ABC Assay: Method Development and Application to Quantify the Role of Three DWV Master Variants in Overwinter Colony Losses of European Honey Bees
Abstract
1. Introduction
2. Materials and Methods
2.1. Primer Design
2.2. Viral Master Variant Plasmid Standards
2.3. Primer Optimisation
2.4. Viral Master Variant cRNA Standards
2.5. Method Validation
2.6. Application of the RT-qPCR (ABC Assay)
2.7. HRM Assay
2.8. Sequencing
3. Results
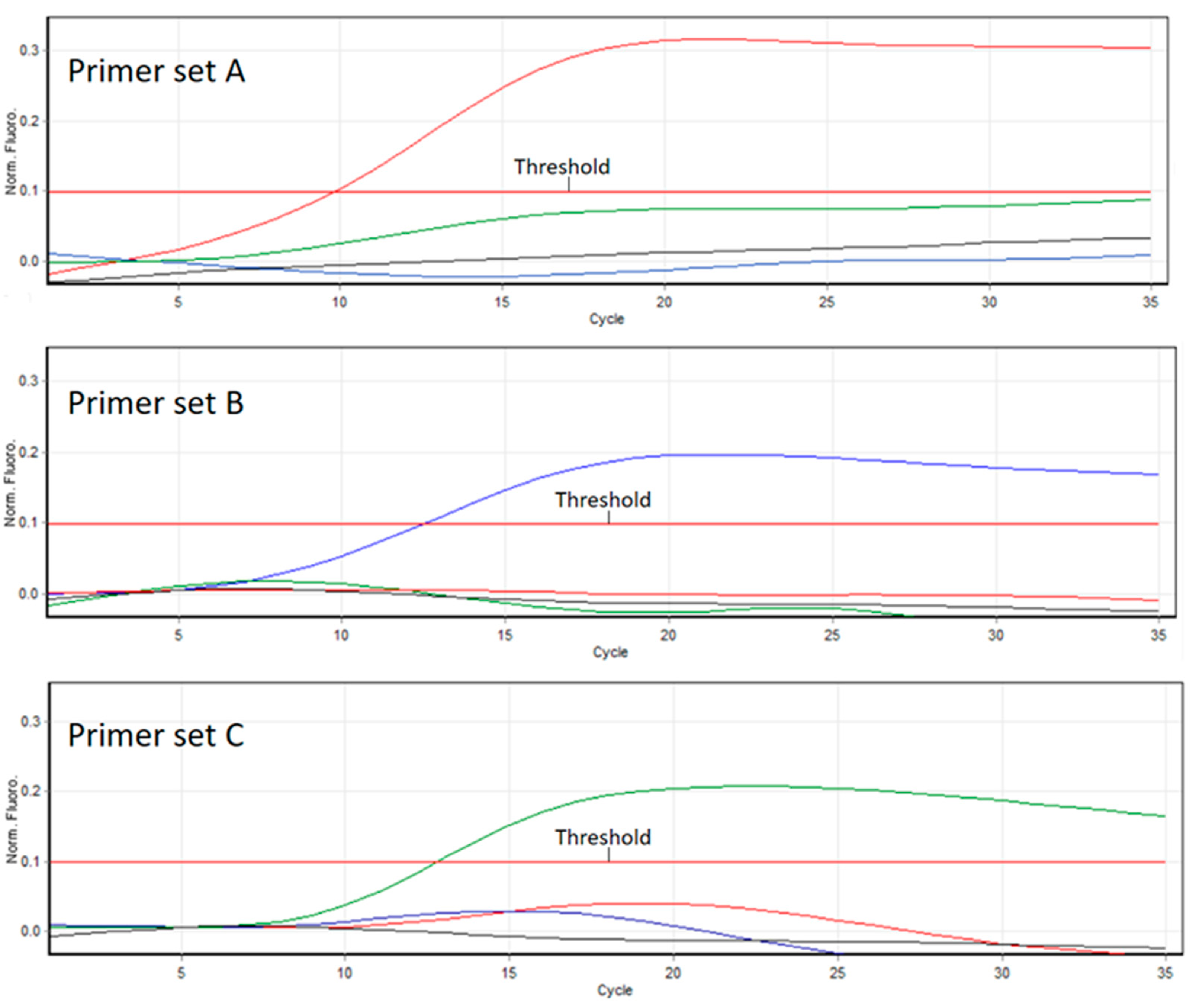
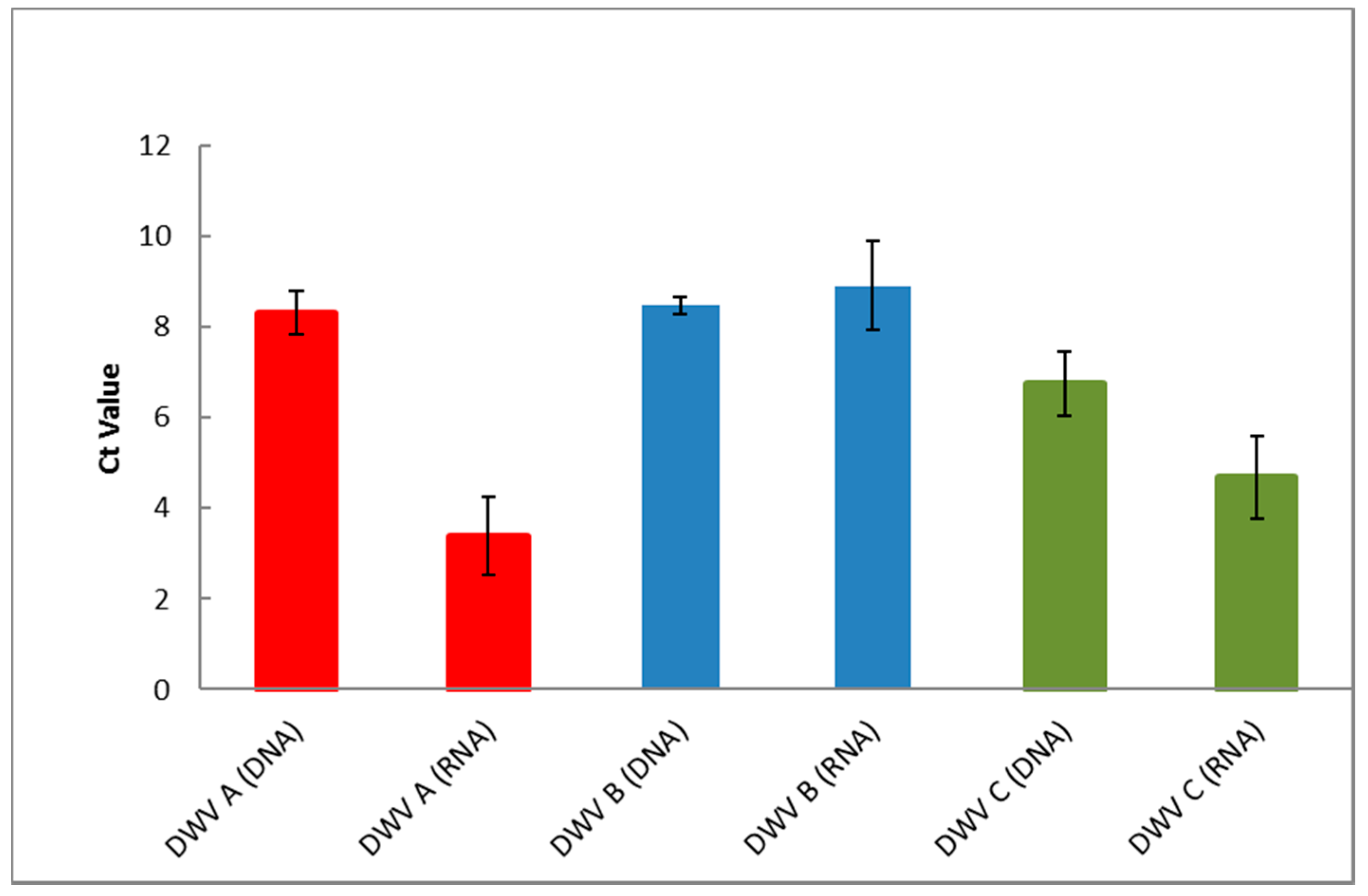
3.1. PCR and RT-PCR Sensitivities
3.2. Competitive PCR
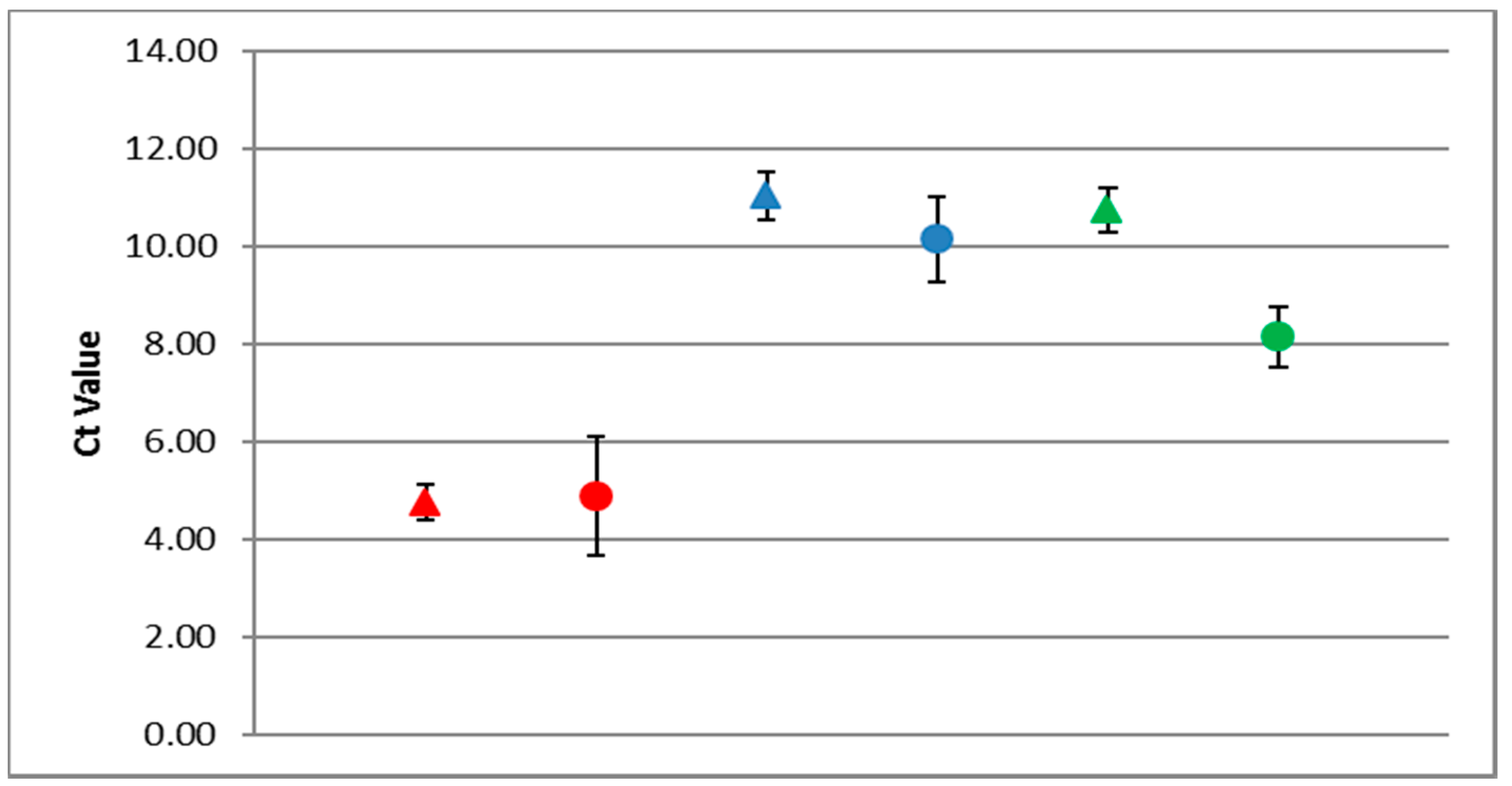
3.3. PCR and RT-PCR Efficiencies
3.4. Robustness of the RT-qPCR (ABC Assay)
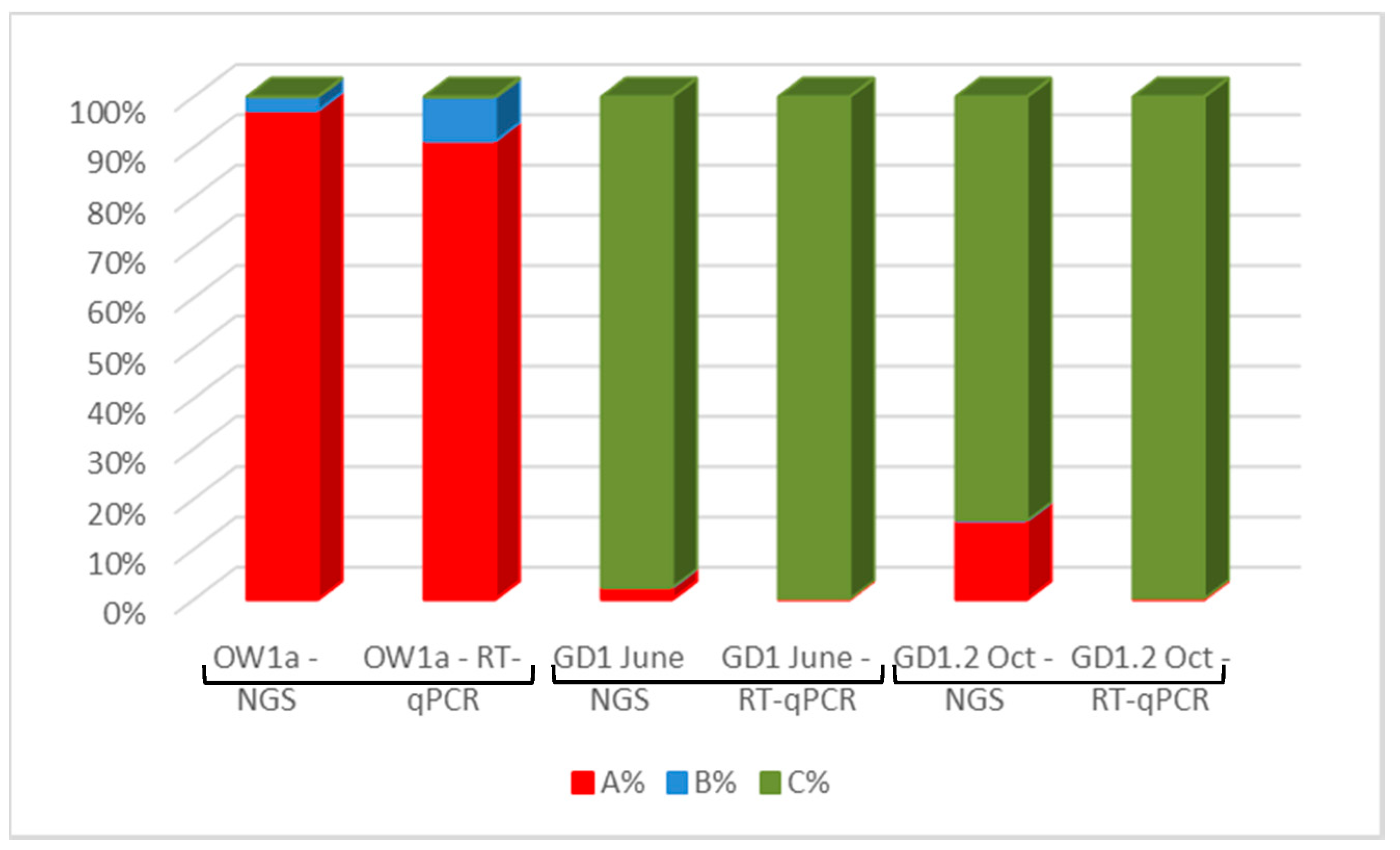
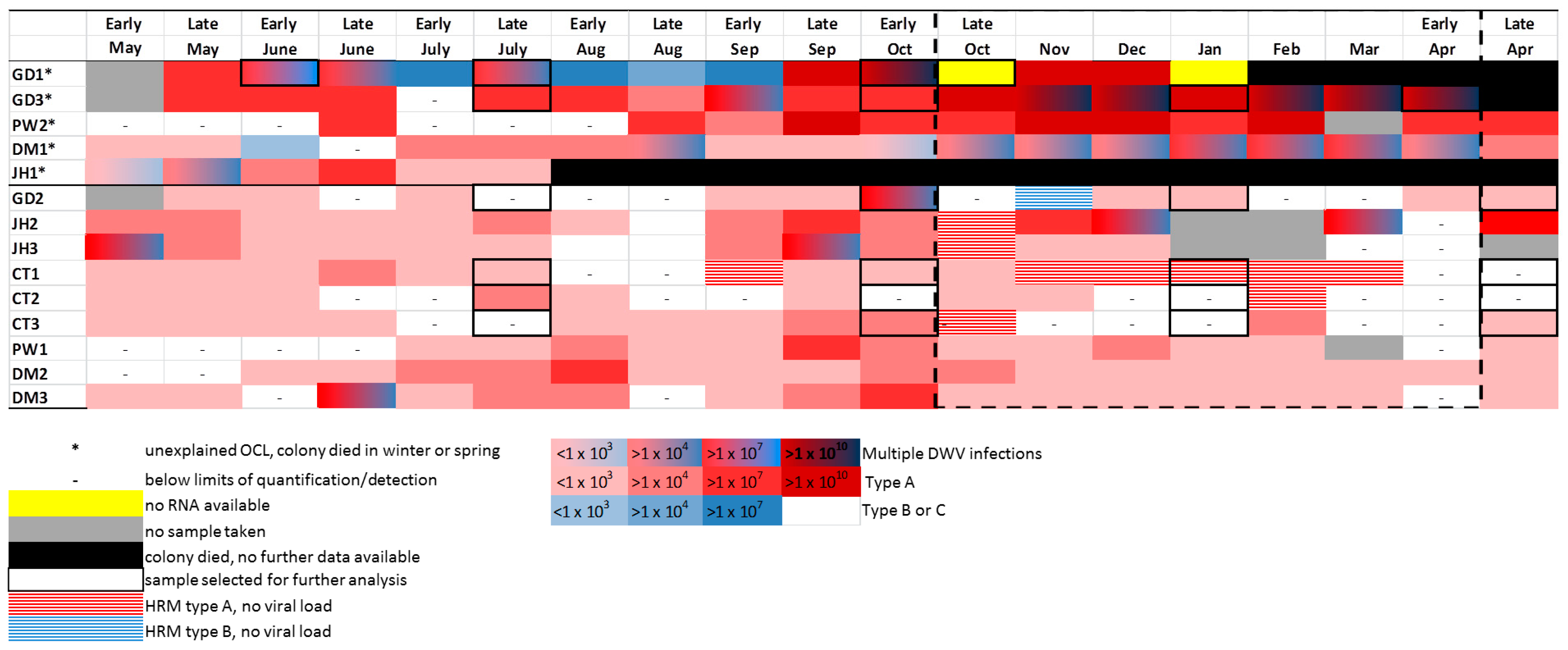
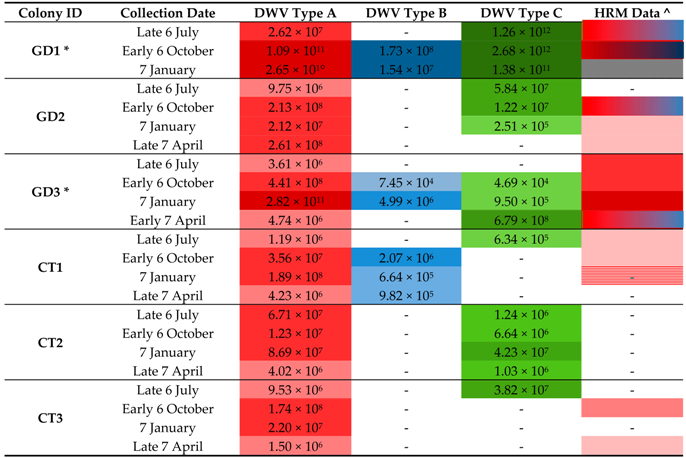
3.5. Detection of DWV in Honey Bees Using the ABC Assay
3.6. DWV Master Variants Implicated in OCL
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Domingo, E.; Holland, J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef] [PubMed]
- Biebricher, C.; Eigen, M. What is a quasispecies? In Quasispecies: Concept and Implications for Virology; Springer: Berlin/Heidelberg, Germany, 2006; pp. 1–31. [Google Scholar]
- Lauring, A.S.; Andino, R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010, 6, e1001005. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Sheldon, J.; Perales, C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 2012, 76, 159–216. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.C.; Schroeder, D.C. The use of RNA-dependent RNA polymerase for the taxonomic assignment of Picorna-like viruses (order Picornavirales) infecting Apis mellifera L. populations. Virol. J. 2008, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, J.R.; Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 2010, 103, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Pettis, J.S.; Corona, M.; Chen, W.P.; Li, C.J.; Spivak, M.; Visscher, P.K.; DeGrandi-Hoffman, G.; Boncristiani, H.; Zhao, Y.; et al. Israeli acute paralysis virus: Epidemiology, pathogenesis and implications for honey bee health. PLoS Pathog. 2014, 10, e1004261. [Google Scholar] [CrossRef] [PubMed]
- Evison, S.E.; Roberts, K.E.; Laurenson, L.; Pietravalle, S.; Hui, J.; Biesmeijer, J.C.; Smith, J.E.; Budge, G.; Hughes, W.O. Pervasiveness of parasites in pollinators. PLoS ONE 2012, 7, e30641. [Google Scholar] [CrossRef] [PubMed]
- Manley, R.; Boots, M.; Wilfert, L. Review: Emerging viral disease risk to pollinating insects: Ecological, evolutionary and anthropogenic factors. J. Appl. Ecol. 2015, 52, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Bowen-Walker, P.L.; Martin, S.J.; Gunn, A. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite varroa jacobsoni oud. J. Invertebr. Pathol. 1999, 73, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Dainat, B.; Ken, T.; Berthoud, H.; Neumann, P. The ectoparasitic mite Tropilaelaps mercedesae (Acari, Laelapidae) as a vector of honeybee viruses. Insects Soc. 2009, 56, 40–43. [Google Scholar] [CrossRef]
- Eyer, M.; Chen, Y.P.; Schäfer, M.O.; Pettis, J.; Neumann, P. Small hive beetle, Aethina tumida, as a potential biological vector of honeybee viruses. Apidologie 2009, 40, 419–428. [Google Scholar] [CrossRef]
