Characterization of Naturally Occurring NS5A and NS5B Polymorphisms in Patients Infected with HCV Genotype 3a Treated with Direct-Acting Antiviral Agents
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and Patients
2.2. Study Assessments
2.3. Nucleic Acid Extraction and PCR Amplifications
2.4. Sequencing
2.5. Data Analysis
2.6. Phylogenetic Analysis
3. Results
3.1. Baseline Characteristics
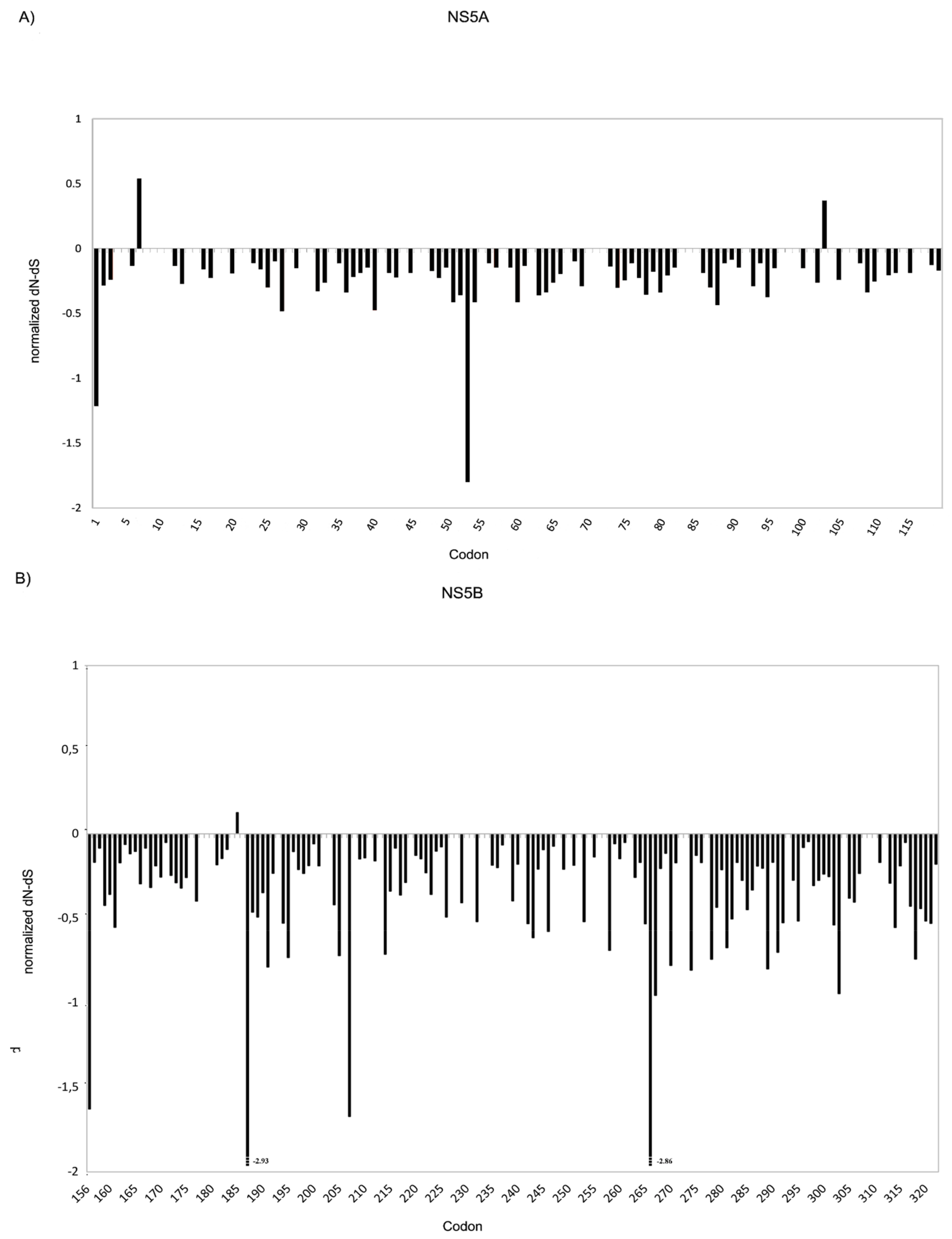
3.2. Genetic Variability at Baseline
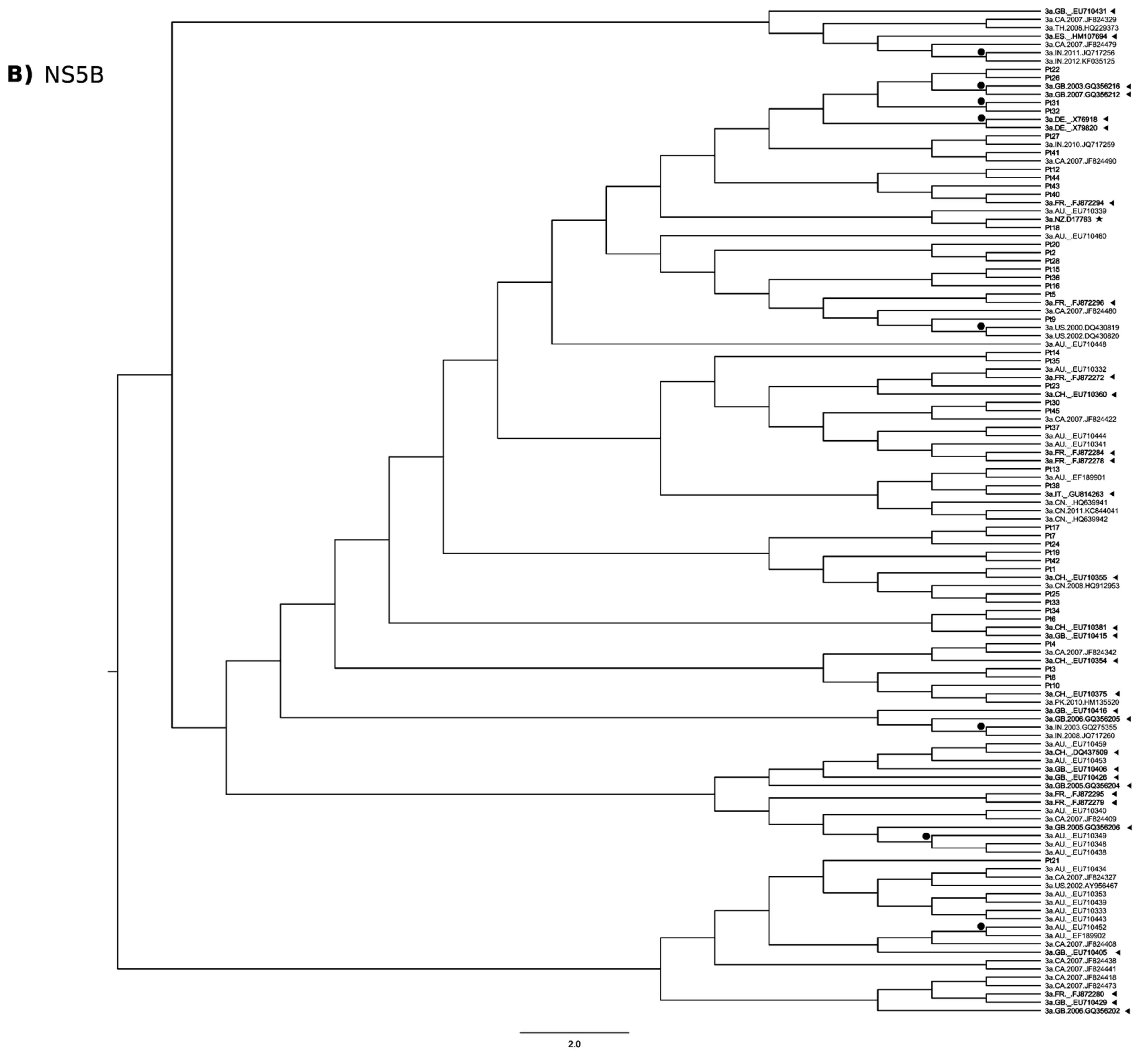
3.3. Phylogenetic Analysis
3.4. Analysis of Emergent Substitutions at Therapy Failure
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Smith, D.B.; Bukh, J.; Kuiken, C.; Muerhoff, A.S.; Rice, C.M.; Stapleton, J.T.; Simmonds, P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatolology 2014, 59, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Humphreys, I.; Flaxman, A.; Brown, A.; Cooke, G.S.; Pybus, O.G.; Barnes, E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 2015, 61, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, F.; Kramer, J.R.; Ilyas, J.; Duan, Z.; El-Serag, H.B. HCV genotype 3 is associated with an increased risk of cirrhosis and hepatocellular cancer in a national sample of U.S. Veterans with HCV. Hepatology 2014, 60, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Pawlotsky, J.