Complex Virus–Host Interactions Involved in the Regulation of Classical Swine Fever Virus Replication: A Minireview
Abstract
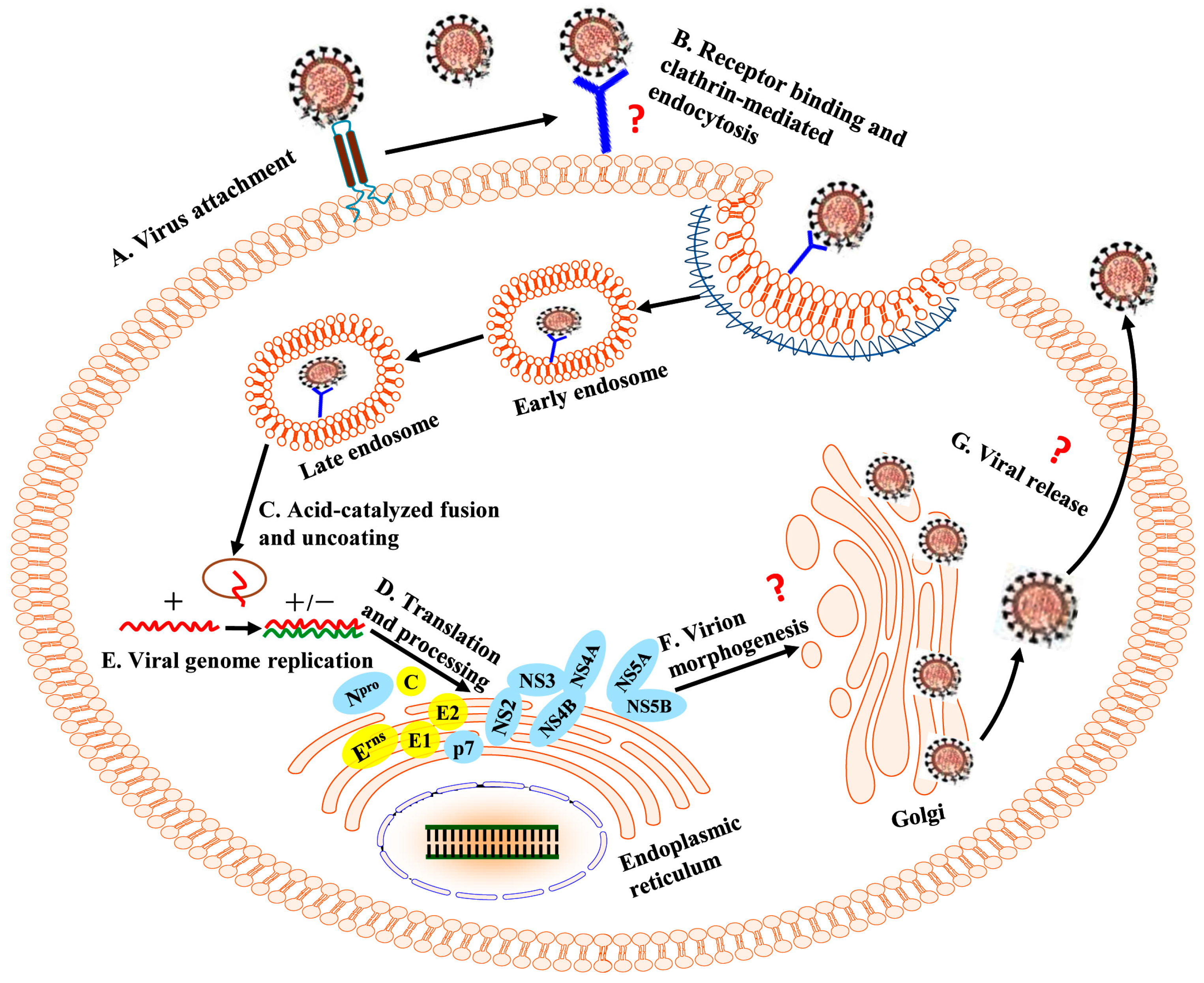
:1. Introduction
2. The CSFV Envelope Proteins Mediate Virus Attachment and Entry
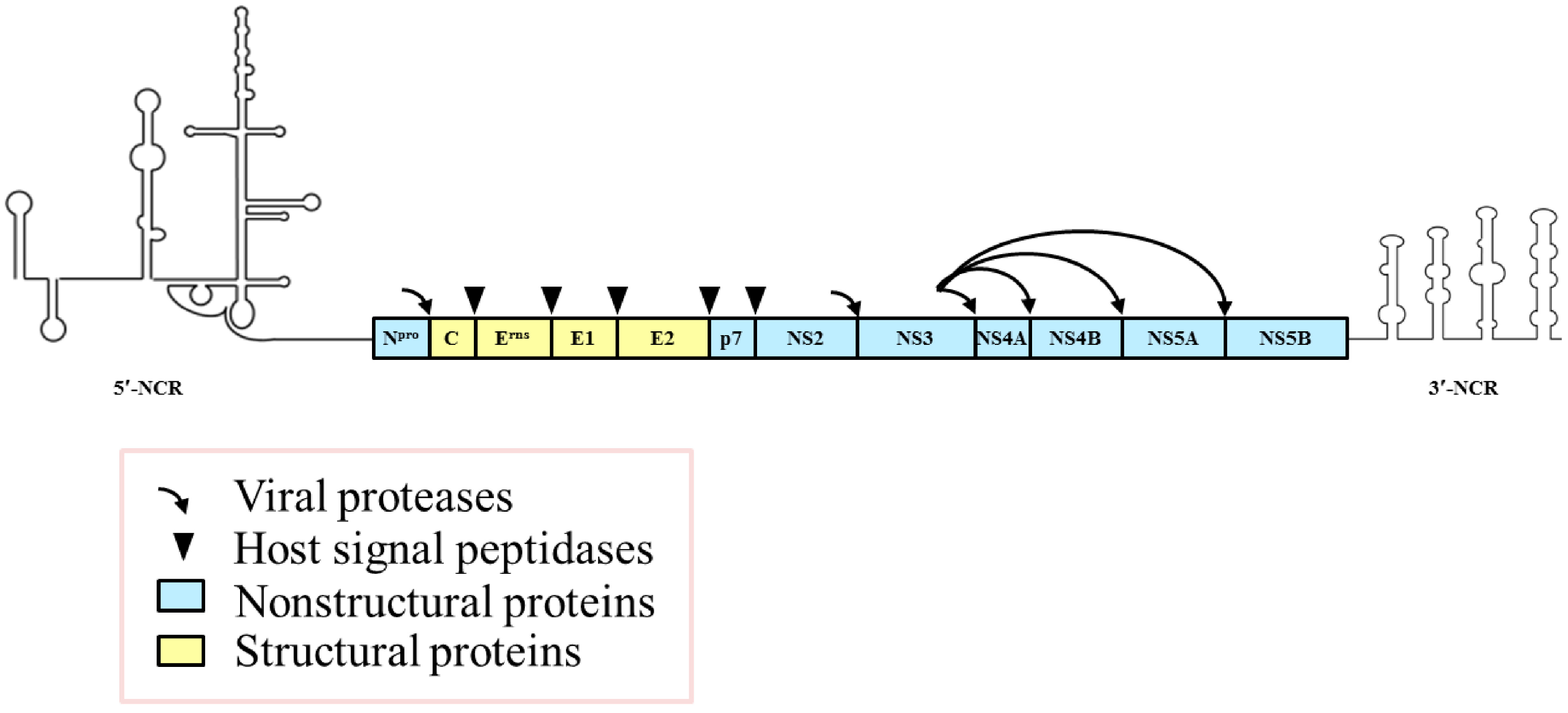
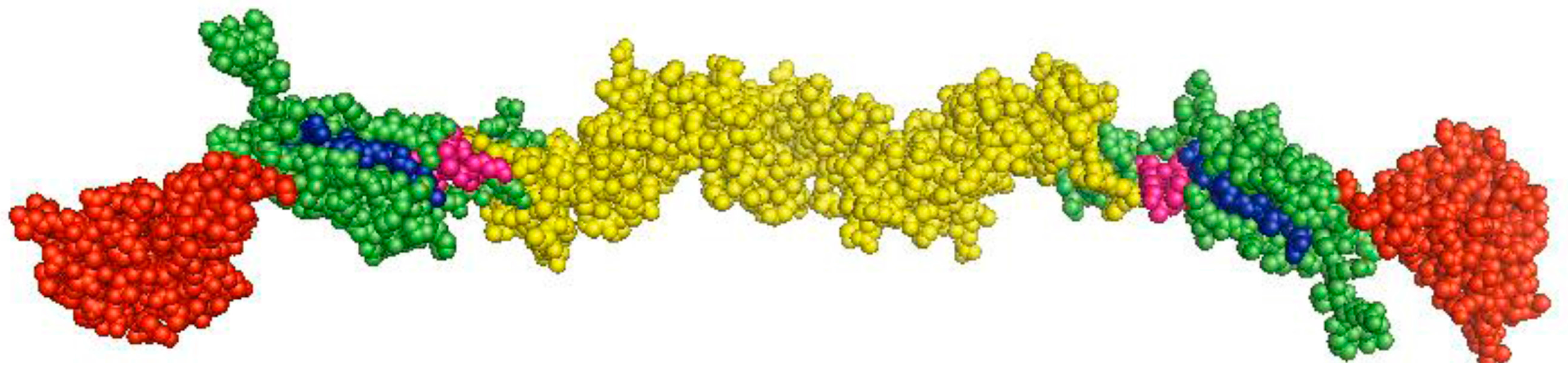
3. Modulation of Viral Genomic Replication and Translation by NCRs and Nonstructural Proteins (NSPs)
4. Interactions between CSFV and Host Cellular Proteins Are Necessary for the CSFV Life Cycle
4.1. Host Factors Modulate the Production of Progeny Virus
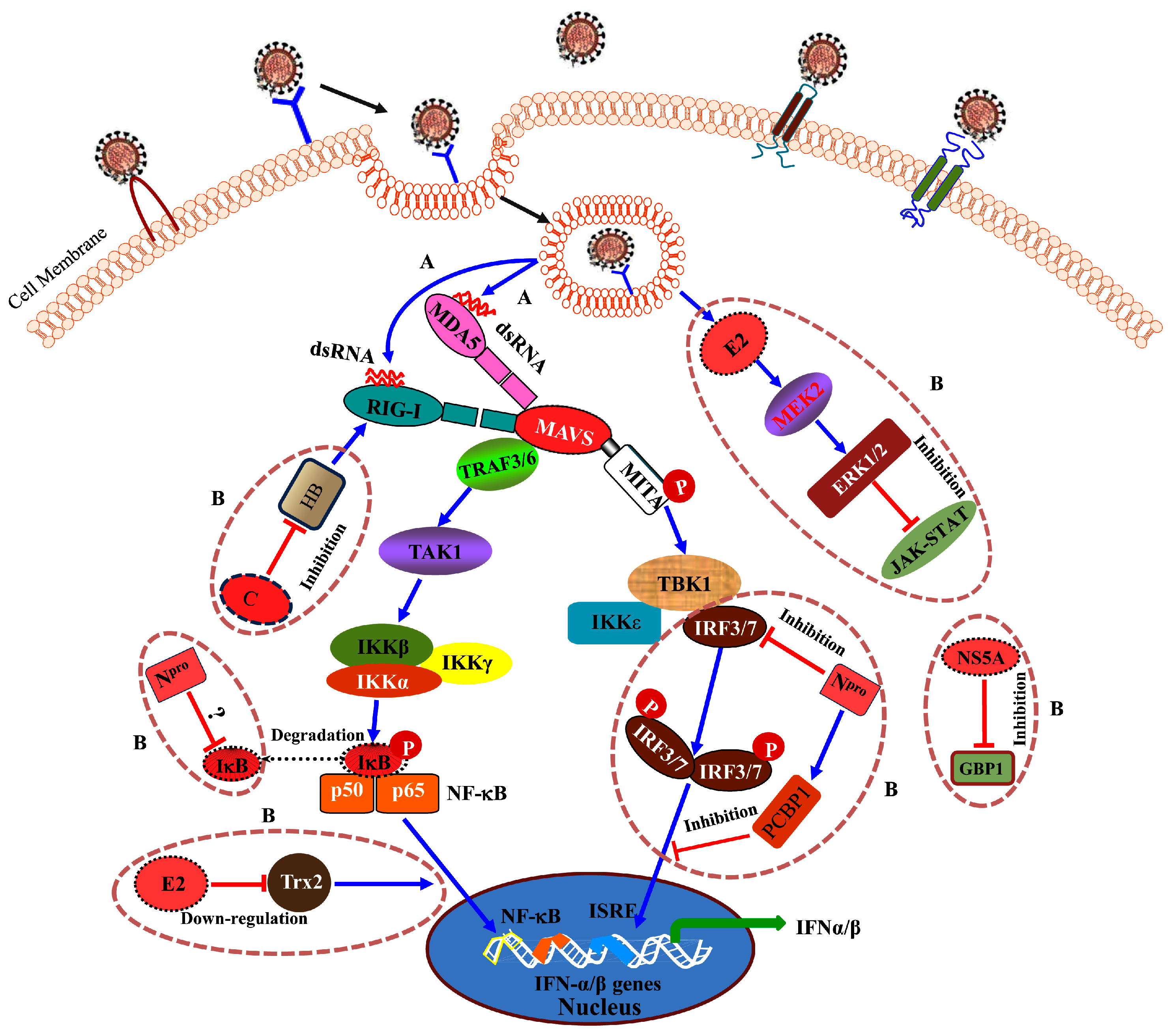
4.2. Viral Proteins Block the Host Innate Immunity
4.3. Disruption of Some Virus-Host Interactions Affects the Viral Virulence in Pigs
5. Changes of Cell Apoptosis and Cell Cycle Induced by CSFV Infection
6. Concluding Remarks and Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ji, W.; Guo, Z.; Ding, N.Z.; He, C.Q. Studying classical swine fever virus: Making the best of a bad virus. Virus Res. 2015, 197, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Lu, Z.; Li, H.; Yu, X.; Liu, X.; Li, Y.; Zhang, H.; Yin, Z. Phylogenetic comparison of classical swine fever virus in China. Virus Res. 2001, 81, 29–37. [Google Scholar] [CrossRef]
- Luo, Y.; Ji, S.; Liu, Y.; Lei, J.L.; Xia, S.L.; Wang, Y.; Du, M.L.; Shao, L.; Meng, X.Y.; Zhou, M.; et al. Isolation and characterization of a moderately virulent classical swine fever virus emerging in China. Transbound. Emerg. Dis. 2016. [Google Scholar] [CrossRef] [PubMed]
- Tu, C. Classical swine fever: International trend, Chinese status and control measures. Sci. Agri. Sin. 2003, 36, 955–960. [Google Scholar]
- Zhu, Y.; Shi, Z.; Drew, T.W.; Wang, Q.; Qiu, H.; Guo, H.; Tu, C. Antigenic differentiation of classical swine fever viruses in China by monoclonal antibodies. Virus Res. 2009, 142, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Pletnev, A.; Gould, E.; Heinz, F.X.; Meyers, G.; Thiel, H.J.; Bukh, J.; Stiasny, K.; Collett, M.S.; Becher, P.; Simmonds, P.; et al. Flaviviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Academic Press: London, UK, 2011; pp. 1003–1020. [Google Scholar]
- Lindenbach, B.D.; Murray, C.L.; Thiel, H.J. Flaviviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Cohen, J.I., Griffin, D.E., Lamb, R.A., Martin, M.A., Racaniello, V.R., Roizman, B., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2, pp. 712–746. [Google Scholar]
- Summerfield, A.; Knötig, S.M.; McCullough, K.C. Lymphocyte apoptosis during classical swine fever: Implication of activation-induced cell death. J. Virol. 1998, 72, 1853–1861. [Google Scholar] [PubMed]
- Summerfield, A.; Knötig, S.M.; Tschudin, R.; McCullough, K.C. Pathogenesis of granulocytopenia and bone marrow atrophy during classical swine fever involves apoptosis and necrosis of uninfected cells. Virology 2000, 272, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Branza-Nichita, N.; Lazar, C.; Dwek, R.A.; Zitzmann, N. Role of N-glycan trimming in the folding and secretion of the pestivirus protein Erns. Biochem. Biophys. Res. Commun. 2004, 319, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Thiel, H.J.; Stark, R.; Weiland, E.; Rümenapf, T.; Meyers, G. Hog cholera virus: Molecular composition of virions from a pestivirus. J. Virol. 1991, 65, 4705–4712. [Google Scholar] [PubMed]
- Lazar, C.; Zitzmann, N.; Dwek, R.A.; Branza-Nichita, N. The pestivirus Erns glycoprotein interacts with E2 in both infected cells and mature virions. Virology 2003, 2, 696–705. [Google Scholar] [CrossRef]
