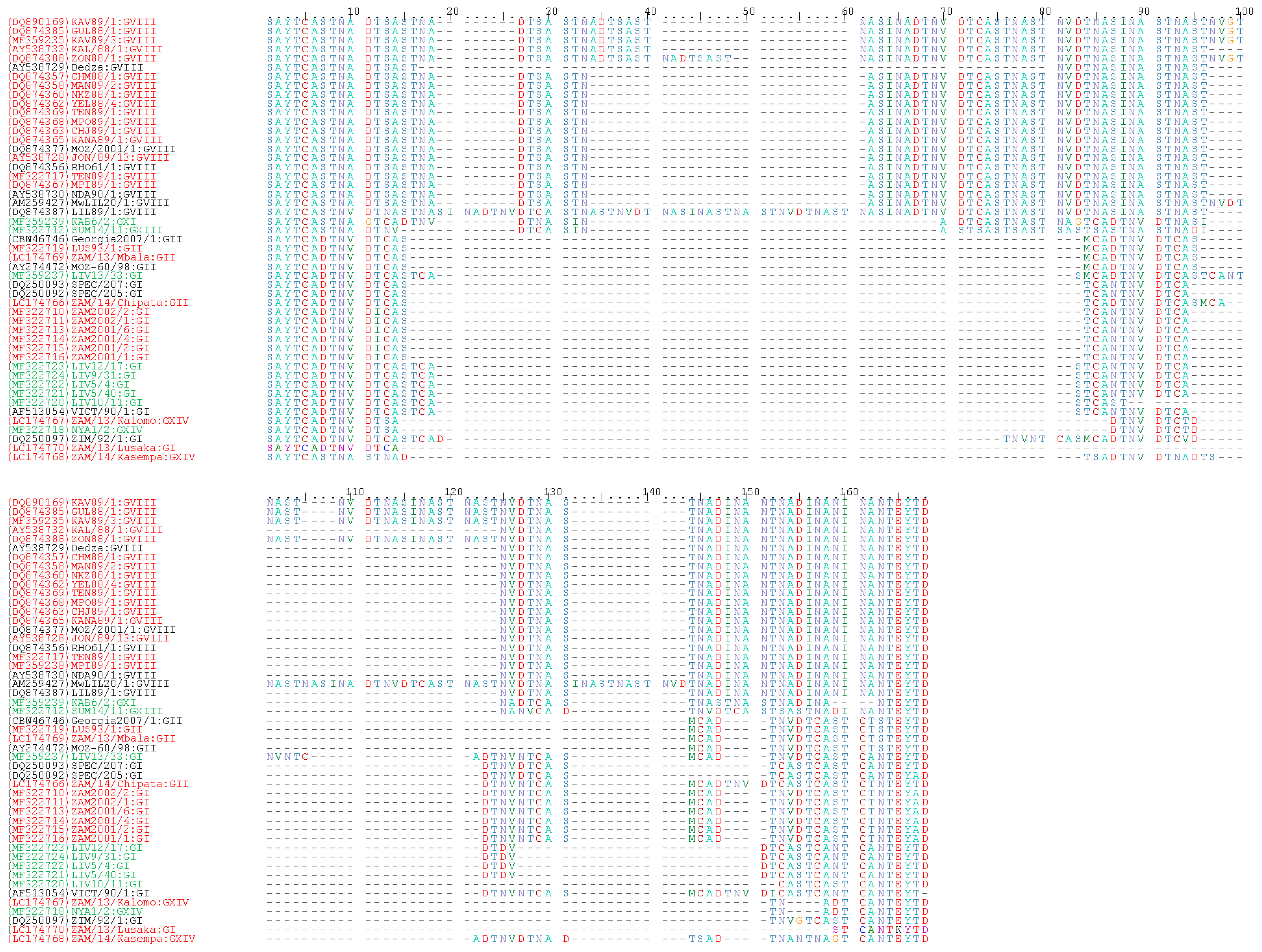
4. Genetic Analysis of ASFVs Detected in Zambia
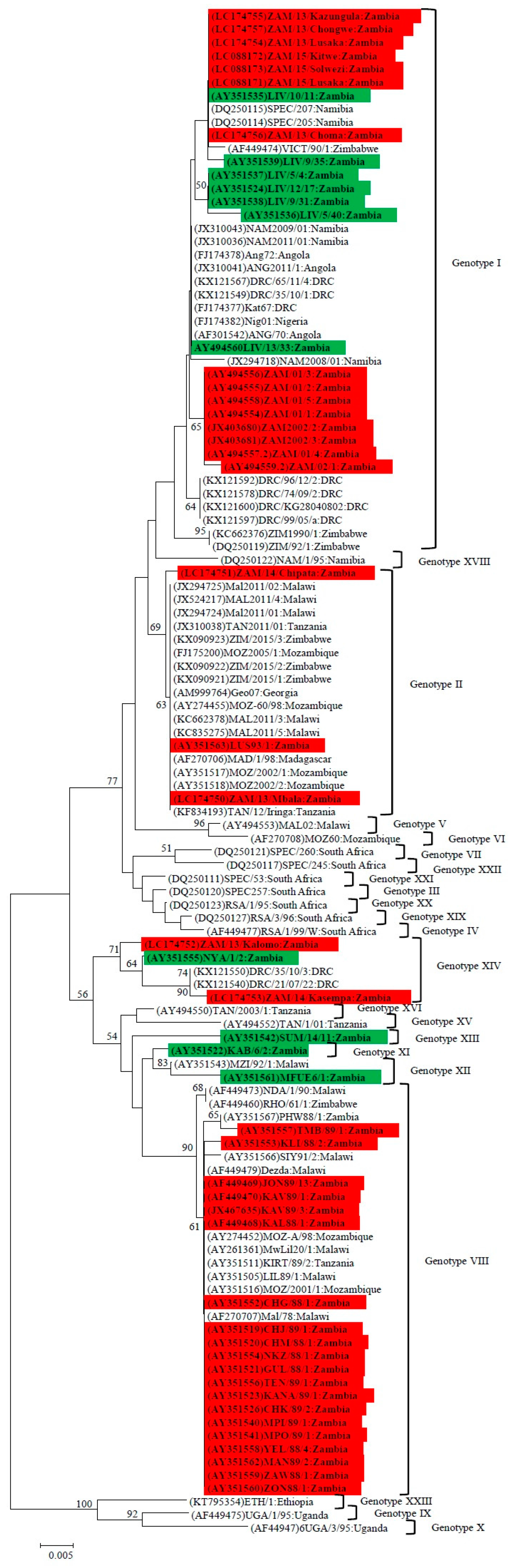
4.1. Phylogenetic Analysis of the p72 (B646L) Gene
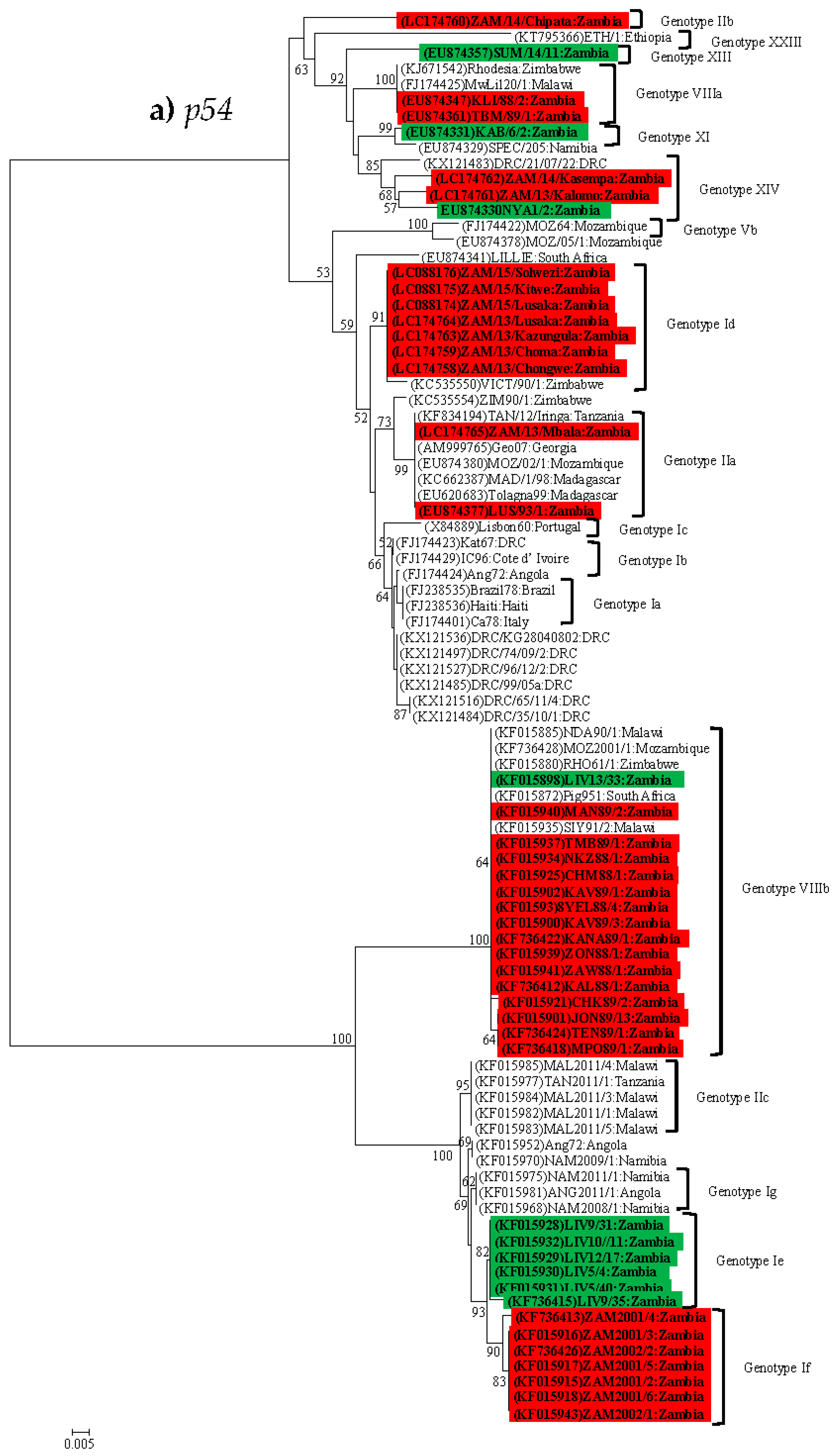
4.2. Phylogenetic Analysis of the p54 (E183L) Gene
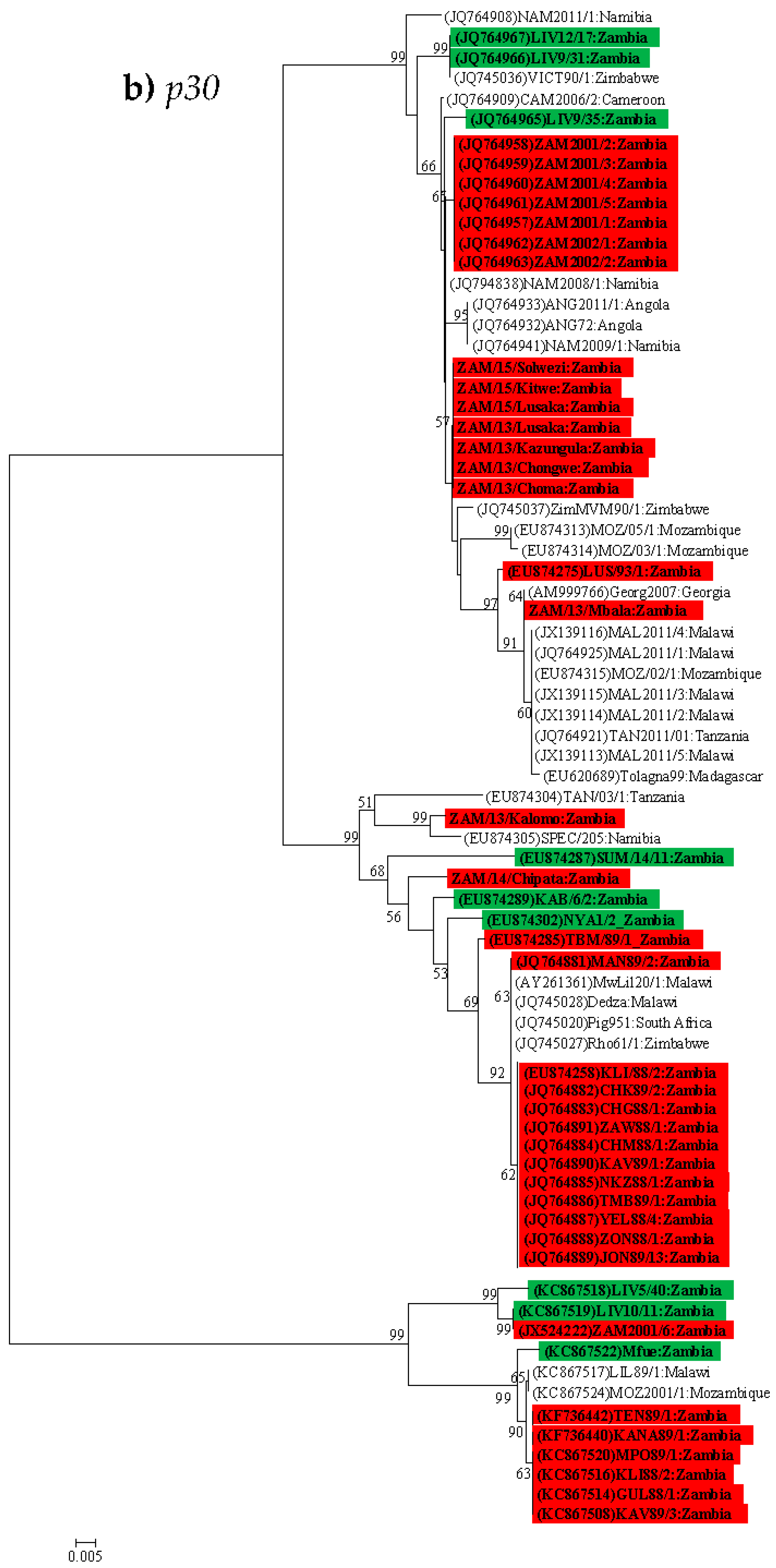
4.3. Phylogenetic Analysis of the p30 (CP204L) Gene
4.4. Sequence Analysis of the CVR within the B602L Gene
5. Discussion
6. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dixon, L.K.; Escribano, J.M.; Martins, C.; Rock, D.L.; Salas, M.L.; Wilkinson, P.J. Asfarviridae. In Virus Taxonomy. VIIIth Report of the ICTV; Fauquet, C.M., Mayo, M.A., Maniloff, J., Desselberger, U., Ball, L.A., Eds.; Elsevier/Academic Press: London, UK, 2005; pp. 135–143. [Google Scholar]
- Chapman, D.A.; Tcherepanov, V.; Upton, C.; Dixon, L.K. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J. Gen. Virol. 2008, 89, 397–408. [Google Scholar] [CrossRef] [PubMed]
- De Villiers, E.P.; Gallardo, C.; Arias, M.; Da Silva, M.; Upton, C.; Martin, R.; Bishop, R.P. Phylogenomic analysis of 11 complete African swine fever virus genome sequences. Virology 2010, 400, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.E. On a form of swine fever occurring in British East Africa (Kenya Colony). J. Comp. Pathol. Ther. 1921, 34, 159–191. [Google Scholar] [CrossRef]
