Characterization of a Novel Bat Adenovirus Isolated from Straw-Colored Fruit Bat (Eidolon helvum)
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Samples
2.2. Virus Isolation
2.3. Transmission Electron Microscopy
2.4. Indirect Immunofluorescence Assay
2.5. Virus Purification and DNA Extraction for Next-Generation Sequencing
2.6. Library Preparation and Full-Length Genomic Sequencing
2.7. Phylogenetic Analysis and Other Bioinformatics Analysis
2.8. Analysis of In Vitro Cell Tropism of EhAdV 06-106
2.9. Nested PCR for Molecular Epizootiology and Direct Sequencing
2.10. Analysis of the 5′ End of the Pol mRNA
3. Results
3.1. Isolation of a Novel Adenovirus from E. helvum Captured in Zambia
3.2. Whole Genome Analysis of EhAdV 06-106
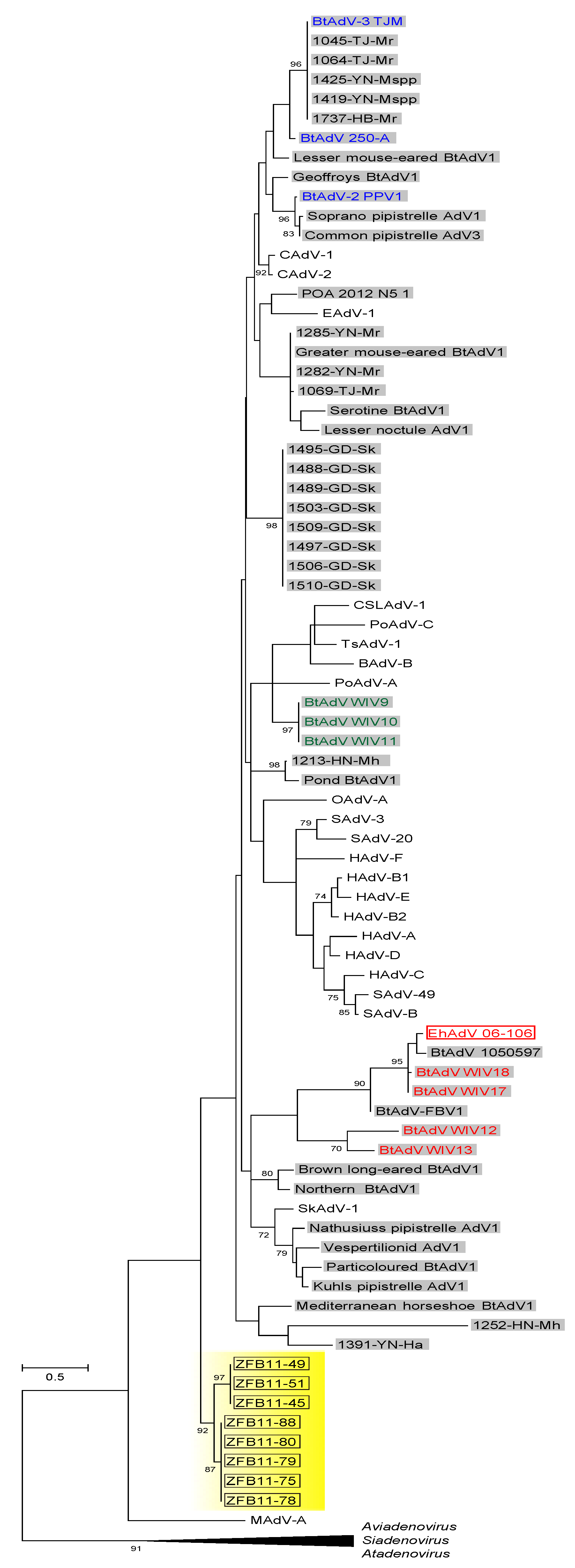
3.3. Phylogenetic Analyses of the Pol and Hexon Proteins
3.4. In Vitro Cell Tropism of EhAdV 06-106
3.5. Prevalence of Bat Adenoviruses in Zambia
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- De Jong, C.; Field, H.; Newman, S.H.; Epstein, J.H. Emerging infectious dieses. In Investigating the Role of Bats in Emerging Zoonoses: Balancing Ecology, Conservation and Public Health Interests; Newman, S.H., Field, H.E., de Jong, C.E., Epstein, J.H., Eds.; FAO: Rome, Italy, 2011; pp. 1–13. ISBN 978-92-5-107028-4. [Google Scholar]
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006, 19, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Muleya, W.; Sasaki, M.; Orba, Y.; Ishii, A.; Thomas, Y.; Nakagawa, E.; Ogawa, H.; Hang’ombe, B.; Namangala, B.; Mweene, A.; et al. Molecular epidemiology of paramyxoviruses in frugivorous Eidolon helvum bats in Zambia. J. Vet. Med. Sci. 2014, 76, 611–614. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Koizumi, N.; Ohnuma, A.; Mutemwa, A.; Hang’ombe, B.M.; Mweene, A.S.; Takada, A.; Sugimoto, C.; Suzuki, Y.; Kida, H.; et al. Molecular epidemiology of pathogenic Leptospira spp. in the straw-colored fruit bat (Eidolon helvum) migrating to Zambia from the Democratic Republic of Congo. Infect. Genet. Evol. 2015, 32, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Miyamoto, H.; Nakayama, E.; Yoshida, R.; Nakamura, I.; Sawa, H.; Ishii, A.; Thomas, Y.; Nakagawa, E.; Matsuno, K.; et al. Seroepidemiological Prevalence of Multiple Species of Filoviruses in Fruit Bats (Eidolon helvum) Migrating in Africa. J. Infect. Dis. 2015, 212, S101–S108. [Google Scholar] [CrossRef] [PubMed]