- Martin, S.J.; Highfield, A.C.; Brettell, L.; Villalobos, E.M.; Budge, G.E.; Powell, M.; Nikaido, S.; Schroeder, D.C. Global honey bee viral landscape altered by a parasitic mite. Science 2012, 336, 1304–1306. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Wilfert, L.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Diversity in a honey bee pathogen: First report of a third master variant of the Deformed Wing Virus quasispecies. ISME J. 2016, 10, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014, 10, e1004230. [Google Scholar] [CrossRef] [PubMed]
- Lanzi, G.; de Miranda, J.R.; Boniotti, M.B.; Cameron, C.E.; Lavazza, A.; Capucci, L.; Camazine, S.M.; Rossi, C. Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L.). J. Virol. 2006, 80, 4998–5009. [Google Scholar] [CrossRef] [PubMed]
- Ongus, J.R.; Peters, D.; Bonmatin, J.M.; Bengsch, E.; Vlak, J.M.; van Oers, M.M. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 2004, 85, 3747–3755. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, D.C.; Martin, S.J. Deformed wing virus: The main suspect in unexplained honeybee deaths worldwide. Virulence 2012, 3, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M.; Nielsen, S.L.; Kryger, P. Varroa-virus interaction in collapsing honey bee colonies. PLoS ONE 2013, 8, e57540. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Brettell, L.E.; Martin, S.J.; Dixon, D.; Jones, I.M.; Schroeder, D.C. Superinfection exclusion and the long-term survival of honey bees in Varroa-infested colonies. ISME J. 2016, 10, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- McMahon, D.P.; Natsopoulou, M.E.; Doublet, V.; Fürst, M.; Weging, S.; Brown, M.J.; Gogol-Doring, A.; Paxton, R.J. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. R. Soc. B 2016, 283. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.; Jironkin, A.; Chandler, D.; Burroughs, N.; Evans, D.J.; Ryabov, E.V. Recombinants between Deformed wing virus and Varroa destructor virus-1 may prevail in Varroa destructor-infested honeybee colonies. J. Gen. Virol. 2011, 92, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Dalmon, A.; Desbiez, C.; Coulon, M.; Thomasson, M.; Le Conte, Y.; Alaux, C.; Vallon, J.; Moury, B. Evidence for positive selection and recombination hotspots in Deformed wing virus (DWV). Sci. Rep. 2017, 7, 41045. [Google Scholar] [CrossRef] [PubMed]
- Tentcheva, D.; Gauthier, L.; Jouve, S.; Canabady-Rochelle, L.; Dainat, B.; Cousserans, F.; Colin, M.; Ball, B.; Bergoin, M. Polymerase Chain Reaction detection of deformed wing virus (DWV) in Apis mellifera and Varroa destructor. Apidologie 2004, 35, 431–439. [Google Scholar] [CrossRef]
- Highfield, A.C.; El Nagar, A.; Mackinder, L.C.; Laure, M.L.N.; Hall, M.J.; Martin, S.J.; Schroeder, D.C. Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, L.; Tentcheva, D.; Tournaire, M.; Dainat, B.; Cousserans, F.; Colin, M.E.; Bergoin, M. Viral load estimation in asymptomatic honey bee colonies using the quantitative RT-PCR technique. Apidologie 2007, 38, 426–435. [Google Scholar] [CrossRef]