- Jori, F.; Vial, L.; Penrith, M.L.; Pérez-Sánchez, R.; Etter, E.; Albina, E.; Michaud, V.; Roger, F. Review of the sylvatic cycle of African swine fever in sub-Saharan Africa and the Indian Ocean. Virus Res. 2013, 173, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Penrith, M.L.; Vosloo, W.; Jori, F.; Bastos, A.D. African swine fever virus eradication in Africa. Virus Res. 2013, 173, 228–246. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, M.C.; de la Torre Reoyo, A.; Fernández-Pinero, J.; Iglasias, I.; Jesús Munoz, M.; Luisa Arias, M. African swine fever: A global view of the current challenge. PHM 2015, 1, 21. [Google Scholar] [CrossRef] [PubMed]
- Costard, S.; Wieland, B.; De Glanville, W.; Jori, F.; Rowlands, R.; Vosloo, W.; Roger, F.; Pfeiffer, D.U.; Dixon, L.K. African swine fever: How can global spread be prevented? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 2683–2696. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, R.J.; Michaud, V.; Heath, L.; Hutchings, G.; Oura, C.; Vosloo, W.; Dwarka, R.; Onashvili, T.; Albina, E.; Dixon, L.K. African swine fever virus isolate, Georgia, 2007. Emerg. Infect. Dis. 2008, 14, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Malogolovkin, A.; Yelsukova, A.; Gallardo, C.; Tsybanov, S.; Kolbasov, D. Molecular characterization of African swine fever virus isolates originating from outbreaks in the Russian Federation between 2007 and 2011. Vet. Microbiol. 2012, 158, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Fernandez-Pinero, J.; Pelayo, V.; Gazaev, I.; Markowska-Daniel, I.; Pridotkas, G.; Nieto, R.; Fernandez-Pacheco, P.; Bokhan, S.; Nevolko, O.; et al. Genetic variation among African swine fever genotype II viruses, eastern and central Europe. Emerg. Infect. Dis. 2014, 20, 1544–1547. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA) (EFSA Panel on Animal Health and Welfare). Scientific opinion on African swine fever. EFSA J. 2015, 13, 4163–4192. [Google Scholar] [CrossRef]
- Wilkinson, P.J.; Pegram, R.G.; Perry, B.D.; Lemche, J.; Schels, H.F. The distribution of African swine fever virus isolated from Ornithodoros moubata in Zambia. Epidemiol. Infect. 1988, 101, 547–564. [Google Scholar] [CrossRef] [PubMed]
- Samui, K.L.; Nambota, A.M.; Mweene, A.S.; Onuma, M. African swine fever in Zambia: Potential financial and production consequences for the financial sector. Jpn. J. Vet. Res. 1996, 44, 119–124. [Google Scholar] [PubMed]
- Samui, K.L.; Mwanaumo, B.; Chizyuka, H.G.B. African swine fever in Zambia—Report on the first outbreak outside the endemic zone. In Proceedings of the 6th International Symposium on Veterinary Epidemiology and Economics, Ottawa, ON, Canada, 12–16 August 1991; pp. 453–455. [Google Scholar]
- Simulundu, E.; Chambaro, H.M.; Sinkala, Y.; Kajihara, M.; Ogawa, H.; Mori, A.; Ndebe, J.; Dautu, G.; Mataa, L.; Lubaba, C.H.; et al. Co-circulation of multiple genotypes of African swine fever viruses among domestic pigs in Zambia (2013–2015). Transbound. Emerg. Dis. 2017. [Google Scholar] [CrossRef] [PubMed]