- Harrach, B.; Benkö, M.; Both, G.; Brown, M.; Davison, A.; Echavarría, M.; Hess, M.; Jones, M.; Kajon, A.; Lehmkuhl, H.; et al. Family Adenoviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A., Adams, M., Carstens, E., Lefkowitz, E., Eds.; Elsevier: San Diego, CA, USA, 2011; pp. 125–141. ISBN 978-0-12-384684-6. [Google Scholar]
- Berk, A.J. Adenoviridae . In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2, pp. 1704–1731. ISBN 978-1451105636. [Google Scholar]
- Benkö, M.; Harrach, B.; Kremer, E.J. Do nonhuman primate or bat adenoviruses pose a risk for human health? Future Microbiol. 2014, 9, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Buonavoglia, C.; Martella, V. Canine respiratory viruses. Vet. Res. 2007, 38, 355–373. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV) Virus Taxonomy: 2016 Release. Available online: http://talk.ictvonline.org/taxonomy/ (accessed on 20 September 2017).
- Maeda, K.; Hondo, E.; Terakawa, J.; Kiso, Y.; Nakaichi, N.; Endoh, D.; Sakai, K.; Morikawa, S.; Mizutani, T. Isolation of novel adenovirus from fruit bat (Pteropus dasymallus yayeyamae). Emerg. Infect. Dis. 2008, 14, 347–349. [Google Scholar] [CrossRef] [PubMed]
- Vidovszky, M.; Kohl, C.; Boldogh, S.; Gőrfől, T.; Wibbelt, G.; Kurth, A.; Harrach, B. Random sampling of the Central European bat fauna reveals the existence of numerous hitherto unknown adenoviruses. Acta Vet. Hung. 2015, 63, 508–525. [Google Scholar] [CrossRef] [PubMed]
- Jánoska, M.; Vidovszky, M.; Molnár, V.; Liptovszky, M.; Harrach, B.; Benkő, M. Novel adenoviruses and herpesviruses detected in bats. Vet. J. 2011, 189, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, X.; Zhang, H.; Zhou, P.; Zhu, Y.; Zhang, Y.; Yuan, J.; Wang, L.F.; Shi, Z. Host range, prevalence, and genetic diversity of adenoviruses in bats. J. Virol. 2010, 84, 3889–3897. [Google Scholar] [CrossRef] [PubMed]
- Raut, C.G.; Yadav, P.D.; Towner, J.S.; Amman, B.R.; Erickson, B.R.; Cannon, D.L.; Sivaram, A.; Basu, A.; Nichol, S.T.; Mishra, A.C.; et al. Isolation of a novel adenovirus from Rousettus leschenaultii bats from India. Intervirology 2012, 55, 488–490. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, M.; Mühldorfer, K.; Speck, S.; Wibbelt, G.; Kurth, A. New adenovirus in bats, Germany. Emerg. Infect. Dis. 2009, 15, 2052–2055. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV) Virus Taxonomy History: Bat Mastadenovirus A. Available online: http://talk.ictvonline.org/taxonomy/p/taxonomy-history?taxnode_id=20161725 (accessed on 8 September 2017).