- Krey, T.; Bontems, F.; Vonrhein, C.; Vaney, M.C.; Bricogne, G.; Rümenapf, T.; Rey, F.A. Crystal structure of the pestivirus envelope glycoprotein Erns and mechanistic analysis of its ribonuclease activity. Structure 2012, 20, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Aberle, D.; Muhle-Goll, C.; Bürck, J.; Wolf, M.; Reißer, S.; Luy, B.; Wenzel, W.; Ulrich, A.S.; Meyers, G. Structure of the membrane anchor of pestivirus glycoprotein Erns, a long tilted amphipathic helix. PLoS Pathog. 2014, 10, e1003973. [Google Scholar] [CrossRef] [PubMed]
- Hulst, M.M.; van Gennip, H.G.; Moormann, R.J. Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein Erns. J. Virol. 2000, 74, 9553–9561. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; He, W.R.; Shen, L.; Dong, H.; Yu, J.; Wang, X.; Yu, S.; Li, Y.; Li, S.; Luo, Y.; et al. The laminin receptor is a cellular attachment receptor for classical swine fever virus. J. Virol. 2015, 89, 4894–4906. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Nie, Y.; Wang, P.; Ding, M.; Deng, H. Characterization of classical swine fever virus entry by using pseudotyped viruses: E1 and E2 are sufficient to mediate viral entry. Virology 2004, 330, 332–341. [Google Scholar] [CrossRef] [PubMed]
- El Omari, K.; Iourin, O.; Harlos, K.; Grimes, J.M.; Stuart, D.I. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013, 3, 30–35. [Google Scholar] [CrossRef] [PubMed]
- König, M.; Lengsfeld, T.; Pauly, T.; Stark, R.; Thiel, H.J. Classical swine fever virus: Independent induction of protective immunity by two structural glycoproteins. J. Virol. 1995, 69, 6479–6486. [Google Scholar] [PubMed]
- Yu, X.; Tu, C.; Li, H.; Hu, R.; Chen, C.; Li, Z.; Zhang, M.; Yin, Z. DNA-mediated protection against classical swine fever virus. Vaccine 2001, 19, 1520–1525. [Google Scholar] [CrossRef]
- Li, N.; Qiu, H.J.; Zhao, J.J.; Li, Y.; Wang, M.J.; Lu, B.W.; Han, C.G.; Hou, Q.; Wang, Z.H.; Gao, H.; et al. A Semliki forest virus replicon vectored DNA vaccine expressing the E2 glycoprotein of classical swine fever virus protects pigs from lethal challenge. Vaccine 2007, 25, 2907–2912. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, H.Y.; Tian, D.Y.; Han, Q.Y.; Zhang, X.; Li, N.; Qiu, H.J. A novel alphavirus replicon-vectored vaccine delivered by adenovirus induces sterile immunity against classical swine fever. Vaccine 2011, 29, 8364–8372. [Google Scholar] [CrossRef] [PubMed]
- Gavrilov, B.K.; Rogers, K.; Fernández-Sainz, I.J.; Holinka, L.G.; Borca, M.V.; Risatti, G.R. Effects of glycosylation on antigenicity and immunogenicity of classical swine fever virus envelope proteins. Virology 2011, 420, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Wensvoort, G. Topographical and functional mapping of epitopes on hog cholera virus with monoclonal antibodies. J. Gen. Virol. 1989, 70, 2865–2876. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Lin, F.; Mallory, M.; Clavijo, A. Deletions of structural glycoprotein E2 of classical swine fever virus strain Alfort/187 resolve a linear epitope of monoclonal antibody WH303 and the minimal N-terminal domain essential for binding immunoglobulin G antibodies of a pig hyperimmune serum. J. Virol. 2000, 24, 11619–11625. [Google Scholar] [CrossRef]
