Abstract
For decades, bacteriophages (phages) have been used for Brucella species identification in the diagnosis and epidemiology of brucellosis. Traditional Brucella phage typing is a multi-day procedure including the isolation of a pure culture, a step that can take up to three weeks. In this study, we focused on the use of brucellaphages for sensitive detection of the pathogen in clinical and other complex samples, and developed an indirect method of Brucella detection using real-time quantitative PCR monitoring of brucellaphage DNA amplification via replication on live Brucella cells. This assay allowed the detection of single bacteria (down to 1 colony-forming unit per milliliter) within 72 h without DNA extraction and purification steps. The technique was equally efficient with Brucella abortus pure culture and with mixed cultures of B. abortus and α-proteobacterial near neighbors that can be misidentified as Brucella spp., Ochrobactrum anthropi and Afipia felis. The addition of a simple short sample preparation step enabled the indirect phage-based detection of B. abortus in spiked blood, with the same high sensitivity. This indirect phage-based detection assay enables the rapid and sensitive detection of live B. abortus in mixed cultures and in blood samples, and can potentially be applied for detection in other clinical samples and other complex sample types.
1. Introduction
Brucellosis is a global bacterial zoonotic disease of high social and economic importance. It is debilitating and disabling for hundreds of thousands of people and causes high reproductive losses in food animals every year [1,2]. The causative agents of livestock and human brucellosis are small Gram-negative facultative intracellular coccobacilli in the genus Brucella, which are slow growing and typically form visible colonies after 48–72 h of incubation on agar media. Six classic terrestrial and two marine pathogenic species of Brucella have been identified, though the discovery of a number of new species over the last decade has continued to expand our understanding of the genus. Four of the classic terrestrial Brucella species can cause human brucellosis: Brucella melitensis (primary hosts: goats and sheep), Brucella abortus (cattle), Brucella suis (swine), and Brucella canis (dogs). The marine mammal pathogen species Brucella ceti and Brucella pinnipedialis have also been reported to infect humans [3,4]. During the Cold War, Brucella spp. were weaponized [5], and nowadays the U.S. Centers for Diseases Control (CDC) still categorizes B. melitensis, B. abortus and B. suis as Category B Select Agents [6].
Since the symptoms of human brucellosis are nonspecific, clinical diagnosis is problematic and must be confirmed by laboratory tests. Laboratory diagnosis is also challenging, especially early in infection [7,8]. The CDC outlines the following three laboratory criteria for definitive diagnosis of human brucellosis: (i) isolation of Brucella spp. from the blood or reticuloendothelial tissue biopsies (bone marrow); (ii) fourfold or greater rise in Brucella agglutination titer between acute- and convalescent-phase serum specimens obtained two weeks or more apart and studied at the same laboratory; and (iii) demonstration by immunofluorescence of Brucella spp. in a clinical specimen [9]. Every criterion is sufficient for confirmation of the disease, but isolation and phenotypic characterization of Brucella spp. is still considered to be the gold standard and ultimate proof of brucellosis [7].
Despite the use of selective media to improve the isolation and identification of Brucella [10,11,12,13], and the successful use of automated culture systems [8,14,15], Brucella isolation remains challenging. The process is laborious (often including sample preparation), lengthy (usually taking 3–45 days of cultivation and additional time for subsequent phenotypic identification), associated with risk of laboratory-acquired infections, and has a relatively low sensitivity: 0–90%, depending on the phase of infection, the least efficient in chronic cases [7,8,14,16].
Unlike bacterial culture, serological testing is rapid, less hazardous, less expensive, and more sensitive; and is thus used preferentially in clinical practice. However, these methodologies also have several disadvantages: (a) a positive serologic assay (detection of Brucella total antibody titer of ≥1:160) is only considered presumptive unless there is a fourfold or greater titer rise; (b) serologic tests are often negative at early stages of infection; (c) diagnostic titers do not necessarily correlate with active infection; (d) assays should be standardized for different animal species and human populations, depending on the levels of endemicity; (e) there are numerous reports of false-positive serologic tests caused by cross-reactivity to epitopes in O-polysaccharide shared between Brucella and Yersinia enterocolitica O:9, Salmonella urbana, Vibrio cholerae, Francisella tularensis, Escherichia coli O:157 and Stenotrophomonas maltophilia. In addition, the use of smooth live attenuated Brucella vaccine strains in animals complicates serologic differentiation between natural and vaccine-induced infection because of the presence of lipopolysaccharide O antigen [7,8].
Detection of Brucella DNA in clinical samples using various PCR assays has proved to be rapid (down to few hours), efficient at different stages of the disease, more specific than serological tests, and more sensitive than blood cultures. The use of different DNA targets provides identification at the level of genus (which is sufficient to start antibacterial therapy) or at the level of species (which is important for epizootological and epidemiological analysis). Real-time PCR allows for high-throughput screening of clinical specimens and improves analytical sensitivity. Multiplex PCR assays enable genotyping and strain characterization [8,17,18,19,20,21]. However, the generally low concentrations of Brucella and the presence of PCR inhibitors in blood and tissues still complicate the molecular diagnosis of brucellosis, and thus PCR tests require sample preparation, including bacterial DNA extraction, concentration, and purification. The DNA diagnostic criteria for active infection are yet to be established, because there are many cases when Brucella DNA is repeatedly detected for months or even years after antibacterial therapy without recovery of live bacteria and in the absence of any symptoms of brucellosis. The interpretation of these results is unclear because PCR assays cannot distinguish between live and dead, or culturable and unculturable bacteria [8,20,22,23,24].
Routine bacteriophage lysis tests have been used for diagnosis of brucellosis for more than 60 years [25]. Brucellaphages Tbilisi (Tb), Firenze (Fz), Weybridge (Wb), S708, Berkeley (Bk), R/C, and Izatnagar (Iz) have very similar morphology and genome structure, but differ in their specificity [26,27,28,29,30]. The use of this set of phages at two different concentrations allows the definitive identification of Brucella at both genus and species level, including differentiation between smooth and rough strains of B. abortus, B. suis, B. melitensis, B. neotomae, B. canis, and B. ovis [26,31,32,33]. Phage-based identification is a multi-day assay, including the isolation of a pure Brucella culture, and a plaque assay using the double-layer agar technique, requiring 48–72 h of incubation [26,32].
We have recently proposed a rapid (4–6 h) and simple indirect bacteriophage-based approach for the detection and identification of Yersinia pestis, the causative agent of plague [34]. The test was based on monitoring amplification of Y. pestis-specific phages in the presence of the live bacteria using real-time quantitative PCR (qPCR). The detection limit was as low as a single bacterium per 1-µL sample (103 colony-forming units (CFU)/mL) and could even be reduced by sample concentration. The method worked well on simulated clinical blood, serum and tissue samples, and did not require DNA isolation [34,35]. This technology was demonstrated to have the same efficiency whether using a laboratory bench-top qPCR machine or a portable instrument designated for field work [35].
The purpose of this study was to develop a qPCR assay targeting brucellaphages—such as Tb, S708 or Bk—for indirect identification of B. abortus and related Brucella spp. Phage propagation was delayed, but robust, and enabled the detection of single B. abortus cells per milliliter of sample within 72 h. Higher concentrations of bacteria resulted in shortened turnaround time (24–48 h). The method also allowed reliable detection of single B. abortus cells in simulated blood samples within 72 h and identification of Brucella in mixed cultures containing relative α-proteobacteria Ochrobactrum anthropi or Afipia felis.
2. Results
2.1. Brucellaphage Propagation and DNA Extraction
Diagnostic brucellaphages Tb, S708, Fz, Wb, and Bk were propagated to high titers (Table 1). It was determined that the use of conical Erlenmeyer flasks with vented closures and shaking at 60 rpm are critical for efficient phage propagation. Flasks were filled to no more than 1/5 of their nominal volume to avoid spillage and to assure the adequate aeration. The bacteriophage titer of each new lysate was determined, and a portion of each lysate was used for phage DNA extraction and purification. Table 1 presents phage concentration in infectious particles and phage genome equivalents per milliliter. The latter was determined using the calibration curve established by qPCR (see the next section).