- Misinzo, G.; Kwavi, D.E.; Sikombe, C.D.; Makange, M.; Peter, E.; Muhairwa, A.P.; Madege, M.J. Molecular characterization of African swine fever virus from domestic pigs in northern Tanzania during an outbreak in 2013. Trop. Anim. Health Prod. 2014, 46, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Misinzo, G.; Kasanga, C.J.; Mpelumbe-Ngeleja, C.; Masambu, J.; Kitambi, A.; Van Doorsselaere, J. African swine fever virus, Tanzania, 2010–2012. Emerg. Infect. Dis. 2012, 18, 2081–2083. [Google Scholar] [CrossRef] [PubMed]
- Owolodun, O.A.; Bastos, A.D.; Antiabong, J.F.; Ogedengbe, M.E.; Ekong, P.S.; Yakubu, B. Molecular characterisation of African swine fever viruses from Nigeria (2003–2006) recovers multiple virus variants and reaffirms CVR epidemiological utility. Virus Genes 2010, 41, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Lubisi, B.A.; Dwarka, R.M.; Meenowa, D.; Jaumally, R. An investigation into the first outbreak of African swine fever in the Republic of Mauritius. Transbound. Emerg. Dis. 2009, 56, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Bastos, A.D.; Penrith, M.L.; Cruciere, C.; Edrich, J.L.; Hutchings, G.; Roger, F.; Couacy-Hymann, E.G.R.T.; Thomson, G.R. Genotyping field strains of African swine fever virus by partial p72 gene characterization. Arch. Virol. 2003, 148, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Lubisi, B.A.; Bastos, A.D.; Dwarka, R.M.; Vosloo, W. Molecular epidemiology of African swine fever in East Africa. Arch. Virol. 2005, 150, 2439–2452. [Google Scholar] [CrossRef] [PubMed]