-M.; Feld, J.J.; Zeuzem, S.; Hoofnagle, J.H. From non-A, non-B hepatitis to hepatitis C virus cure. J. Hepatol. 2015, 62, S87–S99. [Google Scholar] [CrossRef] [PubMed]
- Aghemo, A.; Rumi, M.G.; Monico, S.; Prati, G.M.; D’Ambrosio, R.; Donato, M.F.; Colombo, M. The pattern of pegylated interferon-alpha2b and ribavirin treatment failure in cirrhotic patients depends on hepatitis C virus genotype. Antivir. Ther. 2009, 14, 577–584. [Google Scholar] [PubMed]
- European Association for the Study of the Liver. EASL Recommendations on Treatment of Hepatitis C 2016. J. Hepatol. 2017, 66, 153–194. [Google Scholar] [CrossRef]
- Nelson, D.R.; Cooper, J.N.; Lalezari, J.P.; Lawitz, E.; Pockros, P.J.; Gitlin, N.; Freilich, B.F.; Younes, Z.H.; Harlan, W.; Ghalib, R.; et al. All-oral 12-week treatment with daclatasvir plus sofosbuvir in patients with hepatitis C virus genotype 3 infection: ALLY-3 phase III study. Hepatology 2015, 61, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- Leroy, V.; Angus, P.; Bronowicki, J.-P.; Dore, G.J.; Hezode, C.; Pianko, S.; Pol, S.; Stuart, K.; Tse, E.; McPhee, F.; et al. Daclatasvir, sofosbuvir, and ribavirin for hepatitis C virus genotype 3 and advanced liver disease: A randomized phase III study (ALLY-3+). Hepatology 2016, 63, 1430–1441. [Google Scholar] [CrossRef] [PubMed]