Table 1.
Brucellaphage propagation: quantification by live counts and quantitative PCR (qPCR).
2.2. Performance Testing of qPCR with Phage DNA and Intact Phage Particles
The quantitative parameters of bacteriophage-based qPCR detection were first tested by using serial 10-fold dilutions of purified DNA from brucellaphage S708. These experiments were done in three separate runs, each in duplicate, and yielded a log linear function between DNA concentrations and threshold cycle number (Ct), spanning a 9-log dilution series, from 3.5 ng down to 0.35 fg of DNA (Figure 1a). We calculated that this corresponds to about 10 to 1.0 × 109 S708 genome equivalents, based on the fact that its genome size is 38,253 bp [31]. To compare qPCR results obtained using purified DNA and intact phage particles, a series of 100-fold dilutions of phage S708 lysate was prepared in saline magnesium (SM) buffer, and qPCR reactions were run with different concentrations of phage particles ranging from 100 to 108 PFU per 1 µL of phage lysate (per 10 µL of qPCR sample). The number of viable particles per sample was confirmed by plaque assays. The results of enumeration of phage particles by qPCR (Figure 1b) were one to two logs higher in comparison with plaque counts, and in case of phage Bk this difference was up to four logs (see also Table 1). This could be explained by partial inactivation of brucellaphages with chloroform [36], and the results may suggest that Bk is the phage most sensitive to chloroform. However, since qPCR of brucellaphage S708 DNA indicated that the actual number of phage genomes per sample is at least 10 times greater than the number obtained by plaque count, the assay sensitivity was not compromised. Very similar results were obtained with genomic DNA of other brucellaphages (data not shown).
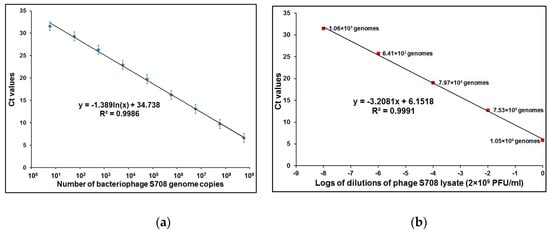
Figure 1.
qPCR of purified DNA of brucellaphage S708 (a) and S708 lysate (b). Calibration curve (a) was established using qPCR of 1 µL aliquots of nine different 10-fold dilutions of purified DNA from brucellaphage S708, ranging from 0.35 fg to 3.5 ng. Calibration curve (b) was established using qPCR of 1 µL aliquots of several 100-fold dilutions of phage S708 lysate, ranging from ~100 to ~108 PFU. The number of viable particles per sample was confirmed by plaque assays Ct: cycle threshold.
2.3. Sensitivity of Phage-Based qPCR Detection of Brucella abortus in Broth Culture
To define the sensitivity of indirect phage-mediated detection of B. abortus, first a series of experiments was conducted in which diagnostic brucellaphage S708 was propagated on B. abortus cultures at different concentrations ranging from 1 × 108 to about 1 CFU/mL. The use of several starting concentrations of the S708 phage showed that 103 PFU/mL is the minimum titer, providing clear and consistent phage amplification at any bacterial concentration (data not shown). This starting phage titer was then used in all further experiments. The increase in phage titer was detected by qPCR (Figure 2), applying Ct values to the calibration curve (Figure 1a). The lowest amount of phage providing reliable positive qPCR signal was determined to be 100 phage genome equivalents per 20 µL sample (containing 1 µL of phage lysate). This number was normalized to 1 (Figure 2). Our data showed that the detection of B. abortus is robust and there are clear differences in phage yields at different concentrations of B. abortus over the course of time. We were unable to detect any rise in phage titer over 6–8 h of incubation. The first detectable phage burst was observed after 24 h of incubation at higher initial concentrations of B. abortus ranging from 108 to 106 CFU/mL. This increase in phage concentration was unexpectedly large: over five orders of magnitude. After 48 h of incubation, a similar 5 times log burst was noted at lower B. abortus concentrations (105–103 CFU/mL). Finally, a 72 h incubation resulted in the robust phage titer increase at the lowest concentrations of B. abortus (102–100 CFU/mL). Thus, the sensitivity of S708-mediated qPCR assay was ~100 CFU/mL. The lower limits of detection were also determined for several other brucellaphages: Tb, Fz, Wb, and Bk. The results are presented in Figure 3. Each phage from the diagnostic panel produced a robust burst detected by qPCR. The highest titer rise was observed with the Tb and Bk phages. The vigorous amplification of Bk that enabled efficient indirect detection of B. abortus was of particular interest, since this phage can propagate on a number of Brucella spp. [32,37,38], and thus its use in this approach should increase the spectrum of detected Brucella spp. Based on this finding, we used the Bk phage in most further experiments.

Figure 2.
Dynamics of phage S708 growth on B. abortus S19 detected by qPCR. An overnight culture of B. abortus S19 was diluted to OD600 = 0.1 in Brucella Broth. 20 mL of each serial bacterial dilution from 100 to 10−8 (circa 108 to 100 CFU/mL, respectively) was made in Brucella Broth. Diagnostic bacteriophage S708 was added to each aliquot to a final concentration of 103 PFU/mL, and the sample was incubated in a shaker incubator at 37 °C and 60 rpm. 500 µL samples were withdrawn after 0, 6, 24, 48, and 72 h of incubation, treated with 10% chloroform and centrifuged to remove cell debris. 1 µL of each sample, neat or diluted 1:100 with Brucella Broth, was used for qPCR analysis. Reaction mixes were prepared in a total volume of 10 µL including 5 µL of the master mix and 0.3 µM of each primer. The cycling parameters were: 95 °C, 5 min; 40× (95 °C, 20 s; 60 °C, 60 s) with fluorescence measurement at the end of each cycle.
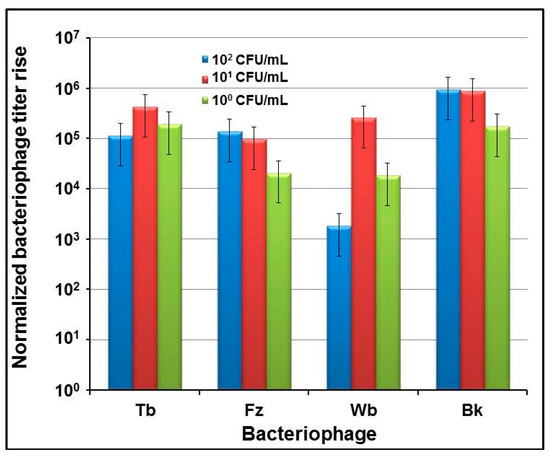
Figure 3.
Amplification of brucellaphages Tb, Fz, Wb, and Bk on low concentrations of B. abortus S19. Phage lysates were diluted in saline magnesium (SM) buffer and added at 103 PFU/mL to the dilutions of bacterial culture in Brucella Broth at concentrations of ~1 × 102; ~1 × 101 and ~1 × 100 CFU/mL, which were confirmed by live bacterial counts. qPCR was performed after 72 h of incubation of each test culture with the corresponding phage.
2.4. Specificity of qPCR Tests with the Bk Bacteriophage
Specificity of the phage-based detection method was tested on six bacterial species belonging to different taxonomic groups, including two representatives of α-proteobacteria close to Brucella, O. anthropi and A. felis, as well as Y. enterocolitica serovar O:9 carrying antigens cross-reactive with Brucella (see Table 2). We did not observe any propagation of the Bk phage after 72 h incubation with any of the tested bacterial cultures except B. abortus (data not shown), suggesting that the assay is specific. Moreover, the Bk phage was shown to robustly amplify on B. abortus in the mixed cultures with faster-growing O. anthropi or Y. enterocolitica O:9 (Figure 4), though some degree of inhibition of the phage propagation in comparison with the control Brucella culture was observed, especially in the presence of Y. enterocolitica O:9. However, the phage titer rise was still remarkably high, and easily detectable by qPCR.