- Boshoff, C.I.; Bastos, A.D.; Gerber, L.J.; Vosloo, W. Genetic characterisation of African swine fever viruses from outbreaks in southern Africa (1973–1999). Vet. Microbiol. 2007, 121, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Achenbach, J.E.; Gallardo, C.; Nieto-Pelegrín, E.; Rivera-Arroyo, B.; Degefa-Negi, T.; Arias, M.; Jenberie, S. Identification of a new genotype of African swine fever virus in domestic pigs from Ethiopia. Transbound. Emerg. Dis. 2016. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Mwaengo, D.M.; Macharia, J.M.; Arias, M.; Taracha, E.A.; Soler, A.; Okoth, E.; Martin, E.; Kasiti, J.; Bishop, R.P. Enhanced discrimination of African swine fever virus isolates through nucleotide sequencing of the p54, p72, and pB602L (CVR) genes. Virus Genes 2009, 38, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Lubisi, B.A.; Bastos, A.D.; Dwarka, R.M.; Vosloo, W. Intra-genotypic resolution of African swine fever viruses from an East African domestic pig cycle: A combined p72-CVR approach. Virus Genes 2007, 35, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Mweene, A.S.; Pandey, G.S.; Sinyangwe, P.; Nambota, A.; Samui, K.; Kida, H. Viral diseases of livestock in Zambia. Jpn. J. Vet. Res. 1996, 44, 89–105. [Google Scholar] [PubMed]
- World Organisation for Animal Health (OIE). Annual Animal Disease Status-2001. The World Animal Health Information System Data before 2005 (Handistatus). Available online: http://web.oie.int/hs2/zi_pays.asp?c_pays=219&annee=2001 (accessed on 22 June 2017).