- AASLD HCV-Guidance_April_2016 AASLD. Available online: http://hcvguidelines.org/sites/default/files/HCV-Guidance_April_2016_e1.pdf (accessed on 1 May 2016).
- Lu, L.; Li, C.; Yuan, J.; Lu, T.; Okamoto, H.; Murphy, D.G. Full-length genome sequences of five hepatitis C virus isolates representing subtypes 3g, 3h, 3i and 3k, and a unique genotype 3 variant. J. Gen. Virol. 2013, 94, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Morice, Y.; Cantaloube, J.-F.; Beaucourt, S.; Barbotte, L.; De Gendt, S.; Goncales, F.L.; Butterworth, L.; Cooksley, G.; Gish, R.G.; Beaugrand, M.; et al. Molecular epidemiology of hepatitis C virus subtype 3a in injecting drug users. J. Med. Virol. 2006, 78, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Saraswat, V.; Norris, S.; de Knegt, R.J.; Sanchez Avila, J.F.; Sonderup, M.; Zuckerman, E.; Arkkila, P.; Stedman, C.; Acharya, S.; Aho, I.; et al. Historical epidemiology of hepatitis C virus (HCV) in select countries—Volume 2. J. Viral Hepat. 2015, 22 (Suppl. 1), 6–25. [Google Scholar] [CrossRef] [PubMed]
- Italian Medicines Agency AIFA. Available online: http://www.agenziafarmaco.gov.it/ (accessed on 3 May 2017).