- Liu, S.; Yu, X.; Wang, C.; Wu, J.; Kong, X.; Tu, C. Quadruple antigenic epitope peptide producing immune protection against classical swine fever virus. Vaccine 2006, 24, 7175–7180. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhang, B.Q.; Shen, Z.; Chen, Y.H. Antigens containing TAVSPTTLR tandem repeats could be used in assaying antibodies to classical swine fever virus. Acta Virol. 2009, 53, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Li, G.X.; Zhou, Y.J.; Yu, H.; Li, L.; Wang, Y.X.; Tong, W.; Hou, J.W.; Xu, Y.Z.; Zhu, J.P.; Xu, A.T.; et al. A novel dendrimeric peptide induces high level neutralizing antibodies against classical swine fever virus in rabbits. Vet. Microbiol. 2012, 156, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Van der Wal, F.J.; Jelsma, T.; Fijten, H.; Achterberg, R.P.; Loeffen, W.L. Towards a peptide-based suspension array for the detection of pestivirus antibodies in swine. J. Virol. Methods 2016, 235, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Kanai, R.; Modis, Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc. Natl. Acad. Sci. USA 2013, 110, 6805–6810. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sainz, I.J.; Largo, E.; Gladue, D.P.; Fletcher, P.; O’Donnell, V.; Holinka, L.G.; Carey, L.B.; Lu, X.; Nieva, J.L.; Borca, M.V. Effect of specific amino acid substitutions in the putative fusion peptide of structural glycoprotein E2 on classical swine fever virus replication. Virology 2014, 456–457, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Holinka, L.G.; Largo, E.; Gladue, D.P.; O’Donnell, V.; Risatti, G.R.; Nieva, J.L.; Borca, M.V. Alteration of a second putative fusion peptide of structural glycoprotein E2 of classical swine fever virus alters virus replication and virulence in swine. J. Virol. 2016, 90, 10299–10308. [Google Scholar] [CrossRef] [PubMed]
- Maurer, K.; Krey, T.; Moennig, V.; Thiel, H.J.; Rümenapf, T. CD46 is a cellular receptor for bovine viral diarrhea virus. J. Virol. 2004, 78, 1792–1799. [Google Scholar] [CrossRef] [PubMed]
- Dräger, C.; Beer, M.; Blome, S. Porcine complement regulatory protein CD46 and heparan sulfates are the major factors for classical swine fever virus attachment in vitro. Arch. Virol. 2015, 160, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Björklund, H.; Stadejek, T.; Belák, S. Molecular characterization of the 3′ non-coding region of classical swine fever virus vaccine strains. Virus Genes 1998, 16, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Risager, P.C.; Fahnøe, U.; Gullberg, M.; Rasmussen, T.B.; Belsham, G.J. Analysis of classical swine fever virus RNA replication determinants using replicons. J. Gen. Virol. 2013, 94, 1739–1748. [Google Scholar] [CrossRef] [PubMed]
- Lackner, T.; Thiel, H.J.; Tautz, N. Dissection of a viral autoprotease elucidates a function of a cellular chaperone in proteolysis. Proc. Natl. Acad. Sci. USA 2006, 103, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Moulin, H.R.; Seuberlich, T.; Bauhofer, O.; Bennett, L.C.; Tratschin, J.D.; Hofmann, M.A.; Ruggli, N. Nonstructural proteins NS2-3 and NS4A of classical swine fever virus: Essential features for infectious particle formation. Virology 2007, 365, 376–389. [Google Scholar] [CrossRef] [PubMed]
- Agapov, E.V.; Murray, C.L.; Frolov, I.; Qu, L.; Myers, T.M.; Rice, C.M. Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus. J. Virol. 2004, 78, 2414–2425. [Google Scholar] [CrossRef] [PubMed]
- Klemens, O.; Dubrau, D.; Tautz, N. Characterization of the determinants of NS2-3-independent virion morphogenesis of Pestiviruses. J. Virol. 2015, 89, 11668–11680. [Google Scholar] [CrossRef] [PubMed]
- Tamura, J.K.; Warrener, P.; Collett, M.S. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology 1993, 193, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Warrener, P.; Collett, M.S. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J. Virol. 1995, 69, 1720–1726. [Google Scholar] [PubMed]
- Wiskerchen, M.; Collett, M.S. Pestivirus gene expression: Protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology 1991, 184, 341–350. [Google Scholar] [CrossRef]