- Kohl, C.; Vidovszky, M.Z.; Mühldorfer, K.; Dabrowski, P.W.; Radonic, A.; Nitsche, A.; Wibbelt, G.; Kurth, A.; Harrach, B. Genome analysis of bat adenovirus 2: Indications of interspecies transmission. J. Virol. 2012, 86, 1888–1892. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Yang, X.L.; Ge, X.Y.; Peng, C.; Zhang, Y.Z.; Zhang, L.B.; Shi, Z.L. Novel bat adenoviruses with an extremely large E3 gene. J. Gen. Virol. 2016, 97, 1625–1635. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Yang, X.L.; Ge, X.Y.; Peng, C.; Liu, H.Z.; Zhang, Y.Z.; Zhang, L.B.; Shi, Z.L. Novel bat adenoviruses with low G + C content shed new light on the evolution of adenoviruses. J. Gen. Virol. 2017, 98, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Hinojosa, G.; Gulland, F.M.; Goldstein, T.; Venn-Watson, S.; Rivera, R.; Waltzek, T.B.; Salemi, M.; Wellehan, J.F., Jr. Phylogenomic characterization of California sea lion adenovirus-1. Infect. Genet. Evol. 2015, 31, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Nao, N.; Yamagishi, J.; Miyamoto, H.; Igarashi, M.; Manzoor, R.; Ohnuma, A.; Tsuda, Y.; Furuyama, W.; Shigeno, A.; Kajihara, M.; et al. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. MBio 2017, 8, e02298-16. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, J.; Miyamoto, H.; Kajihara, M.; Ogawa, H.; Maeda, K.; Sakoda, Y.; Yoshida, R.; Takada, A. Characterization of the envelope glycoprotein of a novel filovirus, lloviu virus. J. Virol. 2014, 88, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Chroboczek, J.; Bieber, F.; Jacrot, B. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology 1992, 186, 280–285. [Google Scholar] [CrossRef]