- Lindström, I.; Kjellin, M.; Palanisamy, N.; Bondeson, K.; Wesslén, L.; Lannergard, A.; Lennerstrand, J. Prevalence of polymorphisms with significant resistance to NS5A inhibitors in treatment-naive patients with hepatitis C virus genotypes 1a and 3a in Sweden. Infect. Dis. Lond. Engl. 2015, 47, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Pilon, R.; Leonard, L.; Kim, J.; Vallee, D.; De Rubeis, E.; Jolly, A.M.; Wylie, J.; Pelude, L.; Sandstrom, P. Transmission patterns of HIV and hepatitis C virus among networks of people who inject drugs. PLoS ONE 2011, 6, e22245. [Google Scholar] [CrossRef] [PubMed]
- Pawlotsky, J.-M. Hepatitis C Virus Resistance to Direct-Acting Antiviral Drugs in Interferon-Free Regimens. Gastroenterology 2016, 151, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Simen, B.B.; Simons, J.F.; Hullsiek, K.H.; Novak, R.M.; MacArthur, R.D.; Baxter, J.D.; Huang, C.; Lubeski, C.; Turenchalk, G.S.; Braverman, M.S.; et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment–naive patients significantly impact treatment outcomes. J. Infect. Dis. 2009, 199, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Kuiken, C.; Yusim, K.; Boykin, L.; Richardson, R. The Los Alamos hepatitis C sequence database. Bioinforma. Oxf. Engl. 2005, 21, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Frost, S.D.W. Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 2005, 21, 2531–2533. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinforma. Oxf. Engl. 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Quer, J.; Gregori, J.; Rodríguez-Frias, F.; Buti, M.; Madejon, A.; Perez-del-Pulgar, S.; Garcia-Cehic, D.; Casillas, R.; Blasi, M.; Homs, M.; et al. High-resolution hepatitis C virus subtyping using NS5B deep sequencing and phylogeny, an alternative to current methods. J. Clin. Microbiol. 2015, 53, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 1989, 5, 164–166. [Google Scholar]
- Bartolini, B.; Giombini, E.; Zaccaro, P.; Selleri, M.; Rozera, G.; Abbate, I.; Comandini, U.V.; Ippolito, G.; Solmone, M.; Capobianchi, M.R. Extent of HCV NS3 protease variability and resistance-associated mutations assessed by next generation sequencing in HCV monoinfected and HIV/HCV coinfected patients. Virus Res. 2013, 177, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Sulkowski, M.S.; Gardiner, D.F.; Rodriguez-Torres, M.; Reddy, K.R.; Hassanein, T.; Jacobson, I.; Lawitz, E.; Lok, A.S.; Hinestrosa, F.; Thuluvath, P.J.; et al. AI444040 Study Group Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N. Engl. J. Med. 2014, 370, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Han, Z.; Hartman-Neumann, S.; DeGray, B.; Ueland, J.; Vellucci, V.; Hernandez, D.; McPhee, F. Characterization of NS5A polymorphisms and their impact on response rates in patients with HCV genotype 2 treated with daclatasvir-based regimens. J. Antimicrob. Chemother. 2016, 71, 3495–3505. [Google Scholar] [CrossRef] [PubMed]