- World Organisation for Animal Health (OIE). Annual Animal Disease Status-2002. The World Animal Health Information System Data before 2005 (Handistatus). Available online: http://web.oie.int/hs2/zi_pays.asp?c_pays=219&annee=2002 (accessed on 22 June 2017).
- Phiri, I.K.; Dorny, P.; Gabriël, S.; Willingham, A.L.; Speybroeck, N.; Vercruysse, J. The prevalence of porcine cysticercosis in Eastern and Southern provinces of Zambia. Vet. Parasitol. 2002, 108, 31–39. [Google Scholar] [CrossRef]
- African Swine Fever, Zambia. Promed. 24 September 2001. Available online: http://www.promedmail.org (accessed on 22 June 2017). archive number: 20010924.2321.
- African Swine Fever, Zambia. Promed. 25 September 2002. Available online: http://www.promedmail.org (accessed on 22 June 2017). archive number: 20020925.5394.
- Food and Agriculture Organization of the United Nations (FAO)/EMPRES Early Warning Messages, 2004: Outbreak of African Swine Fever in Zambia (April 2004). Report Date May 2004. Available online: http://www.fao.org/AG/AGAInfo/programmes/en/empres/earlywarning/ew25.html (accessed on 22 June 2017).
- World Organisation for Animal Health (OIE), 2006: African Swine Fever in Zambia. Immediate Notification Ref OIE 5372, Report Date: 14/04/2006. Available online: http://www.oie.int/wahis_2/temp/reports/en_imm_0000005372_20060414_171516.pdf (accessed on 22 June 2017).
- World Organisation for Animal Health (OIE), 2007: African Swine Fever in Zambia. Immediate Notification Ref OIE 6579, Report Date: 12/12/2007. Available online: http://www.oie.int/wahis_2/temp/reports/en_imm_0000006579_20071212_171139.pdf (accessed on 22 June 2017).
- World Organisation for Animal Health (OIE), 2009: African swine fever in Zambia. Follow-up Report No. 2 Ref OIE 8023, Report Date: 23/04/2009. Available online: http://www.oie.int/wahis_2/temp/reports/en_fup_0000008023_20090423_122525.pdf (accessed on 22 June 2017).
- African Swine Fever, Zambia. Promed. 9 February 2008. Available online: http://www.promedmail.org (accessed on 22 June 2017). archive number: 20080209.0527.
- Yabe, J.; Hamambulu, P.; Simulundu, E.; Ogawa, H.; Kajihara, M.; Mori-Kajihara, A.; Changula-Chitanga, K.; Mwase, M.; Mweemba-Muwowo, M.; Chambaro, H.M.; et al. Pathological and molecular diagnosis of the 2013 African swine fever outbreak in Lusaka, Zambia. Trop. Anim. Health Prod. 2015, 47, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Thoromo, J.; Simulundu, E.; Chambaro, H.M.; Mataa, L.; Lubaba, C.H.; Pandey, G.S.; Takada, A.; Misinzo, G.; Mweene, A.S. Diagnosis and genotyping of African swine fever viruses from 2015 outbreaks in Zambia. Onderstepoort J. Vet. Res. 2016, 83, a1095. [Google Scholar] [CrossRef] [PubMed]
- Gago da Câmara, N.J. História da peste suína em Angola. Pecuária 1933, 1, 25–40. [Google Scholar]
- Mendes, A.M. A história da peste suína africana em Angola. Revista Portuguesa Ciências Veterinárias 1994, 89, 110–120. [Google Scholar]
- Penrith, M.-L. History of “swine fever” in Southern Africa. J. S. Afr. Vet. Assoc. 2013, 84. [Google Scholar] [CrossRef] [Green Version]
- El-Sawalhy, A.; Soumaré, B.; Nouala, S.; Mukanda, B.; Wamwayi, H.; Ahmed, I.G. African Swine fever. In Pan African Animal Health Yearbook; Interafrican Bureau for Animal Resources, African Union: Nairobi, Kenya, 2011; pp. 16–17. [Google Scholar]