Table 2.
Bacteriophages and bacterial strains used in this work.

Figure 4.
Brucellaphage Bk propagation on mixed bacterial cultures. Bacterial cultures overnight in Brucella Broth. The cultures were diluted 1:1000 with Brucella Broth and brucellaphage Bk was added to get a final concentration of 103 PFU/mL and approximate multiplicity of infection (MOI) of 1:1000. All cultures were incubated at 37 °C for 72 h with shaking at 200 rpm, treated with 10% chloroform; then a 0.5-mL aliquot was taken and centrifuged to remove cell debris. One microliter of each lysate was used for the qPCR reaction. For testing phage specificity in mixed cultures, equal volumes of diluted overnight non-Brucella and B. abortus S19 cultures were mixed, grown in the presence of the Bk phage and processed as described above.
2.5. Assay Performance with Simulated Clinical Samples
To test if the qPCR phage-based assay can be used for the detection of Brucella spp. in blood, we used different commercially available sheep blood products including whole blood with different anticoagulants such as sodium citrate, ethylenediaminetetraacetic acid (EDTA), heparin, and acid citrate dextrose (ACD), as well as defibrinated blood, plasma and serum (all purchased from LAMPIRE Biological Laboratories, Inc., Pipersville, PA, USA). Blood cells were removed from whole and defibrinated blood by centrifugation, and early-logarithmic-phase culture of B. abortus S19 was added to the resulting serum or plasma to a final concentration of 107 CFU/mL. The mix was diluted 1:10 with Brucella Broth to provide the better growth conditions for Brucella, and brucellaphage S708 was added at 103 PFU/mL. After 72 h of incubation at 37 °C, we were unable to detect any rise in the phage titer by qPCR, regardless of the final dilution of the sample. Similar results were observed when the S708 phage was added to commercially available sheep plasma or serum and when using other brucellaphages with different sheep blood products (data not shown). Since a 5 times log increase in brucellaphage titers was observed before in Brucella Broth, the negative result with all blood products suggested that serum component(s) may inhibit brucellaphage propagation.
In order to address this inhibition, a simple and short (about 10 min) sample preparation step was added that included removal of erythrocytes, lysis of leukocytes and washing of the bacterial cells (see Materials and Methods). Sheep blood with Na2-EDTA was spiked with B. abortus at different concentrations, and the loss of bacteria over centrifugation steps was monitored. We observed not more than a 20% decrease of Brucella live count in the final water suspension in comparison with the initial blood sample. Bacteriophage Bk was selected for this test because of its broad host range [32,37,38]. After 72 h of incubation, robust phage amplification was revealed by qPCR even at the highest bacterial dilution, corresponding to single Brucella cells per milliliter of blood (Figure 5).
Figure 5.
Amplification of the Bk phage after 72 h growth on different concentrations of B. abortus S19 in simulated infected blood samples. The tests were performed in Na2- ethylenediaminetetraacetic acid (EDTA)-treated whole sheep blood. An overnight culture of B. abortus S19 was diluted to OD600 = 0.1 in Brucella Broth. Serial bacterial dilutions 10−2 (~3 × 106 CFU/mL), 10−4 (~3 × 104 CFU/mL), 10−6 (~3 × 102 CFU/mL), 10−7 (~3 × 101 CFU/mL), and 10−8 (~3 × 100 CFU/mL) were made in sheep blood. To avoid inhibition of phage infection by components of sheep serum, the samples were processed using a lysis-centrifugation procedure (see Section 4.6). Determination of live brucellae was performed by plating 100 µL of 10−6 dilution of the washed blood sample. The entire processed blood sample was inoculated into 20 mL of Brucella Broth. Phage was added at 103 PFU/mL, and the samples were incubated at 37 °C with shaking at 60 rpm for 72 h, then treated with 10% chloroform and centrifuged to remove cell debris. One microliter of each sample was used for qPCR analysis.
3. Discussion
Laboratory diagnosis of brucellosis is a challenging and often time-consuming task. The best proof of the infection is isolation of Brucella culture from clinical samples followed by standard bacteriological characterization. Culturing Brucella poses among the highest risks of laboratory-acquired aerosol infection, while the isolation rate of these fastidious bacteria from blood is relatively low, and subsequent phenotypic bacteriological identification is time-consuming. Thus, current laboratory diagnosis is mainly based on serologic and molecular methods, but none of these approaches can be used on its own for consistent and reliable diagnosis. For example, serologic tests are ineffective early in the infection, sometimes have limited specificity because of cross-reactive antibodies and, in multiple observations, no correlation was found between antibody titers and clinical picture, including seronegative brucellosis patients and seropositive tests after effective antibiotic treatment. Many PCR assays targeting Brucella DNA have been developed, but these cannot discriminate between live and dead bacteria, and there are still no established DNA diagnostic criteria for active brucellosis, since Brucella DNA can be detected long after antibiotic treatment in the absence of culture of live bacteria or any brucellosis symptoms. Thus, the question of whether a positive DNA test in clinical samples should be followed by prolonged antibacterial therapy is often debatable [7,8,24]. In addition, despite the plethora of sensitive and specific PCR assays that have been developed against Brucella pathogens, the detection of infection in clinical specimens using PCR is still not widely accepted to be reliable, and we have experience with multiple DNA extraction techniques and highly sensitive real-time PCR assays failing to detect Brucella in human blood samples from which Brucella spp. were cultured [39]. All these facts indicate the need to develop new rapid and reliable diagnostic methods for brucellosis.
The purpose of this work was to develop a bacteriophage-based assay for highly sensitive and specific detection of live Brucella in liquid cultures and in blood, and potentially in other key biological samples. Before the development of a wide array of automated systems for blood culture identification and PCR-based molecular tests, typing with a panel of diagnostic brucellaphages was the tool of choice for species determination of Brucella spp. [27,32,33]. The use of phages for this purpose is still included in the Manual of Diagnostic Tests and Vaccines for Terrestrial Animals [40] and is applied globally as a complementary method [41,42,43,44]. The intrinsic ability of phage to specifically infect and amplify in live host bacteria is a feature very important for phage-based diagnostics. Standard phage amplification assays based on plaque formation (also called phage titer increase tests) have been used since the early 1960s for the detection of Shigella, Salmonella, Listeria and other pathogenic bacteria [45,46]. These assays remain important for the detection of slow growing bacteria, such as Mycobacterium tuberculosis [47,48] and Mycobacterium paratuberculosis [49,50]. Phage amplification assays have been improved using several quantitative approaches for phage detection, including high-performance liquid chromatography [51], qPCR [34,35,52,53], competitive ELISA [54], and lateral flow immunoassay [55]. Another group of methods measure the extent of phage-based bacterial lysis using fluorescent or amperometric detection of released products, like adenylate kinase [56] and β-galactosidase [57]. Finally, several genetically engineered reporter phages have been successfully used for indirect fluorescent detection of M. tuberculosis [58,59], Y. pestis [60] and B. anthracis [61].
We demonstrated previously that one particle of a plague diagnostic phage φA1122 is reproduced in 57 copies (burst size) upon its propagation in one Y. pestis cell during 30 min [34]. An assay for qPCR monitoring of amplification of this specific phage in the presence of Y. pestis was developed that is rapid (4 h), highly sensitive (one CFU per sample) and specific for indirect detection of the plague bacterium, and which performed well with various clinical samples [34,35]. Since phages propagate only on viable and culturable cells [62], the method allows the identification of only live bacteria that gives some additional important clinically and epidemiologically relevant information in comparison with assays targeting bacterial DNA. Another advantage of indirect phage-based detection is that there is no need in phage DNA extraction and purification [34,35]. qPCR targeting phage γ has been employed for rapid and sensitive indirect detection of Bacillus anthracis, though assay specificity was not tested using near-relative Bacillus cereus [53].