- McPhee, F.; Hernandez, D.; Zhou, N. Effect of minor populations of NS5A and NS5B resistance-associated variants on HCV genotype-3 response to daclatasvir plus sofosbuvir, with or without ribavirin. Antivir. Ther. 2017, 22, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Le Pogam, S.; Li, L.; Haines, K.; Piso, K.; Baronas, V.; Yan, J.-M.; So, S.-S.; Klumpp, K.; Nájera, I. In vivo emergence of a novel mutant L159F/L320F in the NS5B polymerase confers low-level resistance to the HCV polymerase inhibitors mericitabine and sofosbuvir. J. Infect. Dis. 2014, 209, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-W.; Li, H.; Ren, H.; Hu, P. Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): Mining the GenBank HCV genome data. Sci. Rep. 2016, 6, 20310. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.; Zhou, N.; Ueland, J.; Monikowski, A.; McPhee, F. Natural prevalence of NS5A polymorphisms in subjects infected with hepatitis C virus genotype 3 and their effects on the antiviral activity of NS5A inhibitors. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2013, 57, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Jia, L.; O’Boyle, D.R.; Sun, J.-H.; Rigat, K.; Valera, L.; Nower, P.; Huang, X.; Kienzle, B.; Roberts, S.; et al. Comparison of daclatasvir resistance barriers on NS5A from hepatitis C virus genotypes 1 to 6: Implications for cross-genotype activity. Antimicrob. Agents Chemother. 2014, 58, 5155–5163. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Valera, L.; Jia, L.; Kirk, M.J.; Gao, M.; Fridell, R.A. In vitro activity of daclatasvir on hepatitis C virus genotype 3 NS5A. Antimicrob. Agents Chemother. 2013, 57, 611–613. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, S.; Kanda, T.; Wu, S.; Shirasawa, H.; Yokosuka, O. Hepatitis C virus NS5A inhibitors and drug resistance mutations. World J. Gastroenterol. 2014, 20, 2902–2912. [Google Scholar] [CrossRef] [PubMed]

| Total | SOF + DCV | SOF + DCV + RBV | SOF + RBV | SOF + P-R | Not Known | ||
|---|---|---|---|---|---|---|---|
| Number of patients (F/M) | 45 | 7 | 16 | 12 | 9 | 1 | |
| (7/38) | (0/7) | (1/15) | (2/10) | (4/5) | (0/1) | ||
| Median Age, years (range) | 56 | 56 | 56 | 57 | 56 | n.a. | |
| (42–69) | (49–63) | (42–63) | (52–69) | (47–66) | |||
| HCV RNA (Log10 IU/mL) Median (Min–Max) | 5.3 | 5.0 | 5.2 | 5.5 | 5.9 | 5.4 | |
| (1.1–7.0) | (1.1–7.0) | (4.0–6.8) | (3.8–6.7) | (3.2–6.7) | |||
| HIV positive patients (F/M) | 3 | 0 | 1 | 2 | 0 | 0 | |
| (0/3) | (0/0) | (0/1) | (0/2) | (0/0) | (0/0) | ||
| Number of patients without cirrhosis * (F/M) | 9 | 2 | 3 | 3 | 1 | n.a. | |
| (0/9) | (0/2) | (0/3) | (0/3) | (0/1) | |||
| Number of patients with cirrhosis * (F/M) | 35 | 5 | 13 | 9 | 8 | n.a. | |
| (7/28) | (0/5) | (1/12) | (2/7) | (4/4) | |||
| Prior pegIFN experience | Treatment–naive (F/M) | 26 | 5 | 8 | 6 | 6 | 1 |
| (3/23) | (0/5) | (0/8) | (1/5) | (2/4) | (0/1) | ||
| Treatment–experienced (F/M) | 19 | 2 | 8 | 6 | 3 | 0 | |
| (4/15) | (0/2) | (1/7) | (1/5) | (2/1) | (0/0) | ||
| Pt | Fibrosis Stage | Prior IFN Treatment Experience (Response) | DAA | Baseline HCV RNA (LogIU/mL) | Antiviral Regimen (wks) | Response | Known RAS Detected at Failure by Sanger Sequencing | |
|---|---|---|---|---|---|---|---|---|
| NS5A | NS5B | |||||||
| 42 | 3 | Naïve | First DAA regimen | 7.0 | SOF + DCV (12) | Relapser | L31F | none |
| Second DAA regimen | 7.0 | SOF + DCV + RBV (24) | Relapser | L31F | none | |||
| 43 | 4 | Naïve | First DAA regimen | 5.0 | SOF + DCV (24) | Relapser | P58S Y93H | S282T |
| Second DAA regimen | 5.8 | SOF + RBV (48) | Relapser | P58S Y93H | none | |||