- Mulumba–Mfumu, L.K.; Achenbach, J.E.; Mauldin, M.R.; Dixon, L.K.; Tshilenge, C.G.; Thiry, E.; Moreno, N.; Blanco, E.; Saegerman, C.; Lamien, C.E.; et al. Genetic assessment of African swine fever isolates involved in outbreaks in the Democratic Republic of Congo between 2005 and 2012 reveals co-circulation of p72 genotypes I, IX and XIV, including 19 variants. Viruses 2017, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Matson, B.A. An outbreak of African swine fever in Nyasaland. Bull. Epizoot. Dis. Afr. 1960, 8, 305–308. [Google Scholar]
- Abreu, E.F.; de Valadão, F.B.; Limpo Serra, J.J.B.; Ornelas Mário, R.; Sousa Montenegro, A. Peste suína africana em Moçambique. Anais Serviços Veterinárias Moçambique 1962, 8, 105–123. [Google Scholar]
- Mendes, A.M. Algumas doenças dos animais em Angola e Moçambique e sua importância na higiene das carnes. Revista Portuguesa Ciencias Veterinárias 1971, 66, 271–286. [Google Scholar]
- Haresnape, J.M.; Lungu, S.A.; Mamu, F.D. An updated survey of African swine fever in Malawi. Epidemiol. Infect. 1987, 99, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Penrith, M.-L.; Lopes Pereira, C.; Lopes da Silva, M.M.R.; Quembo, C.; Nhamusso, A.; Banze, J. African swine fever in Mozambique: Review, risk factors and considerations for control. Onderstepoort J. Vet. Res. 2007, 74, 149–160. [Google Scholar] [PubMed]
- Quembo, C.J.; Jori, F.; Heath, L.; Pérez-Sánchez, R.; Vosloo, W. Investigation into the epidemiology of African swine fever virus at the wildlife-domestic interface of the Gorongosa National Park, Central Mozambique. Transbound. Emerg. Dis. 2016, 63, 443–451. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health (OIE), 2009: African Swine Fever in Namibia. Immediate Notification Ref OIE 8011, Report Date: 15/04/2009. Available online: http://www.oie.int/wahis_2/temp/reports/en_imm_0000008011_20090415_123338.pdf (accessed on 26 July 2017).
- Van Heerden, J.; Malan, K.; Gadaga, B.M.; Spargo, R.M. Reemergence of African swine fever in Zimbabwe, 2015. Emerg. Infect. Dis. 2017, 23, 860–861. [Google Scholar] [CrossRef] [PubMed]
- Wambura, P.N.; Masambu, J.; Msami, H. Molecular diagnosis and epidemiology of African swine fever outbreaks in Tanzania. Vet. Res. Commun. 2006, 30, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Misinzo, G.; Magambo, J.; Masambu, J.; Yongolo, M.G.; Van Doorsselaere, J.; Nauwynck, H.J. Genetic characterization of African swine fever viruses from a 2008 outbreak in Tanzania. Transbound. Emerg. Dis. 2011, 58, 86–92. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health (OIE). African swine fever. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; World Organisation for Animal Health: Paris, France, 2013; Volume 2, Chapter 2.8.1.; Available online: http://www.oie.int/international-standard-setting/terrestrial-manual/access-online/ (accessed on 23 June 2017).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health (OIE), 2008: African Swine Fever in Namibia. Follow-up Report No. 1 Ref OIE 7512, Report Date: 12/11/2008. Available online: http://www.oie.int/wahis_2/temp/reports/en_fup_0000007512_20081112_170845.pdf (accessed on 23 June 2017).
- Sanna, G.; Dei Giudici, S.; Bacciu, D.; Angioi, P.P.; Giammarioli, M.; De Mia, G.M.; Oggiano, A. Improved strategy for molecular characterization of African swine fever viruses from Sardinia, based on analysis of p30, CD2V and I73R/I329L variable regions. Transbound. Emerg. Dis. 2016, 64, 1280–1286. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health (OIE), 2017: African Swine Fever in Zambia. Immediate Notification Ref OIE 23633, Report Date: 26/04/2017. Available online: http://www.oie.int/wahis_2/temp/reports/en_imm_0000023633_20170426_155931.pdf (accessed on 23 July 2017).