- Hackenbrack, N.; Rogers, M.B.; Ashley, R.E.; Keel, M.K.; Kubiski, S.V.; Bryan, J.A.; Ghedin, E.; Holmes, E.C.; Hafenstein, S.L.; Allison, A.B. Evolution and cryo-electron microscopy capsid structure of a north American bat adenovirus and its relationship to other Mastadenoviruses. J. Virol. 2017, 91, e01504-16. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J.; Benko, M.; Harrach, B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003, 84, 2895–2908. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.S.; Leggett, R.M.; Bexfield, N.H.; Alston, M.; Daly, G.; Todd, S.; Tachedjian, M.; Holmes, C.E.; Crameri, S.; Wang, L.F.; et al. Metagenomic study of the viruses of African straw-coloured fruit bats: Detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology 2013, 441, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.S.; Idamakanti, N.; Zakhartchouk, A.N.; Baxi, M.K.; Lee, J.B.; Pyne, C.; Babiuk, L.A.; Tikoo, S.K. Nucleotide sequence, genome organization, and transcription map of bovine adenovirus type 3. J. Virol. 1998, 72, 1394–1402. [Google Scholar] [PubMed]
- DeFrees, S.L.; Wilson, D.E. Eidolon helvum. Mamm. Species 1988, 312, 1–5. [Google Scholar] [CrossRef]
- Richter, H.V.; Cumming, G.S. First application of satellite telemetry to track African straw-coloured fruit bat migration. J. Zool. 2008, 275, 172–176. [Google Scholar] [CrossRef]
- Benkö, M.; Harrach, B. A proposal for a new (third) genus within the family Adenoviridae. Arch. Virol. 1998, 143, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Benkö, M.; Harrach, B. Molecular evolution of adenoviruses. Curr. Top. Microbiol. Immunol. 2003, 272, 3–35. [Google Scholar] [CrossRef] [PubMed]