| 44 | 4 | P-R (Null responder) | First DAA regimen | 5.9 | SOF + RBV (24) | Relapser | none | none |
| Second DAA regimen | 6.3 | SOF + DCV + RBV (24) | SVR12 | n.a. | n.a. | |||
| 45 | 4 | P-R (Null responder) | First DAA regimen | 4.6 | SOF + RBV (24) | Relapser | none | none |
| Second DAA regimen | 4.1 | SOF + DCV + RBV (24) | SVR12 | n.a. | n.a. | |||
| Non-Structural Protein 5A (NS5A) | ||||||||||||||||
| Reference | D40.5 | D92.9 | R95.2 | T52.4 | D97.6 | S83.3 | L100.0 | A4.8 | K97.6 | A9.5 | L97.6 | K97.6 | M97.6 | A90.5 | L97.6 | I97.6 |
| aa | 2 | 3 | 6 | 7 | 10 | 14 | 16 | 17 | 20 | 21 | 23 | 26 | 28 | 30 | 34 | 37 |
| Variants % | G61.9 | E7.1 | H4.8 | A2.4 | E2.4 | I2.4 | V2.4 | S90.5 | R2.4 | T92.9 | I2.4 | R2.4 | L2.4 | K9.5 | I2.4 | L2.4 |
| N2.4 | D23.8 | L2.4 | T2.4 | |||||||||||||
| I14.3 | T16.7 | Y2.4 | ||||||||||||||
| V16.7 | ||||||||||||||||
| Reference | K95.2 | Y97.6 | K97.6 | V97.6 | M100.0 | S97.6 | T97.6 | P92.9 | A2.4 | I97.6 | T88.1 | V100.0 | L97.6 | A92.9 | P97.6 | T90.5 |
| aa | 41 | 43 | 44 | 46 | 53 | 54 | 55 | 58 | 62 | 63 | 64 | 67 | 74 | 75 | 77 | 79 |
| Variants % | R4.8 | F2.4 | R2.4 | A2.4 | V2.4 | A2.4 | A2.4 | A2.4 | P2.4 | L2.4 | A7.1 | I2.4 | I2.4 | V9.5 | S2.4 | A2.4 |
| L2.4 | S2.4 | S85.7 | S4.8 | M2.4 | M2.4 | |||||||||||
| T2.4 | T14.3 | R2.4 | ||||||||||||||
| S2.4 | ||||||||||||||||
| Reference | M97.6 | H73.8 | Y95.2 | S66.7 | S54.8 | P97.6 | T83.3 | W97.6 | N90.5 | S97.6 | ||||||
| aa | 83 | 85 | 93 | 98 | 103 | 104 | 107 | 111 | 116 | 117 | ||||||
| Variants % | T4.8 | C2.4 | D4.8 | G35.7 | A4.8 | L2.4 | A2.4 | L2.4 | S9.5 | G2.4 | ||||||
| R2.4 | H2.4 | P45.2 | K2.4 | |||||||||||||
| Y28.6 | N2.4 | N2.4 | ||||||||||||||
| S7.1 | ||||||||||||||||
| Non-Structural Protein 5B (NS5B) | ||||||||||||||||
| Reference | P95.2 | G100.0 | V100 | Q97.6 | I90.5 | E88.1 | T95.2 | G97.6 | P31.0 | K81.0 | S95.2 | T92.9 | Q97.6 | V95.2 | I97.6 | N90.5 |
| aa | 156 | 166 | 169 | 180 | 184 | 185 | 186 | 188 | 189 | 206 | 210 | 213 | 231 | 235 | 239 | 244 |
| Variants % | A4.8 | R2.4 | I2.4 | K2.4 | L4.8 | A11.9 | A2.4 | S2.4 | A4.8 | E2.4 | A4.8 | A4.8 | R2.4 | G2.4 | V2.4 | D11.9 |
| T4.8 | G2.4 | V2.4 | S64.3 | Q14.3 | N2.4 | M4.8 | ||||||||||
| T2.4 | ||||||||||||||||
| Reference | E97.6 | R90.5 | S97.6 | S97.6 | F97.6 | K97.6 | A85.7 | Q95.2 | C100.0 | I95.2 | T100.0 | K76.2 | N0.0 | R97.6 | N97.6 | F97.6 |
| aa | 246 | 250 | 254 | 255 | 267 | 270 | 272 | 273 | 274 | 293 | 300 | 304 | 307 | 309 | 310 | 313 |
| Variants % | Q2.4 | K11.9 | T2.4 | A2.4 | Y2.4 | R2.4 | D16.7 | P4.8 | W2.4 | L4.8 | S2.4 | R26.2 | G97.6 | Q2.4 | D4.8 | L2.4 |
| S2.4 | ||||||||||||||||
| Non-Structural Protein 5A (NS5A) | ||||||
| Ref. | Amino-Acid Position | Variant | Baseline n° Reads (Frequency, %) | T1 n° Reads (Frequency, %) | T2 n° Reads (Frequency, %) | |
| Pt42 | A | 30 | K | 847 (92.0) | n.d. | n.d. |
| R | 72 (7.8) | 12 (1.3) | n.d. | |||
| T | n.d. | 884 (97.2) | 1526 (98.4) | |||
| L | 31 | F | n.d. | 902 (99.2) | 1527 (98.5) | |
| L | 34 | I | n.d. | 898 (98.8) | 1536 (99.1) | |
| V | 52 | M | n.d. | 585 (64.4) | 1530 (98.741) | |
| A | 62 | S | 915 (99.3) | 894 (98.3) | 1539 (99.3) | |
| T | 87 | P | n.d. | 672 (74.0) | n.d. | |
| Y | 93 | F | 10 (1.1) | n.d. | n.d. | |