It was previously shown that lysis of B. abortus by phage Tb is delayed but very robust, producing about 121 phage particles from one bacterium [36]. In the present study we employed brucellaphage amplification monitored by qPCR for indirect detection of B. abortus. Five brucellaphages from the typing panel, Tb, S708, Fz, Wb, and Bk (see Table 2), were studied as potential reporters. Testing quantitative parameters of qPCR with purified DNA of phages Tb, S708, Wb, and Bk showed reliable standard curves down to 0.35–0.5 fg (10–15 genome equivalents; as an example, see Figure 1a); based on Tb burst size this is equivalent to 0.05–0.1 cells of B. abortus. Conducting the qPCR with dilutions of phage lysates (without DNA purification) yielded similar curves (Figure 1b). However, calculated phage genome equivalents were somewhat higher than the numbers of live phage particles. For example, 1 µL of 10−6 dilution of S708 (with initial concentration of 2 × 109 PFU/mL) is expected to contain only two live phage particles, whereas qPCR data showed a signal equivalent to 641 phage genomes (Figure 1b). This discrepancy can be explained by the previous observation that the treatment of brucellaphage lysates with 10% chloroform decreases the number of live phage [36]. For this reason the authors [36] proposed that brucellaphage lysates be treated with toluene instead of chloroform. However, since this discrepancy did not impair assay efficiency, for the purposes of this study chloroform was used in all experiments to standardize the procedure with previously developed assays [34,35] and to reduce the risk of laboratory exposure to live B. abortus S19, which is known to cause human infection [63].
The indirect phage-based Y. pestis detection assay developed previously in our laboratory allowed the detection of a single bacterium per sample in 4 h [34,35]. However, in the case of B. abortus and S708 there was no phage burst detected by qPCR after 6 or 8 h of incubation (Figure 2). The initial burst was observed only at the highest concentrations of B. abortus (108–106) and after 18 to 24 h of phage propagation. However, the increase in phage titer in this burst was remarkable, about five to six orders of magnitude above the starting titer. After 48 h the phage burst was noted at lower bacterial concentrations, 105–103 CFU/mL, and after 72-h incubation the marked burst was observed at the lowest concentrations (102–100 CFU/mL). Remarkably, the final yield of bacteriophage after 72 h of incubation was almost the same at any given concentration of Brucella starting culture (Figure 2). Thus the lower limit of detection for indirect S708 phage-based qPCR-assisted detection of B. abortus was about one cell per mL of test culture, indicating the high sensitivity of the assay. Testing of four additional brucellaphages capable of propagating on B. abortus (Tb, Fz, Wb, and Bk) showed that all these phages can robustly and consistently amplify on low concentrations of B. abortus down to a single bacterium per milliliter of culture (the detection limit was the same, about 100 CFU/mL) after 72 h (Figure 3), and thus can be efficient reporters in the indirect detection approach. Since brucellaphage Bk (both at high titer and diluted) has been shown to lyse smooth cultures of many Brucella spp. including B. abortus, B. suis, B. melitensis, B. neotomae [32,37], and B. microti [38], we therefore expect that this phage will be an efficient reporter for the indirect detection of the majority of Brucella strains that cause human brucellosis.
In order to test assay specificity, we used brucellaphage Bk and six bacterial species of different taxonomic groups (Table 2). Of them, O. anthropi is the nearest phylogenetic relative of Brucella, an agent of opportunistic infections and a frequent cause of misdiagnosis [64,65,66]. Y. enterocolitica serovar O:9 was found to have common antigens with Brucella that result in cross-reactivity when doing serologic tests for brucellosis and yersiniosis [67,68,69]. The genus Afipia also belongs to α-proteobacteria, order Rhizobiales, along with Brucella and Ochrobactrum, and it has been reported that Afipia clevelandensis has antigens that are cross-reactive with Brucella and Y. enterocolitica O:9 [70]. The lack of propagation of the Bk phage after 72 h incubation with all of the bacterial cultures tested other than the positive control indicates that the assay is specific for B. abortus. Moreover, the Bk phage was demonstrated to efficiently amplify on B. abortus in the mixed cultures containing faster growing O. anthropi or Y. enterocolitica O:9 (Figure 4).
The clinical relevance of the bacteriophage-based qPCR assay was evaluated first with sheep blood, plasma and serum artificially contaminated with B. abortus. We did not detect any Bk phage propagation in any of these blood products. Since all these products contain sheep serum, it appears there is a serum component (or components) that inhibits brucellaphage propagation. Thus a simple and short (10 min) lysis-centrifugation procedure was introduced to concentrate and wash B. abortus cells from sheep blood samples before subsequent Bk phage amplification. The processed samples then provided robust brucellaphage propagation within 72 h, with a lower limit of detection of approximately three B. abortus cells per milliliter of blood (Figure 5). The analytical sensitivity of previously described qPCR assays targeting different Brucella genes that used DNA purification methods depended upon the gene target and varied from 200 to 1 fg of DNA per reaction, equivalent to 4 × 104 to 5 × 102 CFU/mL [71,72,73,74,75,76]. In one report, one of six tested commercial DNA extraction kits allowed detection of 100 CFU/mL of B. abortus in suspension [24]. Detection of Brucella by qPCR from whole blood [77] and serum [78] provided the detection levels of about 2 × 103 and 7 × 103 CFU/mL, respectively, and the best result obtained from serum samples was 5 CFU/mL [79]. Our method improves sensitivity because it allowed the consistent detection of single B. abortus cells per milliliter of both culture suspensions and blood.
Since phages amplify only on viable bacterial cells [62], our approach can be considered as an analog of Brucella culture methods, but more rapid and much more sensitive. While phage-based detection takes longer to perform than PCR assays targeting Brucella DNA, our method has improved sensitivity and adds information about the presence of live Brucella in clinical samples that is important for diagnosis, therapy and prognosis of brucellosis. The highly sensitive, specific and rapid method of indirect detection of B. abortus using qPCR monitoring of phage amplification that we developed proved effective with pure and mixed cultures and with blood spiked with Brucella cells. We expect this assay to be readily adaptable not only to blood samples from humans and livestock but also to other relevant biological samples, and expect this assay to detect not only B. abortus but also other pathogenic Brucella species.
4. Materials and Methods
4.1. Bacterial Strains, Bacteriophages and Growth Media
All bacteriophages used in this work were commercially acquired from Félix d’Hérelle Reference Center for Bacterial Viruses (Université Laval, Quebec City, QC, Canada). BBLBrucella Broth, BBLBrucella Agar, Tripticase Soy Broth, and Tripticase Soy Agar (BD Biosciences, San Jose, CA, USA) were used for bacterial and phage growth. Brucellaphages and bacterial strains used in this work are listed in Table 2.
Brucellaphages were propagated on attenuated animal vaccine strain B. abortus S19, as described earlier [31], with some modifications as follows. Phage stock lysate was added to 150 mL of early logarithmic phase liquid bacterial culture at OD600 of <0.1 and multiplicity of infection (MOI) of 0.1 and incubated for 48 h at 37 °C in a 1000 mL vented plastic Erlenmeyer flask with shaking at 60 rpm. After incubation, phage lysate was allowed to stand undisturbed at 4 °C for 24 h. The lysate was treated with chloroform at a final concentration of 10% for 1 h in order to destroy all remaining live bacteria. The bacterial debris was removed by centrifugation at 5000× g for 15 min, and the resulting lysate was filtered through 0.22 µm filter. Phage titers were determined by spotting 10-fold dilutions of lysates on semi-solid agar overlay containing B. abortus S19.