- Kukielka, D.; Esperón, F.; Higes, M.; Sánchez-Vizcaíno, J.M. A sensitive one-step real-time RT-PCR method for detection of deformed wing virus and black queen cell virus in honeybee Apis mellifera. J. Virol. Methods 2008, 147, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Genersch, E. Development of a rapid and sensitive RT-PCR method for the detection of deformed wing virus, a pathogen of the honeybee (Apis mellifera). Vet. J. 2005, 169, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Carreck, N.L.; Ball, B.V.; Martin, S.J. Honey bee colony collapse and changes in viral prevalence associated with Varroa destructor. J. Apic. Res. 2010, 49, 93–94. [Google Scholar] [CrossRef]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Dead or alive: Deformed wing virus and Varroa destructor reduce the life span of winter honeybees. Appl. Environ. Microbiol. 2011, 78, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Leong, D.T.; Gupta, A.; Bai, H.F.; Wan, G.; Yoong, L.F.; Too, H.P.; Chew, F.T.; Hutmacher, D.W. Absolute quantification of gene expression in biomaterials research using real-time PCR. Biomaterials 2007, 28, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Brettell, L.E.; Pachori, P.; Villalobos, E.M.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Moku virus; a new Iflavirus found in wasps, honey bees and Varroa. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Brettell, L.E.; Mordecai, G.J.; Schroeder, D.C.; Jones, I.M.; da Silva, J.R.; Vicente-Rubiano, M.; Martin, S.J. A comparison of deformed wing virus in deformed and asymptomatic honey bees. Insects 2017, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Brettell, L.E. The Spread and Evolution of RNA Viruses among Honey Bees and Wider Insect Communities with Particular Emphasis on Deformed Wing Virus (DWV), Ph.D. Thesis, University of Salford, Salford, UK, 2017. [Google Scholar]