- African Swine Fever, Zambia: Muchinga Spread. Promed. 21 July 2017. Available online: http://www.promedmail.org (accessed on 27 July 2017). archive number: 20170724.5200230.
- World Organisation for Animal Health (OIE), 2017: African Swine Fever in Zambia. Follow up Report No. 1 Ref OIE 24520, Report Date: 08/08/2017. Available online: http://www.oie.int/wahis_2/temp/reports/en_fup_0000024520_20170808_160612.pdf (accessed on 9 August 2017).
- El Hicheri, K.; Gomez-Tejedor, C.; Penrith, M.-L.; Davies, G.; Douati, A.; Edoukou, G.; Wojciechowski, K. L’épizootie de peste porcine africaine de 1996 en Côte d’Ivoire. Sci. Tech. Rev. 1998, 17, 660–673. [Google Scholar] [CrossRef]

| Virus Name | p72 Genotype | Cumulative CVR Variants Identified | CVR Sequence | No. of Repeats |
|---|---|---|---|---|
| KAV 89/1 | VIII | 1 | AVSVSVSOVNAVNOVVNVNOVVNVOVOOV | 29 |
| ZON 88/1 | VIII | 2 | AVSVSVSVSOVNAVNOVVNVVOVVNVOVOOV | 31 |
| CHM 88/1 | VIII | 3 | AVSVSOVNAVNOVVNVOVOOV | 21 |
| ZAM/13/Lusaka | I | 4 | BNAF | 4 |
| LIV 13/33 | I | 5 | BNADBNAFTBTDBNAF | 16 |
| ZAM 2002/2 | I | 6 | BNAFNBTDBNAF | 12 |
| LIV 12/17 | I | 7 | BNAAFNBTAFF | 11 |
| LIV 10/11 | I | 8 | BNAAAAAF | 8 |
| ZAM/14/Chipata | II | 9 | BNABNDBTDBNAAG | 14 |
| ZAM/13/Mbala | II | 10 | BNDBNDBNAA | 10 |
| ZAM/13/Kalomo | XIV | 11 | BNWNBVF | 7 |
| ZAM/14/Kasempa | XIV | 12 | AVVWNVWNVWVVF | 13 |
| KAB 6/2 | XI | 13 | AVBNOVAVVBNOVAVVOV | 18 |
| SUM 14/11 | XIII | 14 | AVNAVSSSSVOVBNASOV | 18 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simulundu, E.; Lubaba, C.H.; Van Heerden, J.; Kajihara, M.; Mataa, L.; Chambaro, H.M.; Sinkala, Y.; Munjita, S.M.; Munang’andu, H.M.; Nalubamba, K.S.; et al. The Epidemiology of African Swine Fever in “Nonendemic” Regions of Zambia (1989–2015): Implications for Disease Prevention and Control. Viruses 2017, 9, 236. https://doi.org/10.3390/v9090236
Simulundu E, Lubaba CH, Van Heerden J, Kajihara M, Mataa L, Chambaro HM, Sinkala Y, Munjita SM, Munang’andu HM, Nalubamba KS, et al. The Epidemiology of African Swine Fever in “Nonendemic” Regions of Zambia (1989–2015): Implications for Disease Prevention and Control. Viruses. 2017; 9(9):236. https://doi.org/10.3390/v9090236
Chicago/Turabian StyleSimulundu, Edgar, Caesar H. Lubaba, Juanita Van Heerden, Masahiro Kajihara, Liywalii Mataa, Herman Moses Chambaro, Yona Sinkala, Samuel Munalula Munjita, Hetron Mweemba Munang’andu, King Shimumbo Nalubamba, and et al. 2017. "The Epidemiology of African Swine Fever in “Nonendemic” Regions of Zambia (1989–2015): Implications for Disease Prevention and Control" Viruses 9, no. 9: 236. https://doi.org/10.3390/v9090236
APA StyleSimulundu, E., Lubaba, C. H., Van Heerden, J., Kajihara, M., Mataa, L., Chambaro, H. M., Sinkala, Y., Munjita, S. M., Munang’andu, H. M., Nalubamba, K. S., Samui, K., Pandey, G. S., Takada, A., & Mweene, A. S. (2017). The Epidemiology of African Swine Fever in “Nonendemic” Regions of Zambia (1989–2015): Implications for Disease Prevention and Control. Viruses, 9(9), 236. https://doi.org/10.3390/v9090236