4.2. Phage DNA Isolation
Brucellaphage lysates were concentrated by centrifugation at 13,250× g for 3 h and resuspended in SM buffer (Teknova, Hollister, CA, USA), at 1/100 of the original volume. The concentrated lysate was incubated for 1 h at 37 °C with 60 µg/mL of RNAse A and 20 µg/mL of DNAse I to remove Brucella DNA and RNA. Then phage suspension was treated with proteinase K (50 µg/mL) and sodium dodecyl sulfate (SDS; 0.5%) for 1 h at 56 °C. DNA was extracted by phenol:chloroform method, ethanol precipitated [87] and resuspended in nuclease-free water. DNA purity was confirmed by restriction analysis. DNA concentration was determined on a Nanodrop 2000 machine (Thermo Scientific, Wilmington, DE, USA).
4.3. Phage S708 Amplification on Different Culture Dilutions of B. abortus
An overnight culture of B. abortus S19 was diluted to OD600 = 0.1 in Brucella Broth. 20 mL of each serial bacterial dilution from 100 to 10−8 (ca. 108 to 100 CFU/mL, respectively) were made in Brucella Broth and placed into conical Erlenmeyer flasks covered with ventilated aerosol-proof closure caps. The live bacteria counts were performed by plating 100 µL of 10−6 dilution on Brucella Agar in triplicate for each independent experiment. Diagnostic bacteriophage S708 was added to each flask to a final concentration of 103 PFU/mL, and the sample was incubated in a shaker incubator at 37 °C and 60 rpm. 500 µL samples were withdrawn after 0, 6, 24, 48, and 72 h of incubation. Samples were treated with chloroform added to a final concentration of 10% and centrifuged in an Eppendorf mini-centrifuge for 3 min to remove Brucella cell debris. One microliter of each sample, neat or diluted 1:100 with Brucella Broth, was used for qPCR analysis (see below).
4.4. Testing Different Brucellaphages for qPCR Limits of Detection
Stock lysates of brucellaphages Tb, Fz, Wb, and Bk were diluted in SM buffer and added at 103 PFU/mL to the culture of B. abortus S19 diluted in Brucella Broth at concentrations of ~1 × 102; ~1 × 101 and ~1 × 100 CFU/mL. Bacterial titer was confirmed by the live bacteria count of cultures plated on Brucella Agar. qPCR was performed after 72 h of incubation of each test culture with corresponding bacteriophage.
4.5. Testing Phage Specificity on Non-Brucella Bacteria
Bacterial cultures used for testing phage specificity (Escherichia coli, Yersinia enterocolitica O:9, Ochrobactrum anthropi, Afipia felis, and B. anthracis, see Table 2) were grown overnight in Brucella Broth. Ten microliters of overnight culture were diluted with 10 mL of Brucella Broth (1:1000), and 10 µL of brucellaphage Bk (106 PFU/mL) were added, to get a final concentration of 103 PFU/mL. As a positive control, we used overnight culture of B. abortus S19 with the same concentration of Bk added. All cultures were incubated at 37 °C for 72 h with shaking at 200 rpm. The resulting culture was treated with 10% chloroform, a 0.5 mL aliquot was withdrawn and centrifuged for 3 min in a mini-centrifuge. One microliter of the resulting lysate was used for qPCR reaction. For testing phage specificity in mixed cultures, the equal volumes of diluted overnight non-Brucella and B. abortus S19 cultures were mixed. Samples were grown in the presence of the Bk phage at 103 PFU/mL for 72 h, followed by chloroform treatment and centrifugation as described above.
4.6. Phage Amplification in Simulated Clinical Samples Containing B. abortus
Simulated clinical tests were performed in Na2-EDTA-treated whole sheep blood (LAMPIRE Biological Laboratories, Inc. Pipersville, PA, USA). Overnight culture of B. abortus S19 was diluted to OD600 = 0.1 in Brucella Broth. Then serial bacterial dilutions 10−2 (~3 × 106 CFU/mL), 10−4 (~3 × 104 CFU/mL), 10−6 (~3 × 102 CFU/mL), 10−7 (~3 × 101 CFU/mL), and 10−8 (~3 × 100 CFU/mL) were made in sheep blood. The live bacteria count was done by plating 100 µL of 10−6 blood dilution on Brucella Agar. To optimize the indirect detection of B. abortus from blood, we used a lysis-centrifugation procedure [88] modified as follows. One milliliter of a blood sample was mixed with 0.5 mL of Red Blood Cell (RBC) Lysis Buffer (Sigma-Aldrich Co. LLC, St. Louis, MO, USA) containing 8.3 g/L of ammonium chloride and 0.01 M Tris-HCl, pH 7.5 ± 0.2. The resulting blood lysates containing intact leukocytes and Brucella cells were centrifuged for 4 min at full speed in a tabletop mini-centrifuge (Eppendorf North America, Hauppauge, NY, USA). The pellets were resuspended again in 1 mL of RBC Lysis Buffer to assure the complete removal of RBC and hemoglobin. After the second round of centrifugation, the pellets were resuspended in 1 mL of sterile water to break leukocytes by osmotic shock in order to simulate the release of intracellular Brucellae. Repeated determination of live Brucella was performed by plating 100 µL of 10−6 dilution of the washed blood sample. All live bacteria counts were done in three independent experiments in triplicate. The entire washed blood sample was inoculated into 20 mL of Brucella Broth in 125 mL Erlenmeyer flask with 0.22-µm filter cap, and bacteriophage was added at 103 PFU/mL. The samples were incubated at 37 °C with shaking at 60 rpm for 72 h, then treated with chloroform, centrifuged and used as indicated above.
4.7. Primer Design
Primers for qPCR were designed by using the Beacon Designer program [89]. The target for brucellaphage-based assay was DNA primase/polymerase gene of phage S708, sequenced in our lab recently [30]. Primers (5′-CATACCAGATGGGTTGATAACTGTTGAG-3′ and 5′-ACTGTTTGTAAATAGACGCCAGAAG-3′) were analyzed using the BLAST (Basic Local Alignment Search Tool) engine at the National Center for Biotechnology Information (NCBI) web site against the nonredundant (nr) database [90], and were shown to be brucellaphage-specific, i.e., to have 100% identity only with DNA of brucellaphages sequenced to date: Tb, Pr [29], S708, Fz, Wb, Bk, R/C [30], and F1 [91]. The primers were also tested for specificity against Brucella genomic DNA (culture suspension of Brucella abortus S19 at about 5 × 108 CFU/mL in distilled water, boiled for 5 min).
4.8. qPCR Assays
In all qPCR experiments, MaximaSYBR Green/ROX qPCR Master Mix 2× (Fermentas Inc., Glen Burnie, MD, USA) was used, according to the vendor’s recommendations. Reaction mixes were prepared in a total volume of 10 µL including 1 µL of DNA template (purified DNA solution or suspension containing live phage particles), 5 µL of the master mix, and 0.3 µM of each primer. Reactions were run on a LightCycler 2.0 (Roche Applied Science, Indianapolis, IN, USA). The cycling parameters were: 95 °C, 5 min; 40× (95 °C, 20 s; 60 °C, 60 s) with fluorescence measurement at the end of each cycle.
4.9. Statistical Analysis
Results of qPCR experiments are shown as the mean values of at least three independent tests. Statistical significance was computed by One Way analysis of variance (ANOVA) free online program at the website of Vassar College [92]. P-values < 0.05 were considered significant.
Acknowledgments
This research was supported in part by the Defense Threat Reduction Agency, Joint Science and Technology Office, Medical S&T Division. The findings and opinions expressed herein belong to the authors and do not necessarily reflect the official views of the Walter Reed Army Institute of Research, the U.S. Army or the Department of Defense.