- Wellehan, J.F.; Johnson, A.J.; Harrach, B.; Benko, M.; Pessier, A.P.; Johnson, C.M.; Garner, M.M.; Childress, A.; Jacobson, E.R. Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J. Virol. 2004, 78, 13366–13369. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, Y.; Takeuchi, O.; Akira, S. TLR9 as a key receptor for the recognition of DNA. Adv. Drug. Deliv. Rev. 2008, 60, 795–804. [Google Scholar] [CrossRef] [PubMed]
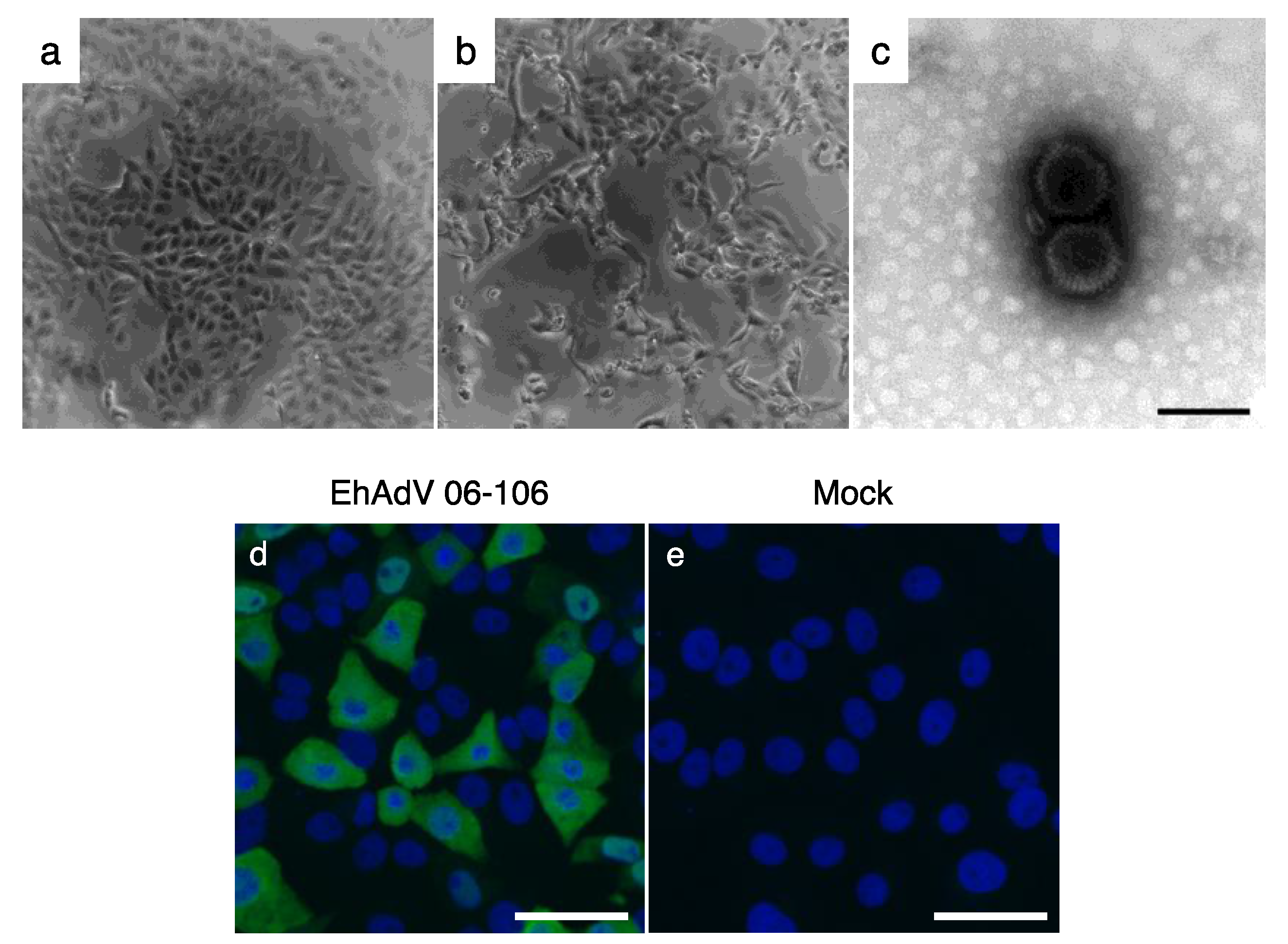

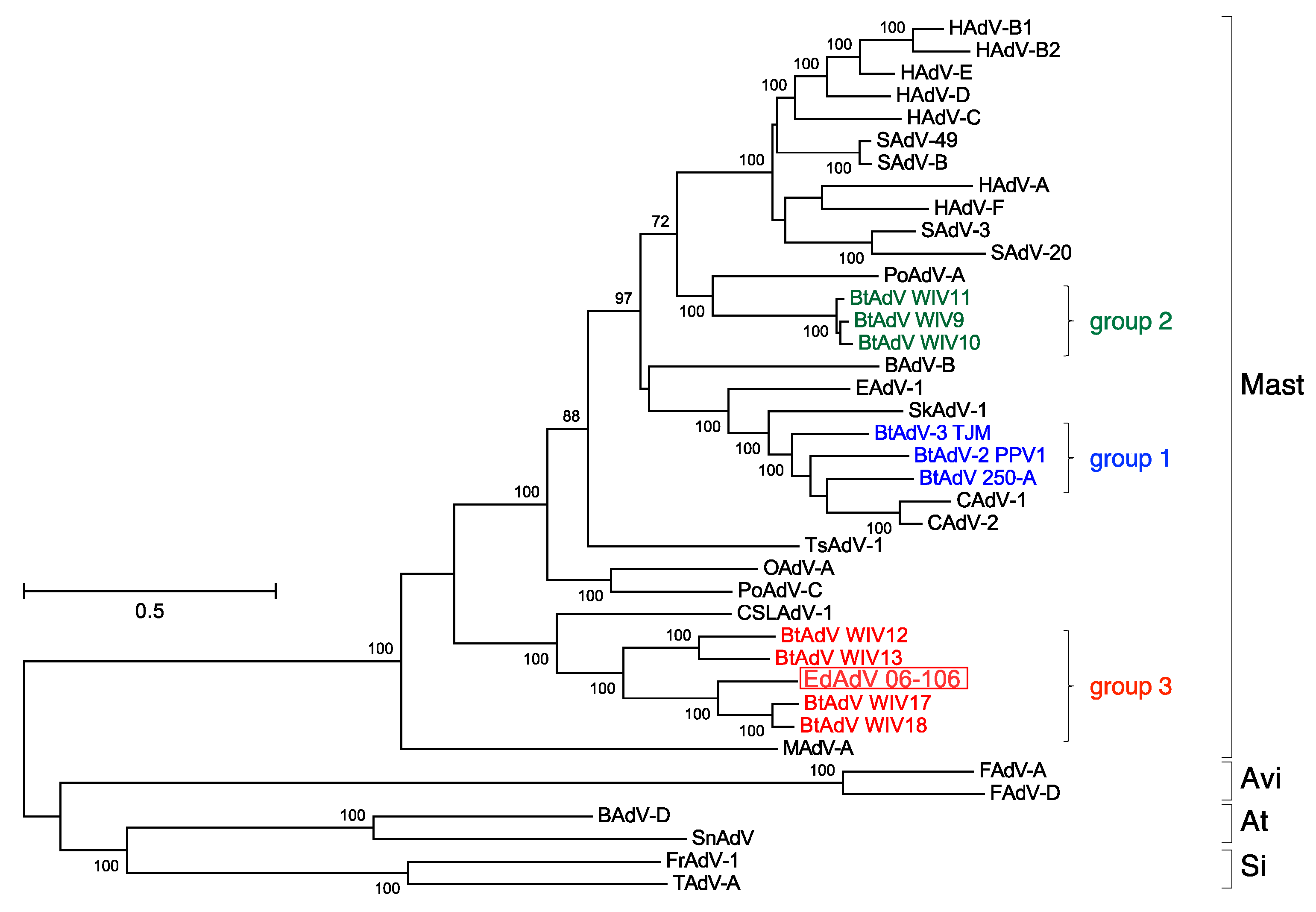
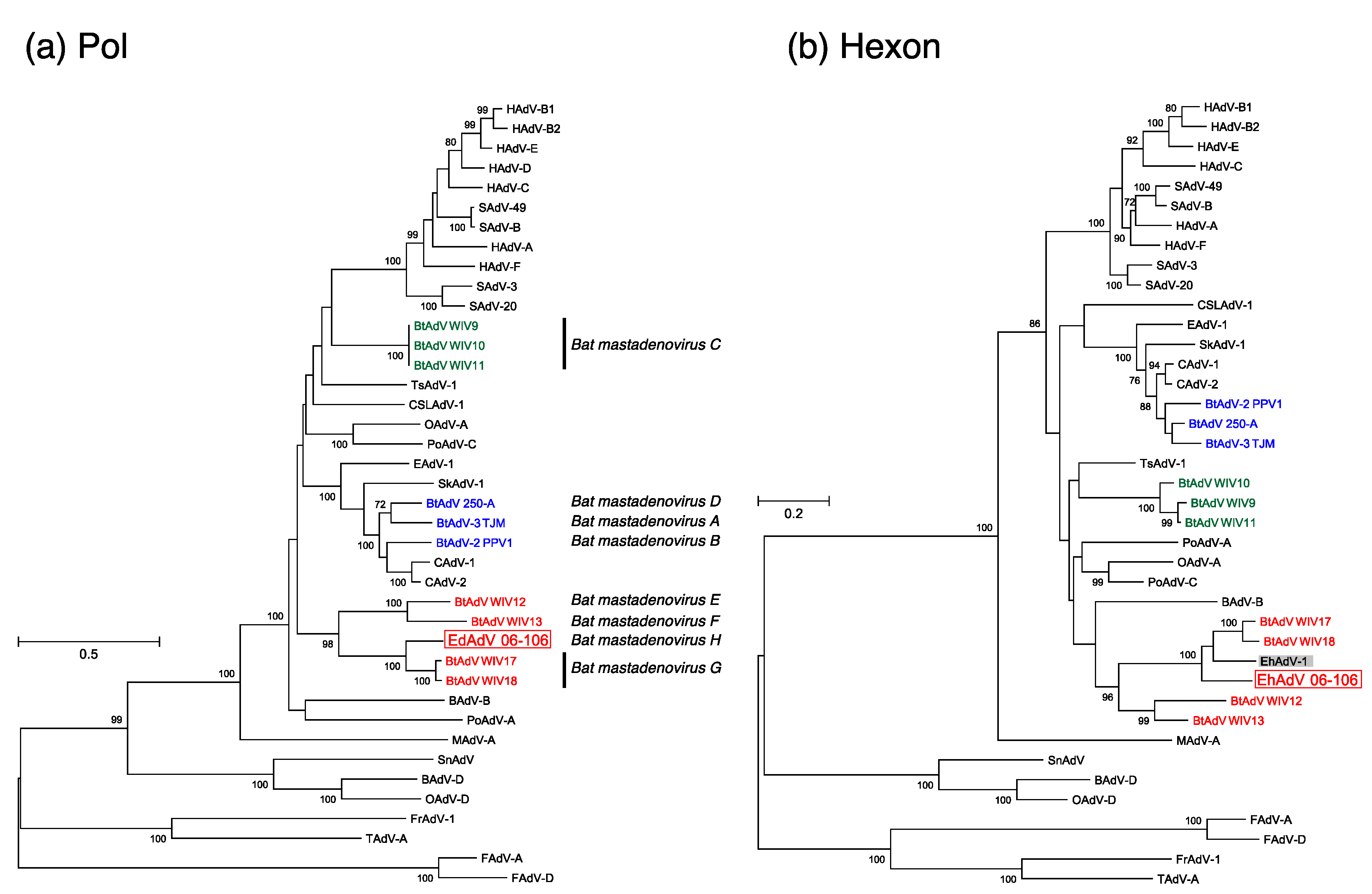
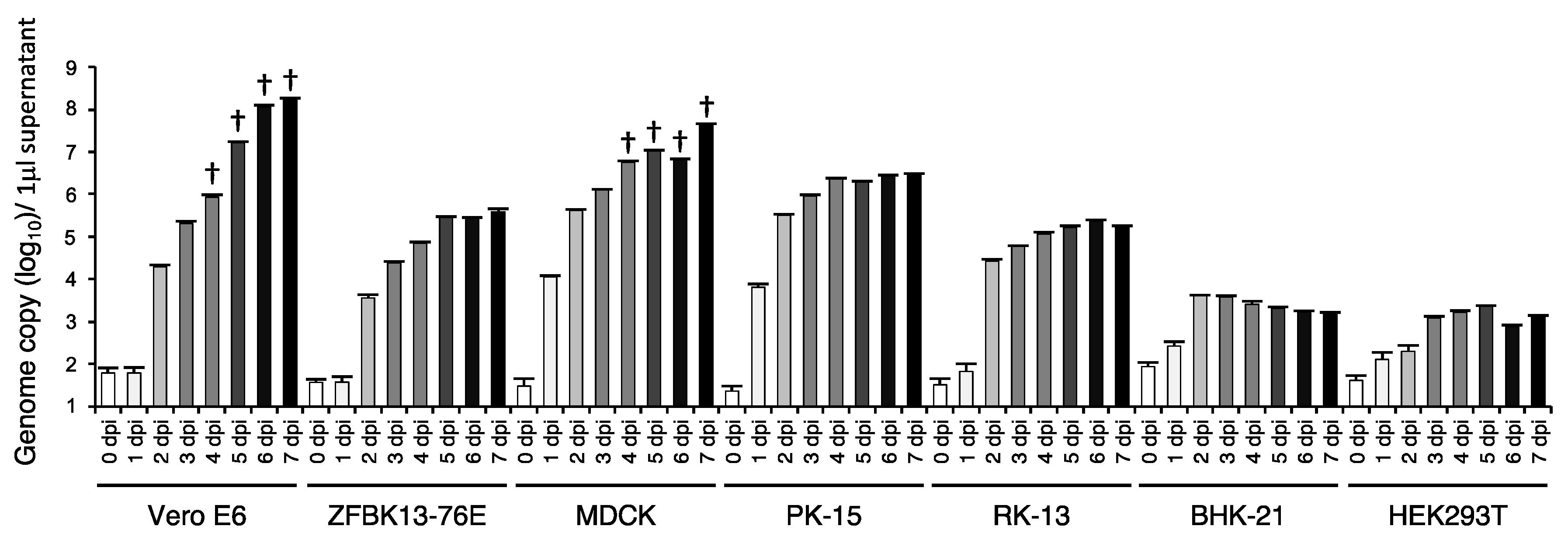
| Group | Strain | ICTV Species * | Host | Country | Full-Genome (bp) | ITR (bp) | E3 Region | G + C Content (%) | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|
| Product | Total Length (bp) † | |||||||||
| Group 1 | PPV1 | Bat mastadenovirus B | Pipistrellus pipistrellus | Germany | 31,616 | 137 | 12.5K, ORF1 | 1524 | 53.5 | [18] |
| TJM | Bat mastadenovirus A | Myotis ricketti | China | 31,681 | 128 | ORFA | 1149 | 56.9 | [14] | |
| 250-A | (Bat mastadenovirus D) | Corynorhinus rafinesquii | United States | 31,484 | 239 | 12.5K, ORFA | 1515 | 49.8 | [28] | |
| Group 2 | WIV9 | (Bat mastadenovirus C) | Rhinolophus sinicus | China | 37,545 | 35 | 12.5K, E3L, E3s | 5145 | 55.5 | [19] |
| WIV10 | (Bat mastadenovirus C) | Rhinolophus sinicus | China | 37,556 | 51 | 12.5K, E3L, E3s | 5133 | 55.2 | [19] | |
| WIV11 | (Bat mastadenovirus C) | Rhinolophus sinicus | China | 38,073 | 51 | 12.5K, E3L, E3s | 5631 | 55.0 | [19] | |