- Evans, J.D.; Schwarz, R.S.; Chen, Y.P.; Budge, G.; Cornman, R.S.; de la Rua, P.; de Miranda, J.R.; Foret, S.; Foster, L.; Gauthier, L.; et al. Standard methods for molecular research in Apis mellifera. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef]
- Nassirpour, R.; Mathur, S.; Gosink, M.M.; Li, Y.; Shoieb, A.M.; Wood, J.; O’Neil, S.P.; Homer, B.L.; Whiteley, L.O. Identification of tubular injury microRNA biomarkers in urine: Comparison of next-generation sequencing and qPCR-based profiling platforms. BMC Genom. 2014, 15, 485. [Google Scholar] [CrossRef] [PubMed]
- Klein, D. Quantification using real-time PCR technology: Applications and limitations. Trends Mol. Med. 2002, 8, 257–260. [Google Scholar] [CrossRef]
- Dieffenbach, C.W.; Lowe, T.M.; Dveksler, G.S. General concepts for PCR primer design. PCR Methods Appl. 1993, 3, S30–S37. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.C.; Ryabov, E.V.; Prince, G.; Mead, A.; Zhang, C.; Baxter, L.A.; Pell, J.K.; Osborne, J.L.; Chandler, D. A strong immune response in young adult honeybees masks their increased susceptibility to infection compared to older bees. PLoS Pathog. 2012, 8, e1003083. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, H.; Imdorf, A.; Haueter, M.; Radloff, S.; Neumann, P. Virus infections and winter losses of honey bee colonies (Apis mellifera). J. Apic. Res. 2010, 49, 60–65. [Google Scholar] [CrossRef]