- Tautz, N.; Kaiser, A.; Thiel, H.J. NS3 serine protease of bovine viral diarrhea virus: Characterization of active site residues, NS4A cofactor domain, and protease-cofactor interactions. Virology 2000, 273, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Dubrau, D.; Tortorici, M.A.; Rey, F.A.; Tautz, N. A positive-strand RNA virus uses alternative protein-protein interactions within a viral protease/cofactor complex to switch between RNA replication and virion morphogenesis. PLoS Pathog. 2017, 13, e1006134. [Google Scholar] [CrossRef] [PubMed]
- Gladue, D.; Gavrilov, B.K.; Holinka, L.G.; Fernández-Sainz, I.J.; Vepkhvadze, N.; Rogers, K.; O’Donnell, V.; Risatti, G.R.; Borca, M.V. Identification of an NTPase motif in classical swine fever virus NS4B protein. Virology 2011, 411, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Sáinz, I.; Gladue, D.P.; Holinka, L.G.; O’Donnell, V.; Gudmundsdottir, I.; Prarat, M.V.; Patch, J.R.; Golde, W.T.; Lu, Z.; Zhu, J.; et al. Mutations in classical swine fever virus NS4B affect virulence in swine. J. Virol. 2010, 3, 1536–1539. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.E.; Gorbalenya, A.E.; Rice, C.M. The NS5A/NS5 proteins of viruses from three genera of the family Flaviviridae are phosphorylated by associated serine/threonine kinases. J. Virol. 1998, 72, 6199–6206. [Google Scholar] [PubMed]
- Pei, J.; Zhao, M.; Ye, Z.; Gou, H.; Wang, J.; Yi, L.; Dong, X.; Liu, W.; Luo, Y.; Liao, M.; et al. Autophagy enhances the replication of classical swine fever virus in vitro. Autophagy 2013, 10, 93–110. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Deng, J.; Ye, Z.; Wang, J.; Gou, H.; Liu, W.; Zhao, M.; Liao, M.; Yi, L.; Chen, J. Absence of autophagy promotes apoptosis by modulating the ROS-dependent RLR signaling pathway in classical swine fever virus-infected cells. Autophagy 2016, 12, 1738–1758. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, M.; Chen, J.; Zhang, W.; Luo, J.; Bao, K.; Nie, M.; Chen, J.; Li, B. Mutational analysis of the GDD sequence motif of classical swine fever virus RNA-dependent RNA polymerases. Virus Genes 2007, 34, 63–65. [Google Scholar] [CrossRef]
- Choi, K.H.; Gallei, A.; Becher, P.; Rossmann, M.G. The structure of bovine viral diarrhea virus RNA-dependent RNA polymerase and its amino-terminal domain. Structure 2006, 14, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Gao, J.; Wang, W.; Wang, Y.; Chen, J.; Chen, J.; Li, B. Specific interaction between the classical swine fever virus NS5B protein and the viral genome. Eur. J. Biochem. 2004, 271, 3888–3896. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Bai, Y.; Xu, H.; Geng, X.; Chen, J.; Wang, Y.; Chen, J.; Li, B. Effect of NS3 and NS5B proteins on classical swine fever virus internal ribosome entry site-mediated translation and its host cellular translation. J. Gen. Virol. 2008, 89, 994–999. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Chen, Y.; Xiao, J.; Xiao, J.; Wang, J.; Li, G.; Chen, J.; Xiao, M. Classical swine fever virus NS5A protein interacts with 3′-untranslated region and regulates viral RNA synthesis. Virus Res. 2012, 3, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xiao, J.; Xiao, J.; Sheng, C.; Wang, J.; Jia, L.; Zhi, Y.; Li, G.; Chen, J.; Xiao, M. Classical swine fever virus NS5A regulates viral RNA replication through binding to NS5B and 3′-UTR. Virology 2012, 432, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Rijnbrand, R.; van der Straaten, T.; van Rijn, P.A.; Spaan, W.J.; Bredenbeek, P.J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J. Virol. 1997, 71, 451–457. [Google Scholar] [PubMed]