| Group 3 | WIV12 | (Bat mastadenovirus E) | Miniopterus schreibersii | China | 29,581 | 73 | 14.7K | 390 | 34.2 | [20] |
| WIV13 | (Bat mastadenovirus F) | Miniopterus schreibersii | China | 29,162 | 61 | 14.7K | 363 | 31.3 | [20] | |
| WIV17 | (Bat mastadenovirus G) | Rousettus Ieschenaultii | China | 29,923 | 178 | 14.7K | 396 | 34.3 | [20] | |
| WIV18 | (Bat mastadenovirus G) | Rousettus Ieschenaultii | China | 29,812 | 177 | 14.7K | 396 | 34.2 | [20] | |
| EhAdV 06-106 | (Bat mastadenovirus H) | Eidolon helvum | Zambia | 30,134 | 102 | 14.7K | 378 | 35.2 | This study | |
| EhAdV 06-106 | Bat Adenovirus WIV17 | Bat Adenovirus WIV18 | |||||
|---|---|---|---|---|---|---|---|
| Gene Product | Genomic Position * | Gene (nt) | Protein (AA) | Protein (AA) | Identity (%) | Protein (AA) | Identity (%) |
| ITR | 1–102 | 102 | N.A. † | N.A. | N.A. | N.A. | N.A. |
| E1A | 487–1090, 1230–1294 | 669 | 222 | 195 | 46.1 | 195 | 44.1 |
| E1B small | 1529–2014 | 486 | 161 | 168 | 53.4 | 168 | 44.7 |
| E1B large | 1909–3180 | 1272 | 423 | 425 | 59.5 | 425 | 60.2 |
| IX | 3240–3506 | 267 | 88 | 83 | 46.9 | 82 | 51.2 |
| Iva2 | 3550–4874c, 5153–5165c | 1338 | 445 | 422 | 82.7 | 422 | 82.7 |
| Pol | 4638–7895c | 3258 | 1085 | 1086 | 80.4 | 1086 | 81.1 |
| pTP | 7892–9682c, 12,334–12,342c | 1780 | 599 | 556 | 78.8 | 556 | 80.2 |
| 52K | 9697–10821 | 1125 | 374 | 373 | 80.4 | 373 | 79.3 |
| pIIIa | 10,742–12,319 | 1578 | 525 | 521 | 82.8 | 521 | 82.4 |
| Penton | 12,377–13,768 | 1392 | 463 | 464 | 82.9 | 464 | 83.8 |
| pVII | 13,771–14,115 | 345 | 114 | 118 | 83.9 | 118 | 83.9 |