| Diversity (nt substitutions per site ×10−4) | 173.6 | 112.8 | 80.8 | |||
| Pt43 | P | 58 | M | n.d. | n.d. | 29 (1.26) |
| S | 2289 (99.1) | 2966 (99.2) | 5401 (99.4) | |||
| A | 62 | S* | 2300 (99.6) | 2983 (99.7) | 5421 (99.7) | |
| Y | 93 | C | 42 (1.8) | n.d. | n.d. | |
| H | n.d. | 2968 (99.2) | 5408 (99.5) | |||
| Diversity (nt substitutions per site ×10−4) | 373.4 | 53.8 | 59.0 | |||
| Non-Structural Protein 5B (NS5B) | ||||||
| Ref. | Amino-Acid Position | Variant | Baseline n° Reads Frequency, %) | T1 n° Reads (Frequency, %) | T2 n° Reads (Frequency, %) | |
| Pt42 | G | 188 | A | 946 (45.1) | n.a. | n.d. |
| K | 212 | R | 944 (45.0) | n.a. | n.d. | |
| N | 244 | D | 364 (17.4) | n.a. | n.d. | |
| K | 304 | R | 540 (51.1) | n.a. | n.d. | |
| A | 306 | V | 5 (0.5) | n.a. | 888 (95.3) | |
| Diversity (nt substitutions per site ×10−4) | 276.4 | n.a. | 62.7 | |||
| Pt43 | I | 160 | V | n.d. | 4 (0.4) | 1120 (94.0) |
| Q | 273 | P | 62 (3.0) | 1971 (99.7) | 2670 (99.0) | |
| S | 282 | T | n.d. | 1892 (95.7) | n.d. | |
| L | 285 | F | n.d. | 1920 (97.2) | 2677 (99.2) | |
| Diversity (nt substitutions per site ×10−4) | 232.2 | 59.6 | 53.1 | |||
| Pt45 | L | 159 | P | n.d. | 5 (0.54) | n.a. |
| I | 184 | T | 15 (0.7) | 2001 (97.32) | n.a. | |
| V | 1312 (60.3) | n.d. | n.a. | |||
| G | 188 | D | 16 (0.7) | 1995 (97.03) | n.a. | |
| P | 189 | S | 2169 (99.6) | 2049 (99.66) | n.a. | |
| K | 206 | E | 1311 (60.2) | n.d. | n.a. | |
| V | 235 | M | 693 (31.8) | n.d. | n.a. | |
| N | 307 | G | 1221 (99.9) | 1134 (99.6) | n.a. | |
| N | 310 | S | 8 (0.6) | 1111 (97.6) | n.a. | |
| Diversity (nt substitutions per site ×10−4) | 384.0 | 96.4 | n.a. | |||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartolini, B.; Giombini, E.; Taibi, C.; Lionetti, R.; Montalbano, M.; Visco-Comandini, U.; D’Offizi, G.; Capobianchi, M.R.; McPhee, F.; Garbuglia, A.R. Characterization of Naturally Occurring NS5A and NS5B Polymorphisms in Patients Infected with HCV Genotype 3a Treated with Direct-Acting Antiviral Agents. Viruses 2017, 9, 212. https://doi.org/10.3390/v9080212
Bartolini B, Giombini E, Taibi C, Lionetti R, Montalbano M, Visco-Comandini U, D’Offizi G, Capobianchi MR, McPhee F, Garbuglia AR. Characterization of Naturally Occurring NS5A and NS5B Polymorphisms in Patients Infected with HCV Genotype 3a Treated with Direct-Acting Antiviral Agents. Viruses. 2017; 9(8):212. https://doi.org/10.3390/v9080212
Chicago/Turabian StyleBartolini, Barbara, Emanuela Giombini, Chiara Taibi, Raffaella Lionetti, Marzia Montalbano, Ubaldo Visco-Comandini, Gianpiero D’Offizi, Maria Rosaria Capobianchi, Fiona McPhee, and Anna Rosa Garbuglia. 2017. "Characterization of Naturally Occurring NS5A and NS5B Polymorphisms in Patients Infected with HCV Genotype 3a Treated with Direct-Acting Antiviral Agents" Viruses 9, no. 8: 212. https://doi.org/10.3390/v9080212
APA StyleBartolini, B., Giombini, E., Taibi, C., Lionetti, R., Montalbano, M., Visco-Comandini, U., D’Offizi, G., Capobianchi, M. R., McPhee, F., & Garbuglia, A. R. (2017). Characterization of Naturally Occurring NS5A and NS5B Polymorphisms in Patients Infected with HCV Genotype 3a Treated with Direct-Acting Antiviral Agents. Viruses, 9(8), 212. https://doi.org/10.3390/v9080212