Author Contributions
K.V.S., A.A.F. and M.P.N. conceived and designed the study. K.V.S. and A.A.F. performed the experiments and analyzed the data. M.P.N. provided reagents and materials. K.V.S., A.A.F. and M.P.N. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dean, A.S.; Crump, L.; Greter, H.; Schelling, E.; Zinsstag, J. Global burden of human brucellosis: A systematic review of disease frequency. PLoS Negl. Trop. Dis. 2012, 6, e1865. [Google Scholar] [CrossRef] [PubMed]
- Moreno, E. Retrospective and prospective perspectives on zoonotic brucellosis. Front. Microbiol. 2014, 5, 213. [Google Scholar] [CrossRef] [PubMed]
- Moreno, E.; Moriyón, I. The genus Brucella. In Prokaryotes—A Handbook on the Biology of Bacteria, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer-Verlag: New York, NY, USA, 2006; Volume 5, pp. 315–456. [Google Scholar]
- Olsen, S.C.; Palmer, M.V. Advancement of knowledge of Brucella over the past 50 years. Vet. Pathol. 2014, 51, 1076–1089. [Google Scholar] [CrossRef] [PubMed]
- Doganay, G.D.; Doganay, M. Brucella as a potential agent of bioterrorism. Recent Pat. Antiinfect. Drug. Discov. 2013, 8, 27–33. [Google Scholar] [CrossRef] [PubMed]
- CDC/USDA Federal Select Agent Program, Select Agents and Toxins List. Available online: https://www.selectagents.gov/selectagentsandtoxinslist.htmL (accessed on 23 January 2017).
- Al Dahouk, S.; Nöckler, K. Implications of laboratory diagnosis on brucellosis therapy. Expert Rev. Anti-Infect. Ther. 2011, 9, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Al Dahouk, S.; Sprague, L.D.; Neubauer, H. New developments in the diagnostic procedures for zoonotic brucellosis in humans. Rev. Sci. Tech. 2013, 32, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Public health consequences of a false-positive laboratory test result for Brucella—Florida, Georgia, and Michigan, 2005. MMWR 2008, 57, 603–605.
- Farrell, I.D. The development of a new selective medium for the isolation of Brucella abortus from contaminated sources. Res. Vet. Sci. 1974, 16, 280–286. [Google Scholar] [PubMed]
- Marín, C.M.; Jiménez-de-Bagüés, M.P.; Barberán, M.; Blasco, J.M. Comparison of two selective media for the isolation of Brucella melitensis from naturally infected sheep and goats. Vet. Rec. 1996, 138, 409–411. [Google Scholar] [CrossRef] [PubMed]
- De Miguel, M.J.; Marín, C.M.; Muñoz, P.M.; Dieste, L.; Grilló, M.J.; Blasco, J.M. Development of a selective culture medium for primary isolation of the main Brucella species. J. Clin. Microbiol. 2011, 49, 1458–1463. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.C.; Almendra, C.; Cardoso, R.; Pereira, M.S.; Beja-Pereira, A.; Luikart, G.; Corrêa de Sá, M.I. Development and evaluation of a selective medium for Brucella suis. Res. Vet. Sci. 2012, 93, 565–567. [Google Scholar] [CrossRef] [PubMed]
- Yagupsky, P. Detection of brucellae in blood cultures. J. Clin. Microbiol. 1999, 37, 3437–3442. [Google Scholar] [PubMed]
- Yagupsky, P.; Peled, N.; Press, J. Use of BACTEC 9240 blood culture system for detection of Brucella melitensis in synovial fluid. J. Clin. Microbiol. 2001, 39, 738–739. [Google Scholar] [CrossRef] [PubMed]
- Traxler, R.M.; Lehman, M.W.; Bosserman, E.A.; Guerra, M.A.; Smith, T.L. A literature review of laboratory-acquired brucellosis. J. Clin. Microbiol. 2013, 51, 3055–3062. [Google Scholar] [CrossRef] [PubMed]
- Bricker, B.J.; Halling, S.M. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 1994, 32, 2660–2666. [Google Scholar] [PubMed]
- Navarro, E.; Casao, M.A.; Solera, J. Diagnosis of human brucellosis using PCR. Expert Rev. Mol. Diagn. 2004, 4, 115–123. [Google Scholar] [CrossRef] [PubMed]
- López-Goñi, I.; García-Yoldi, D.; Marín, C.M.; de Miguel, M.J.; Muñoz, P.M.; Blasco, J.M.; Jacques, I.; Grayon, M.; Cloeckaert, A.; Ferreira, A.C.; et al. Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J. Clin. Microbiol. 2008, 6, 3484–3487. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Z.; Zhang, Y.; Bai, L.; Zhao, Y.; Liu, C.; Ma, A.; Yu, H. Polymerase chain reaction-based assays for the diagnosis of human brucellosis. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Wareth, G.; Melzer, F.; Tomaso, H.; Roesler, U.; Neubauer, H. Detection of Brucella abortus DNA in aborted goats and sheep in Egypt by real-time PCR. BMC Res. Notes 2015, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Vrioni, G.; Pappas, G.; Priavali, E.; Gartzonika, C.; Levidiotou, S. An eternal microbe: Brucella DNA load persists for years after clinical cure. Clin. Infect. Dis. 2008, 46, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Castaño, M.J.; Solera, J. Chronic brucellosis and persistence of Brucella melitensis DNA. J. Clin. Microbiol. 2009, 47, 2084–2089. [Google Scholar] [CrossRef] [PubMed]
- Dauphin, L.A.; Hutchins, R.J.; Bost, L.A.; Bowen, M.D. Evaluation of automated and manual commercial DNA extraction methods for recovery of Brucella DNA from suspensions and spiked swabs. J. Clin. Microbiol. 2009, 47, 3920–3926. [Google Scholar] [CrossRef] [PubMed]
- Droževkina, M.S. The present position in Brucella phage research. Bull. World Health Organ. 1963, 29, 43–57. [Google Scholar] [PubMed]
- Corbel, M.J.; Thomas, E.L. The Brucella-phages: Their Properties, Characterization and Applications, Booklet 2266; Ministry of Agriculture, Fisheries and Food: Pinner, Middlesex, UK, 1980. [Google Scholar]
- Segondy, M.; Allardet-Servent, A.; Caravano, R.; Ramuz, M. Common physical map of four Brucella bacteriophage genomes. FEMS Microbiol. Lett. 1988, 56, 177–181. [Google Scholar] [CrossRef]
- Rigby, C.E.; Cerqueira-Campos, M.L.; Kelly, H.A.; Surujballi, O.P. Properties and partial genetic characterization of Nepean phage and other lytic phages of Brucella species. Can. J. Vet. Res. 1989, 53, 319–325. [Google Scholar] [PubMed]
- Flores, V.; López-Merino, A.; Mendoza-Hernandez, G.; Guarneros, G. Comparative genomic analysis of two brucellaphages of distant origins. Genomics 2012, 99, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Farlow, J.; Filippov, A.A.; Sergueev, K.V.; Hang, J.; Kotorashvili, A.; Nikolich, M.P. Comparative whole genome analysis of six diagnostic brucellaphages. Gene 2014, 541, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.M.; Merz, G.S.; Wilson, J.B. Phage typing reactions on Brucella species. Appl. Microbiol. 1968, 16, 1179–1190. [Google Scholar] [PubMed]
- Corbel, M.J. Brucella phages: Advances in the development of a reliable phage typing system for smooth and non-smooth Brucella isolates. Ann. Inst. Pasteur Microbiol. 1987, 138, 70–75. [Google Scholar] [CrossRef]
- Taran, I.F.; Zanina, V.M.; Liamkin, G.I.; Tsybin, B.P.; Tikhenko, N.I. Comparative evaluation of the spectrum of lytic effects of bacteriophages Tb, Wb, Fi, Bk2 and R on various Brucella species. Zhurnal Mikrobiol. Epidemiol. Immunobiol. 1983, 2, 48–52. [Google Scholar]
- Sergueev, K.V.; He, Y.; Borschel, R.H.; Nikolich, M.P.; Filippov, A.A. Rapid and sensitive detection of Yersinia pestis using amplification of plague diagnostic bacteriophages monitored by real-time PCR. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Sergueev, K.V.; Nikolich, M.P.; Filippov, A.A. Field and clinical applications of advanced bacteriophage-based detection of Yersinia pestis. Adv. Exp. Med. Biol. 2012, 954, 135–141. [Google Scholar] [CrossRef] [PubMed]
- McDuff, C.R.; Jones, L.M.; Wilson, J.B. Characteristics of brucellaphage. J. Bacteriol. 1962, 83, 324–329. [Google Scholar] [PubMed]
- Douglas, J.T.; Elberg, S.S. Isolation of Brucella melitensis phage of broad biotype and species specificity. Infect. Immun. 1976, 14, 306–308. [Google Scholar] [PubMed]
- Al Dahouk, S.; Hofer, E.; Tomaso, H.; Vergnaud, G.; Le Flèche, P.; Cloeckaert, A.; Koylass, M.S.; Whatmore, A.M.; Nöckler, K.; Scholz, H.C. Intraspecies biodiversity of the genetically homologous species Brucella microti. Appl. Environ. Microbiol. 2012, 78, 1534–1543. [Google Scholar] [CrossRef] [PubMed]
- Sanodze, L.; Bautista, C.T.; Garuchava, N.; Chubinidze, S.; Tsertsvadze, E.; Broladze, M.; Chitadze, N.; Sidamonidze, K.; Tsanava, S.; Akhvlediani, T.; et al. Expansion of brucellosis detection in the country of Georgia by screening household members of cases and neighboring community members. BMC Public Health 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2016. Available online: http://www.oie.int/en/international-standard-setting/terrestrial-manual/access-online/ (accessed on 31 March 2017).