| V | 14,159–16,048 | 1890 | 629 | 582 | 63.6 | 571 | 65.9 |
| pX | 16,074–16,280 | 207 | 68 | 66 | 83.3 | 66 | 84.8 |
| pVI | 16,341–16,970 | 630 | 209 | 218 | 81.8 | 218 | 82.8 |
| Hexon | 17,053–19,785 | 2733 | 910 | 910 | 82.1 | 910 | 82.5 |
| Protease | 19,786–20,394 | 609 | 202 | 200 | 77.0 | 200 | 76.0 |
| DBP | 20,457–21,695c | 1239 | 412 | 413 | 72.2 | 413 | 72.0 |
| 100K | 21,710–23,716 | 2007 | 668 | 650 | 77.9 | 650 | 78.5 |
| 33K | 23,595–23,736, 23,897–24,153 | 399 | 132 | 142 | 77.2 | 140 | 75.7 |
| 22K | 23,595–23,921 | 327 | 106 | 118 | 75.9 | 116 | 74.0 |
| pVIII | 24,153–24,794 | 642 | 213 | 213 | 85.9 | 213 | 84.9 |
| E3 14.7K | 24,797–25,174 | 378 | 125 | 131 | 52.7 | 131 | 48.0 |
| U exon | 25,191–25,361c | 171 | 56 | 55 | 70.9 | 55 | 69.0 |
| Fiber | 25,360–27,264 | 1905 | 634 | 593 | 37.6 | 579 | 38.5 |
| E4 ORF6/7 | 27,287–27,514c, 28,217–28,255c | 267 | 88 | 88 | 52.2 | 88 | 52.2 |
| E4 34K | 27,515–28,255 | 741 | 246 | 245 | 62.4 | 245 | 63.2 |
| E4 ORF3 | 28,258–28,662c | 405 | 134 | 141 | 41.0 | 141 | 41.0 |
| E4 ORF2 | 28,674–29,081c | 408 | 135 | 135 | 35.0 | 135 | 35.0 |
| E4 unknown2 | 29,164–29,292c | 129 | 42 | N.A. | N.A. | N.A. | N.A. |
| E4 unknown1 | 29,329–29,556c | 228 | 75 | N.A. | N.A. | N.A. | N.A. |
| ITR | 30,033–30,134 | 102 | N.A. | N.A. | N.A. | N.A. | N.A. |
| Year | Sample ID | Location | No. of Samples | ||
|---|---|---|---|---|---|
| Total | Positive | Positive Rate (%) | |||
| 2010 | ZFB10-01–ZFB10-47 | Kasanka National Park | 47 | 0 | 0 |
| ZFB10-48–ZFB10-52 | Ndola | 4 | 0 | 0 | |
| 2011 | ZFB11-01–ZFB11-38 | Ndola | 38 | 0 | 0 |