- Shi, B.J.; Liu, C.C.; Zhou, J.; Wang, S.Q.; Gao, Z.C.; Zhang, X.M.; Zhou, B.; Chen, P.Y. Entry of classical swine fever virus into PK-15 cells via a pH-, dynamin-, and cholesterol-dependent, clathrin-mediated endocytic pathway that requires Rab5 and Rab7. J. Virol. 2016, 90, 9194–9208. [Google Scholar] [CrossRef] [PubMed]
- Lecot, S.; Belouzard, S.; Dubuisson, J.; Rouillé, Y. Bovine viral diarrhea virus entry is dependent on clathrin-mediated endocytosis. J. Virol. 2005, 79, 10826–10829. [Google Scholar] [CrossRef] [PubMed]
- Manna, D.; Aligo, J.; Xu, C.; Park, W.S.; Koc, H.; Heo, W.D.; Konan, K.V. Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology 2010, 398, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Van der Schaar, H.M.; Rust, M.J.; Chen, C.; van der Ende-Metselaar, H.; Wilschut, J.; Zhuang, X.; Smit, J.M. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 2008, 12, e1000244. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, M.N.; Sukumaran, B.; Pal, U.; Agaisse, H.; Murray, J.L.; Hodge, T.W.; Fikrig, E. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J. Virol. 2007, 81, 4881–4885. [Google Scholar] [CrossRef] [PubMed]
- Foo, K.Y.; Chee, H.Y. Interaction between Flavivirus and cytoskeleton during virus replication. Biomed. Res. Int. 2015, 2015, 427814. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Ling, L.; Liao, Y.; Li, S.; Han, W.; Zhao, B.; Sun, Y.; Qiu, H.J. Beta-actin interacts with the E2 protein and is involved in the early replication of classical swine fever virus. Virus Res. 2014, 179, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.F.; Kurosky, A.; Pryzdial, E.L.; Wasi, S. Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J. Virol. 1995, 69, 4784–4791. [Google Scholar] [PubMed]
- Ryzhova, E.V.; Vos, R.M.; Albright, A.V.; Harrist, A.V.; Harvey, T.; González-Scarano, F. Annexin 2: A novel human immunodeficiency virus type 1 Gag binding protein involved in replication in monocyte-derived macrophages. J. Virol. 2006, 80, 2694–2704. [Google Scholar] [CrossRef] [PubMed]
- LeBouder, F.; Morello, E.; Rimmelzwaan, G.F.; Bosse, F.; Péchoux, C.; Delmas, B.; Riteau, B. Annexin II incorporated into influenza virus particles supports virus replication by converting plasminogen into plasmin. J. Virol. 2008, 82, 6820–6828. [Google Scholar] [CrossRef] [PubMed]
- Backes, P.; Quinkert, D.; Reiss, S.; Binder, M.; Zayas, M.; Rescher, U.; Gerke, V.; Bartenschlager, R.; Lohmann, V. Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 2010, 84, 5775–5789. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Shi, Z.; Guo, H.; Qu, H.; Zhang, Y.; Tu, C. Annexin 2 is a host protein binding to classical swine fever virus E2 glycoprotein and promoting viral growth in PK-15 cells. Virus Res. 2015, 201, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Liu, X.; Jiang, Q.; Xu, B.; Zhou, C.; Wang, Y.; Chen, J.; Xiao, M. Annexin A2 is involved in the production of classical swine fever virus infectious particles. J. Gen. Virol. 2015, 96, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Gladue, D.; O’Donnell, V.; Fernández-Sainz, I.J.; Fletcher, P.; Baker-Branstetter, R.; Holinka, L.; Sanford, B.; Carlson, J.; Lu, Z.; Borca, M.V. Interaction of structural core protein of classical swine fever virus with endoplasmic reticulum-associated degradation pathway protein OS9. Virology 2014, 460, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Kang, K.; Ning, P.; Peng, Y.; Lin, Z.; Cui, H.; Cao, Z.; Wang, J.; Zhang, Y. Heat shock protein 70 is associated with CSFV NS5A protein and enhances viral RNA replication. Virology 2015, 482, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Feng, S.; Wang, J.H.; He, W.R.; Qin, H.Y.; Dong, H.; Li, L.F.; Yu, S.X.; Li, Y.; Qiu, H.J. eEF1A interacts with the NS5A protein and inhibits the growth of classical swine fever virus. Viruses 2015, 7, 4563–4581. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Perez, D.R.; French, R.; Merrick, W.C.; Donis, R.O. The NS5A protein of bovine viral diarrhoea virus interacts with the alpha subunit of translation elongation factor-1. J. Gen. Virol. 2001, 82, 2935–2943. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Yao, Y.; Chen, B.; Wang, Y.; Chen, J.; Xiao, M. RNA helicase is involved in the expression and replication of classical swine fever virus and interacts with untranslated region. Virus Res. 2013, 171, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Bauhofer, O.; Summerfield, A.; Sakoda, Y.; Tratschin, J.D.; Hofmann, M.A.; Ruggli, N. Classical swine fever virus Npro interacts with interferon regulatory factor 3 and induces its proteasomal degradation. J. Virol. 2007, 81, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Fiebach, A.R.; Guzylack-Piriou, L.; Python, S.; Summerfield, A.; Ruggli, N. Classical swine fever virus Npro limits type I interferon induction in plasmacytoid dendritic cells by interacting with interferon regulatory factor 7. J. Virol. 2011, 85, 8002–8011. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, S.; Sun, Y.; Dong, H.; Li, Y.; Zhao, B.; Guo, D.; Weng, C.; Qiu, H.J. Poly(C)-binding protein 1, a novel Npro-interacting protein involved in classical swine fever virus growth. J. Virol. 2013, 87, 2072–2080. [Google Scholar] [CrossRef] [PubMed]
- Doceul, V.; Charleston, B.; Crooke, H.; Reid, E.; Powell, P.P.; Seago, J. The Npro product of classical swine fever virus interacts with Ikappa B alpha, the NF-kappaB inhibitor. J. Gen. Virol. 2008, 89, 1881–1889. [Google Scholar] [CrossRef] [PubMed]
- Johns, H.L.; Doceul, V.; Everett, H.; Crooke, H.; Charleston, B.; Seago, J. The classical swine fever virus N-terminal protease Npro binds to cellular HAX-1. J. Gen. Virol. 2010, 91, 2677–2686. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Dong, H.; Li, S.; Munir, M.; Chen, J.; Luo, Y.; Sun, Y.; Liu, L.; Qiu, H.J. Hemoglobin subunit beta interacts with the capsid protein and antagonizes the growth of classical swine fever virus. J. Virol. 2013, 87, 5707–5717. [Google Scholar] [CrossRef] [PubMed]
- Gladue, D.; Holinka, L.; Fernández-Sainz, I.J.; Prarat, M.; O’Donell, V.; Vepkhvadze, N.; Lu, Z.; Rogers, K.; Risatti, G.; Borca, M.V. Effects of the interactions of classical swine fever virus Core protein with proteins of the SUMOylation pathway on virulence in swine. Virology 2010, 407, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Gladue, D.; Holinka, L.; Fernández-Sainz, I.J.; Prarat, M.; O’Donnell, V.; Vepkhvadze, N.; Lu, Z.; Risatti, G.; Borca, M.V. Interaction between Core protein of classical swine fever virus with cellular IQGAP1 protein appears essential for virulence in swine. Virology 2011, 412, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, J.; He, W.R.; Feng, S.; Li, Y.; Wang, X.; Liao, Y.; Qin, H.Y.; Li, L.F.; Dong, H.; et al. Thioredoxin 2 is a novel E2-interacting protein that inhibits the replication of classical swine fever virus. J. Virol. 2015, 89, 8510–8524. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, S.; Liao, Y.; Zhang, E.; Feng, S.; Yu, S.; Li, L.F.; He, W.R.; Li, Y.; Luo, Y.; et al. Mitogen-activated protein kinase kinase 2, a novel E2-interacting protein, promotes the growth of classical swine fever virus via attenuation of the JAK-STAT signaling pathway. J. Virol. 2016, 90, 10271–10283. [Google Scholar] [CrossRef] [PubMed]