- Worsley, B.; Goodwin, S.; Jahans, K.; Atallah, C. First report of a strain of Brucella melitensis that was widely sensitive to brucellaphages isolated in the United Arab Emirates. Clin. Infect. Dis. 1996, 22, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Köse, S.; Kiliç, S.; Ozbel, Y. Identification of Brucella species isolated from proven brucellosis patients in Izmir, Turkey. J. Basic Microbiol. 2005, 45, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Foster, G.; Osterman, B.S.; Godfroid, J.; Jacques, I.; Cloeckaert, A. Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int. J. Syst. Evolut. Microbiol. 2007, 57, 2688–2693. [Google Scholar] [CrossRef] [PubMed]
- Scholz, H.C.; Nöckler, K.; Göllner, C.; Bahn, P.; Vergnaud, G.; Tomaso, H.; Al Dahouk, S.; Kämpfer, P.; Cloeckaert, A.; Maquart, M.; et al. Brucella inopinata sp. nov., isolated from a breast implant infection. Int. J. Syst. Evol. Microbiol. 2010, 60, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, H. The bacteriophage titer increase reaction. Ann. Inst. Pasteur Lille 1963, 14, 145–158. [Google Scholar] [PubMed]
- Bakulov, I.A.; Kotliarov, V.M.; Kol’pikova, T.I. Sensitivity of the phage titer increase test in detecting Listeria. Zhurnal Mikrobiol. Epidemiol. Immunobiol. 1984, 9, 40–43. [Google Scholar]
- Albert, H.; Heydenrych, A.; Brookes, R.; Mole, R.J.; Harley, B.; Subotsky, E.; Henry, R.; Azevedo, V. Performance of a rapid phage-based test, FASTPlaqueTB, to diagnose pulmonary tuberculosis from sputum specimens in South Africa. Int. J. Tuberc. Lung Dis. 2002, 6, 529–537. [Google Scholar] [PubMed]
- Kiraz, N.; Et, L.; Akgun, Y.; Kasifoglu, N.; Kiremitci, A. Rapid detection of Mycobacterium tuberculosis from sputum specimens using the FASTPlaqueTB test. Int. J. Tuberc. Lung Dis. 2007, 11, 904–908. [Google Scholar] [PubMed]
- Stanley, E.C.; Mole, R.J.; Smith, R.J.; Glenn, S.M.; Barer, M.R.; McGowan, M.; Rees, C.E. Development of a new, combined rapid method using phage and PCR for detection and identification of viable Mycobacterium paratuberculosis bacteria within 48 hours. Appl. Environ. Microbiol. 2007, 73, 1851–1857. [Google Scholar] [CrossRef] [PubMed]
- Swift, B.M.; Denton, E.J.; Mahendran, S.A.; Huxley, J.N.; Rees, C.E. Development of a rapid phage-based method for the detection of viable Mycobacterium avium subsp. paratuberculosis in blood within 48 h. J. Microbiol. Methods 2013, 94, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Hirsh, D.C.; Martin, L.D. Rapid detection of Salmonella spp. by using Felix-O1 bacteriophage and high-performance liquid chromatography. Appl. Environ. Microbiol. 1983, 45, 260–264. [Google Scholar] [PubMed]
- Edelman, D.C.; Barletta, J. Real-time PCR provides improved detection and titer determination of bacteriophage. Biotechniques 2003, 35, 368–375. [Google Scholar] [PubMed]
- Reiman, R.W.; Atchley, D.H.; Voorhees, K.J. Indirect detection of Bacillus anthracis using real-time PCR to detect amplified gamma phage DNA. J. Microbiol. Methods 2007, 68, 651–653. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Chan, M.; Allain, B.; Mandeville, R.; Brooks, B.W. Detection of multiple antibiotic-resistant Salmonella enterica serovar Typhimurium DT104 by phage replication-competitive enzyme-linked immunosorbent assay. J. Food Prot. 2006, 69, 739–742. [Google Scholar] [CrossRef] [PubMed]
- Bhowmick, T.; Mirrett, S.; Reller, L.B.; Price, C.; Qi, C.; Weinstein, M.P.; Kirn, T.J. Controlled multicenter evaluation of a bacteriophage-based method for rapid detection of Staphylococcus aureus in positive blood cultures. J. Clin. Microbiol. 2013, 51, 1226–1230. [Google Scholar] [CrossRef] [PubMed]
- Kannan, P.; Yong, H.Y.; Reiman, L.; Cleaver, C.; Patel, P.; Bhagwat, A.A. Bacteriophage-based rapid and sensitive detection of Escherichia coli O157:H7 isolates from ground beef. Foodborne Pathog. Dis. 2010, 7, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, T.; Schwartz-Mittelmann, A.; Biran, D.; Ron, E.Z.; Rishpon, J. Combined phage typing and amperometric detection of released enzymatic activity for the specific identification and quantification of bacteria. Anal. Chem. 2003, 75, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, W.R., Jr.; Barletta, R.G.; Udani, R.; Chan, J.; Kalkut, G.; Sosne, G.; Kieser, T.; Sarkis, G.J.; Hatfull, G.F.; Bloom, B.R. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science 1993, 260, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Dusthackeer, A.; Kumar, V.; Subbian, S.; Sivaramakrishnan, G.; Zhu, G.; Subramanyam, B.; Hassan, S.; Nagamaiah, S.; Chan, J.; Paranji Rama, N. Construction and evaluation of luciferase reporter phages for the detection of active and non-replicating tubercle bacilli. J. Microbiol. Methods 2008, 73, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Schofield, D.A.; Molineux, I.J.; Westwater, C. Diagnostic bioluminescent phage for detection of Yersinia pestis. J. Clin. Microbiol. 2009, 47, 3887–3894. [Google Scholar] [CrossRef] [PubMed]
- Schofield, D.A.; Westwater, C. Phage-mediated bioluminescent detection of Bacillus anthracis. J. Appl. Microbiol. 2009, 107, 1468–1478. [Google Scholar] [CrossRef] [PubMed]
- Awais, R.; Fukudomi, H.; Miyanaga, K.; Unno, H.; Tanji, Y. A recombinant bacteriophage-based assay for the discriminative detection of culturable and viable but nonculturable Escherichia coli O157:H7. Biotechnol. Prog. 2006, 22, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Wallach, J.C.; Ferrero, M.C.; Victoria Delpino, M.; Fossati, C.A.; Baldi, P.C. Occupational infection due to Brucella abortus S19 among workers involved in vaccine production in Argentina. Clin. Microbiol. Infect. 2008, 14, 805–807. [Google Scholar] [CrossRef] [PubMed]
- Elsaghir, A.A.; James, E.A. Misidentification of Brucella melitensis as Ochrobactrum anthropi by API 20NE. J. Med. Microbiol. 2003, 52, 441–442. [Google Scholar] [CrossRef] [PubMed]
- Horvat, R.T.; El Atrouni, W.; Hammoud, K.; Hawkinson, D.; Cowden, S. Ribosomal RNA sequence analysis of Brucella infection misidentified as Ochrobactrum anthropi infection. J. Clin. Microbiol. 2011, 49, 1165–1168. [Google Scholar] [CrossRef] [PubMed]