| ZFB11-39–ZFB11-95 | Kasanka National Park | 57 | 8 | 14.04 | |
| 2012 | ZFB12-01–ZFB12-60 | Ndola | 60 | 0 | 0 |
| ZFB12-61–ZFB12-110 * | Kasanka National Park | 49 | 0 | 0 | |
| 2013 | ZFB13-01–ZFB13-76 | Ndola | 76 | 0 | 0 |
| ZFB13-77–ZFB13-111 † | Kasanka National Park | 34 | 0 | 0 | |
| Total | 365 | 8 | 2.19 | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogawa, H.; Kajihara, M.; Nao, N.; Shigeno, A.; Fujikura, D.; Hang’ombe, B.M.; Mweene, A.S.; Mutemwa, A.; Squarre, D.; Yamada, M.; et al. Characterization of a Novel Bat Adenovirus Isolated from Straw-Colored Fruit Bat (Eidolon helvum). Viruses 2017, 9, 371. https://doi.org/10.3390/v9120371
Ogawa H, Kajihara M, Nao N, Shigeno A, Fujikura D, Hang’ombe BM, Mweene AS, Mutemwa A, Squarre D, Yamada M, et al. Characterization of a Novel Bat Adenovirus Isolated from Straw-Colored Fruit Bat (Eidolon helvum). Viruses. 2017; 9(12):371. https://doi.org/10.3390/v9120371
Chicago/Turabian StyleOgawa, Hirohito, Masahiro Kajihara, Naganori Nao, Asako Shigeno, Daisuke Fujikura, Bernard M. Hang’ombe, Aaron S. Mweene, Alisheke Mutemwa, David Squarre, Masao Yamada, and et al. 2017. "Characterization of a Novel Bat Adenovirus Isolated from Straw-Colored Fruit Bat (Eidolon helvum)" Viruses 9, no. 12: 371. https://doi.org/10.3390/v9120371
APA StyleOgawa, H., Kajihara, M., Nao, N., Shigeno, A., Fujikura, D., Hang’ombe, B. M., Mweene, A. S., Mutemwa, A., Squarre, D., Yamada, M., Higashi, H., Sawa, H., & Takada, A. (2017). Characterization of a Novel Bat Adenovirus Isolated from Straw-Colored Fruit Bat (Eidolon helvum). Viruses, 9(12), 371. https://doi.org/10.3390/v9120371