- Li, L.F.; Yu, J.H.; Li, Y.; Wang, J.; Li, S.; Zhang, L.K.; Xia, S.L.; Yang, Q.; Wang, X.; Yu, S.; et al. Guanylate-binding protein 1, an interferon-induced GTPase, exerts an antiviral activity against classical swine fever virus depending on its GTPase activity. J. Virol. 2016, 90, 4412–4426. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Sun, J.; Guo, H.; Yang, Z.; Ma, Z.; Tu, C. Down-regulation of cellular protein heme oxygenase 1 inhibits proliferation of classical swine fever virus in PK-15 cells. Virus Res. 2013, 173, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Zögg, T.; Sponring, M.; Schindler, S.; Koll, M.; Schneider, R.; Brandstetter, H.; Auer, B. Crystal structures of the viral protease Npro imply distinct roles for the catalytic water in catalysis. Structure 2013, 21, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Klenk, C.; Liu, B.; Keiner, B.; Cheng, J.K.; Zheng, B.J.; Li, L.; Han, Q.L.; Wang, C.; Li, T.X.; et al. Modification of nonstructural protein 1 of influenza A virus by SUMO1. J. Virol. 2011, 85, 1086–1098. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Chang, C.; Li, L.; Klenk, C.; Cheng, J.; Chen, Y.; Xia, N.; Shu, Y.; Chen, Z.; Gabriel, G.; et al. SUMOylation of influenza A virus nucleoprotein is essential for intracellular trafficking and virus growth. J. Virol. 2014, 88, 9379–9390. [Google Scholar] [CrossRef] [PubMed]
- Higginbotham, J.M.; O’Shea, C.C. Adenovirus E4-ORF3 targets PIAS3 and together with E1B-55K remodels SUMO interactions in the nucleus and at virus genome replication domains. J. Virol. 2015, 89, 10260–10272. [Google Scholar] [CrossRef] [PubMed]
- Richt, J.A.; García-Sastre, A. Attenuated influenza virus vaccines with modified NS1 proteins. Curr. Top. Microbiol. Immunol. 2009, 333, 177–195. [Google Scholar] [PubMed]
- Sanchez-Cordon, P.J.; Romanini, S.; Salguero, F.J.; Nunez, A.; Bautista, M.J.; Jover, A.; Gomez-Villamos, J.C. Apoptosis of thymocytes related to cytokine expression in experimental classical swine fever. J. Comp. Pathol. 2002, 127, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Summerfield, A.; Alves, M.; Ruggli, N.; de Bruin, M.G.; McCullough, K.C. High IFN-alpha responses associated with depletion of lymphocytes and natural IFN-producing cells during classical swine fever. J. Interferon Cytokine Res. 2006, 26, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cordón, P.J.; Núñez, A.; Salguero, F.J.; Pedrera, M.; Fernández de Marco, M.; Gómez-Villamandos, J.C. Lymphocyte apoptosis and thrombocytopenia in spleen during classical swine fever: Role of macrophages and cytokines. Vet. Pathol. 2005, 42, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Bruschke, C.J.; Hulst, M.M.; Moormann, R.J.; van Rijn, P.A.; van Oirschot, J.T. Glycoprotein Erns of pestiviruses induces apoptosis in lymphocytes of several species. J. Virol. 1997, 71, 6692–6696. [Google Scholar] [PubMed]
- Hsu, W.L.; Chen, C.L.; Huang, S.W.; Wu, C.C.; Chen, I.H.; Nadar, M.; Su, Y.P.; Tsai, C.H. The untranslated regions of classic swine fever virus RNA trigger apoptosis. PLoS ONE 2014, 9, e88863. [Google Scholar] [CrossRef] [PubMed]
- Ruggli, N.; Bird, B.H.; Liu, L.; Bauhofer, O.; Tratschin, J.D.; Hofmann, M.A. Npro of classical swine fever virus is an antagonist of double-stranded RNA-mediated apoptosis and IFN-α/β induction. Virology 2005, 340, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.H.; Guo, K.; Kang, K.; Zhang, Y.; He, L.; Wang, J. Classical swine fever virus NS2 protein promotes interleukin-8 expression and inhibits MG132-induced apoptosis. Virus Genes 2011, 42, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.H.; Zhang, Y.M.; Fan, L.; Tong, G.; He, L.; Dai, C. Classic swine fever virus NS2 protein leads to the induction of cell cycle arrest at S-phase and endoplasmic reticulum stress. Virol. J. 2010, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Rust, M.J.; Lakadamyali, M.; Zhang, F.; Zhuang, X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 2004, 11, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Gerold, G.; Bruening, J.; Weigel, B.; Pietschmann, T. Protein interactions during the Flavivirus and Hepacivirus life cycle. Mol. Cell. Proteom. 