- Carrington, M.; Choe, U.; Ubillos, S.; Stanek, D.; Campbell, M.; Wansbrough, L.; Lee, P.; Churchwell, G.; Rosas, K.; Zaki, S.R.; et al. Fatal case of brucellosis misdiagnosed in early stages of Brucella suis infection in a 46-year-old patient with Marfan syndrome. J. Clin. Microbiol. 2012, 50, 2173–2175. [Google Scholar] [CrossRef] [PubMed]
- Ahvonen, P.; Jansson, E.; Aho, K. Marked crossagglutination between brucellae and a subtype of Yersinia enterocolitica. Acta Pathol. Microbiol. Scand. 1969, 75, 291–295. [Google Scholar] [PubMed]
- Bundle, D.R.; Gidney, M.A.J.; Perry, M.B.; Duncan, J.R.; Cherwonogrodzky, J.W. Serological confirmation of Brucella abortus and Yersinia enterocolitica O:9 O-antigens by monoclonal antibodies. Infect. Immun. 1984, 46, 389–393. [Google Scholar] [PubMed]
- Chenais, E.; Bagge, E.; Lambertz, S.T.; Artursson, K. Yersinia enterocolitica serotype O:9 cultured from Swedish sheep showing serologically false-positive reactions for Brucella melitensis. Infect. Ecol. Epidemiol. 2012, 2, 19027. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M.; Brouqui, P.; Raoult, D. Afipia clevelandensis antibodies and cross-reactivity with Brucella spp. and Yersinia enterocolitica O:9. Clin. Diagn. Lab. Immunol. 1997, 4, 748–752. [Google Scholar] [PubMed]
- Probert, W.S.; Schrader, K.N.; Khuong, N.Y.; Bystrom, S.L.; Graves, M.H. Real-time multiplex PCR assay for detection of Brucella spp., B. abortus, and B. melitensis. J. Clin. Microbiol. 2004, 42, 1290–1293. [Google Scholar] [CrossRef] [PubMed]
- Bogdanovich, T.; Skurnik, M.; Lübeck, P.S.; Ahrens, P.; Hoorfar, J. Validated 5′ nuclease PCR assay for rapid identification of the genus Brucella. J. Clin. Microbiol. 2004, 42, 2261–2263. [Google Scholar] [CrossRef] [PubMed]
- Gopaul, K.K.; Koylass, M.S.; Smith, C.J.; Whatmore, A.M. Rapid identification of Brucella isolates to the species level by real time PCR based single nucleotide polymorphism (SNP) analysis. BMC Microbiol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Bounaadja, L.; Albert, D.; Chénais, B.; Hénault, S.; Zygmunt, M.S.; Poliak, S.; Garin-Bastuji, B. Real-time PCR for identification of Brucella spp.: A comparative study of IS711, bcsp31 and per target genes. Vet. Microbiol. 2009, 137, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Winchell, J.M.; Wolff, B.J.; Tiller, R.; Bowen, M.D.; Hoffmaster, A.R. Rapid identification and discrimination of Brucella isolates by use of real-time PCR and high-resolution melt analysis. J. Clin. Microbiol. 2010, 48, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Hänsel, C.; Mertens, K.; Elschner, M.C.; Melzer, F. Novel real-time PCR detection assay for Brucella suis. Vet. Rec. Open 2015, 2. [Google Scholar] [CrossRef] [PubMed]
- Kattar, M.M.; Zalloua, P.A.; Araj, G.F.; Samaha-Kfoury, J.; Shbaklo, H.; Kanj, S.S.; Khalife, S.; Deeb, M. Development and evaluation of real-time polymerase chain reaction assays on whole blood and paraffin-embedded tissues for rapid diagnosis of human brucellosis. Diagn. Microbiol. Infect. Dis. 2007, 59, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Gwida, M.M.; El-Gohary, A.H.; Melzer, F.; Tomaso, H.; Rösler, U.; Wernery, U.; Wernery, R.; Elschner, M.C.; Khan, I.; Eickhoff, M.; et al. Comparison of diagnostic tests for the detection of Brucella spp. in camel sera. BMC Res. Notes 2011, 4. [Google Scholar] [CrossRef] [PubMed]
- Queipo-Ortuño, M.I.; Colmenero, J.D.; Reguera, J.M.; García-Ordoñez, M.A.; Pachón, M.E.; Gonzalez, M.; Morata, P. Rapid diagnosis of human brucellosis by SYBR Green I-based real-time PCR assay and melting curve analysis in serum samples. Clin. Microbiol. Infect. 2005, 11, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Popkhadze, M.Z.; Abashidze, T.G. Characterization of a Brucella Phage isolated at Tbilisi Research Institute for Vaccines and Sera, Abstracts of Inter-Institute Science Conference on Bacteriophagy; Tbilisi University Press: Tbilisi, USSR, 1957; p. 40. [Google Scholar]
- Corbel, M.J.; Thomas, E.L. Description of a new phage lytic for several Brucella species. J. Biol. Stand. 1976, 4, 195–201. [Google Scholar] [CrossRef]
- Graves, R.R. The story of John M. Buck’s and Matilda’s contribution to the cattle industry. J. Am. Vet. Med. Assoc. 1943, 102, 193–195. [Google Scholar]
- Holmes, B.; Popoff, M.; Kiredjian, M.; Kersters, K. Ochrobactrum anthropi gen. nov., sp. nov. from human clinical specimens and previously known as group Vd. Int. J. Syst. Bacteriol. 1988, 38, 406–416. [Google Scholar] [CrossRef]
- Brenner, D.J.; Hollis, D.G.; Moss, C.W.; English, C.K.; Hall, G.S.; Vincent, J.; Radosevic, J.; Birkness, K.A.; Bibb, W.F.; Quinn, F.D.; et al. Proposal of Afipia gen. nov., with Afipia felis sp. nov. (formerly the cat scratch disease bacillus), Afipia clevelandensis sp. nov. (formerly the Cleveland Clinic Foundation strain), Afipia broomeae sp. nov., and three unnamed genospecies. J. Clin. Microbiol. 1991, 29, 2450–2460. [Google Scholar] [PubMed]
- Sterne, M. The use of anthrax vaccines prepared from avirulent (uncapsulated) variants of Bacillus anthracis. Onderstepoort J. Vet. Sci. Anim. Ind. 1939, 13, 307–312. [Google Scholar]
- Appleyard, R.K. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K-12. Genetics 1954, 39, 440–452. [Google Scholar] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Etemadi, H.; Raissadat, A.; Pickett, M.J.; Zafari, Y.; Vahedifar, P. Isolation of Brucella spp. from clinical specimens. J. Clin. Microbiol. 1984, 20, 586. [Google Scholar] [PubMed]
- NetPrimer, Free Primer Analysis Software, Premier Biosoft Int. Available online: http://www.premierbiosoft.com/netprimer/index.html (accessed on 25 January 2017).
- Basic Local Alignment Search Tool (BLAST). Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 25 January 2017).
- Hammerl, J.A.; Al Dahouk, S.; Nöckler, K.; Göllner, C.; Appel, B.; Hertwig, S. F1 and Tbilisi are closely related brucellaphages exhibiting some distinct nucleotide variations which determine the host specificity. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- VassarStats: Website for Statistical Computation/ANOVA/One-Way Analysis of Variance for Independent or Correlated Samples. Available online: http://faculty.vassar.edu/lowry/anova1u.html (accessed on 24 January 2017).
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).