- Genersch, E.; Von Der Ohe, W.; Kaatz, H.; Schroeder, A.; Otten, C.; Büchler, R.; Berg, S.; Ritter, W.; Mühlen, W.; Gisder, S.; et al. The German bee monitoring project: A long term study to understand periodically high winter losses of honey bee colonies. Apidologie 2010, 41, 332–352. [Google Scholar] [CrossRef]
- Thompson, C.E.; Biesmeijer, J.C.; Allnutt, T.R.; Pietravalle, S.; Budge, G.E. Parasite pressures on feral honey bees (Apis mellifera sp.). PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]

| Target | Primer Name | Sequence (5′–3′) | Genome Target (NC004830.2) | Size of Product (bp) | Reference |
|---|---|---|---|---|---|
| DWV | DWVnew-F1 | TACTAGTGCTGGTTTTCCTTT | 8653–8673 | This study | |
| DWV Type A | DWVA-R1 | CTCATTAACTGTGTCGTTGAT | 8808–8788 | 155 | This study |
| DWV Type B | DWVB-R1 | CTCATTAACTGAGTTGTTGTC | 8808–8788 | 155 | This study |
| DWV Type C | DWVC-R1 | ATAAGTTGCGTGGTTGAC | 8805–8788 | 152 | This study |
| DWV q | DWVq-R1 | CTGTGTCGTTGATAATTGAATCTC | 8656–8676 | 145 | [25] |
| DWVq-F1 | TAGTGCTGGTTTTCCTTTGTC | 8800–8777 | |||
| M13 | M13F | GTAAAACGACGGCCA | Na | 361 | [25] |
| M13R | CAGGAAACAGCTATG | ||||
| Actin | ActinR1 | AAGAATTGACCCACCAATCCATAC | Na | 120 | [25] |
| ActinF1 | CCTGGAATCGCAGATAGAATGC |
| Target | Insert |
|---|---|
| DWV A | TATCTTGGAATACTAGTGCTGGTTTTCCTTTGTCTTCATTAAAGCCACCTGGAACATCAGGTAAGCGATGGTTGTTTGATATTGAGCTACAAGATTCGGGATGTTATCTCTTGCGTGGAATGCGTCCCGAACTTGAGATTCAATTATCAACGACACAGTTAATGAGGAAAAAGGGA |
| DWV B | TATCCTGGAATACTAGTGCTGGTTTTCCTTTATCTTCATTAAAACCGCCAGGCTCTTCTGGTAAGCGATGGTTGTTTGATATTGAATTACAAGATTCAGGATGTTATCTTTTGAGAGGGATGAGACCTGAACTTGAGATACAGTTGACAACAACTCAGTTAATGAGGAAGAAGGGA |