2017, 16, 75–91. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, T.; Yao, L.; Liu, B.; Teng, C.; Ouyang, H.S. Classical swine fever virus replicated poorly in cells from MxA transgenic pigs. BMC Vet. Res. 2016, 12, 169. [Google Scholar] [CrossRef] [PubMed]
- Burkard, C.; Lillico, S.G.; Reid, E.; Jackson, B.; Mileham, A.J.; Ait-Ali, T.; Whitelaw, C.B.; Archibald, A.L. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 2017, 13, e1006206. [Google Scholar] [CrossRef] [PubMed]
| Viral Proteins | Interacting Partners or Replication Cycle-Contributing Factors | Functions | Ref. |
|---|---|---|---|
| 5′- and 3′-NCRs | RHA | Modulation of RNA synthesis, replication and translation of CSFV | [75] |
| Npro | IRF-3 | Blockage of IFN-β production | [76] |
| IRF-7 | Blockage of IFN-α production | [77] | |
| PCBP1 | Blockage of IFN-β production | [78] | |
| IκBα | — | [79] | |
| HAX-1 | Cellular resistance to apoptosis | [80] | |
| C | OS9 | Regulation of virus replication | [71] |
| HB | Blockage of IFN-β production | [81] | |
| UBC9 | Involvement of viral virulence | [82] | |
| SUMO-1 | Involvement of viral virulence | [82] | |
| IQGAP1 | Involvement of viral virulence | [83] | |
| Erns | HS | Attachment receptor | [15] |
| LamR | Attachment receptor | [16] | |
| E2 | β-Actin | Regulation of virus replication | [64] |
| Anx2 | Regulation of virus growth | [69] | |
| Trx2 | Inhibition of the NF-κB signaling | [84] | |
| MEK2 | Inhibition of the JAK-STAT signaling | [85] | |
| NS5A | Anx2 | Regulation of viral assembly | [70] |
| HSP70 | Regulation of virus replication | [72] | |
| eEF1A | Inhibition of IRES-mediated translation efficiency | [73] | |
| GBP1 | Regulation of virus replication | [86] | |
| – | CD46 | Involvement of virus attachment | [34] |
| – | Clathrin | Involvement of virus internalization | [58] |
| – | Cholesterol | Involvement of virus internalization | [58] |
| – | Dynamin | Involvement of virus internalization | [58] |
| – | Rab5 | Involvement of virus internalization | [58] |
| – | Rab7 | Involvement of virus internalization | [58] |
| – | HO-1 | Regulation of virus replication | [87] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Wang, J.; Yang, Q.; Naveed Anwar, M.; Yu, S.; Qiu, H.-J. Complex Virus–Host Interactions Involved in the Regulation of Classical Swine Fever Virus Replication: A Minireview. Viruses 2017, 9, 171. https://doi.org/10.3390/v9070171
Li S, Wang J, Yang Q, Naveed Anwar M, Yu S, Qiu H-J. Complex Virus–Host Interactions Involved in the Regulation of Classical Swine Fever Virus Replication: A Minireview. Viruses. 2017; 9(7):171. https://doi.org/10.3390/v9070171
Chicago/Turabian StyleLi, Su, Jinghan Wang, Qian Yang, Muhammad Naveed Anwar, Shaoxiong Yu, and Hua-Ji Qiu. 2017. "Complex Virus–Host Interactions Involved in the Regulation of Classical Swine Fever Virus Replication: A Minireview" Viruses 9, no. 7: 171. https://doi.org/10.3390/v9070171
APA StyleLi, S., Wang, J., Yang, Q., Naveed Anwar, M., Yu, S., & Qiu, H.-J. (2017). Complex Virus–Host Interactions Involved in the Regulation of Classical Swine Fever Virus Replication: A Minireview. Viruses, 9(7), 171. https://doi.org/10.3390/v9070171