| DWV C | TTTCGTGGAATACTAGTGCTGGTTTTCCTTTATCCTCACTGAAACCAGCTGGAACATCAGGAAAAAGGTGGTTATTTGATATTGAATTGCAAGATTCGGGATGTTATCTTTTACGAGGTATGCGTCCCGAATTAGAAATACAATTGTCAACCACGCAACTTATGAGGAAAAAGGGA |
 : no RNA available;
: no RNA available;  : HRM type A, no viral load;
: HRM type A, no viral load;  : Multiple DWV variants;
: Multiple DWV variants;  : DWV type A;
: DWV type A;  : DWV type B (in ABC assay only);
: DWV type B (in ABC assay only);  : DWV type C.
: DWV type C.© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kevill, J.L.; Highfield, A.; Mordecai, G.J.; Martin, S.J.; Schroeder, D.C. ABC Assay: Method Development and Application to Quantify the Role of Three DWV Master Variants in Overwinter Colony Losses of European Honey Bees. Viruses 2017, 9, 314. https://doi.org/10.3390/v9110314
Kevill JL, Highfield A, Mordecai GJ, Martin SJ, Schroeder DC. ABC Assay: Method Development and Application to Quantify the Role of Three DWV Master Variants in Overwinter Colony Losses of European Honey Bees. Viruses. 2017; 9(11):314. https://doi.org/10.3390/v9110314
Chicago/Turabian StyleKevill, Jessica L., Andrea Highfield, Gideon J. Mordecai, Stephen J. Martin, and Declan C. Schroeder. 2017. "ABC Assay: Method Development and Application to Quantify the Role of Three DWV Master Variants in Overwinter Colony Losses of European Honey Bees" Viruses 9, no. 11: 314. https://doi.org/10.3390/v9110314
APA StyleKevill, J. L., Highfield, A., Mordecai, G. J., Martin, S. J., & Schroeder, D. C. (2017). ABC Assay: Method Development and Application to Quantify the Role of Three DWV Master Variants in Overwinter Colony Losses of European Honey Bees. Viruses, 9(11), 314. https://doi.org/10.3390/v9110314





