Binding Specificities of the Telomere Phage ϕKO2 Prophage Repressor CB and Lytic Repressor Cro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Phages, Plasmids, and Growth Conditions
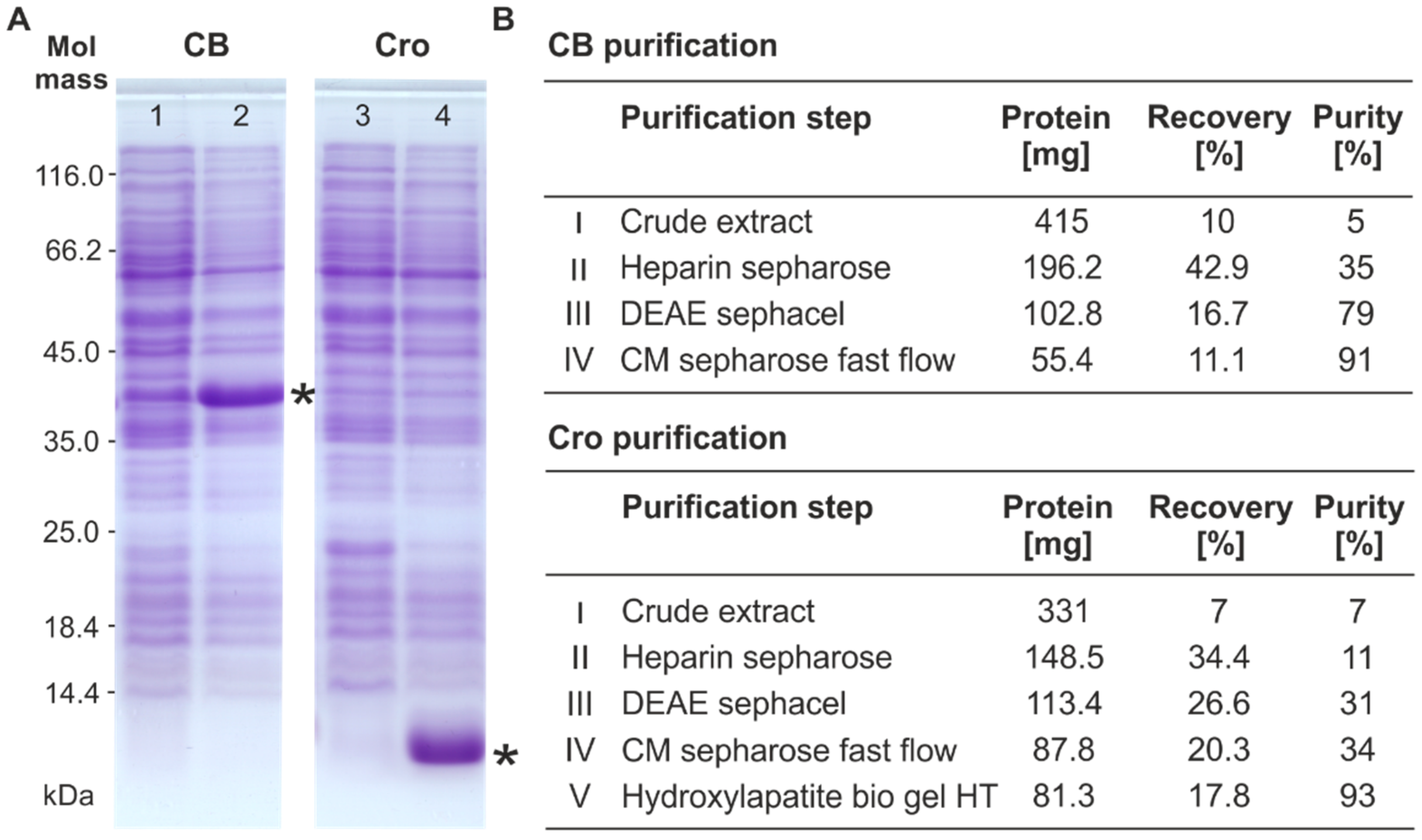
2.2. Overproduction and Purification of the Repressor Proteins
2.3. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Mass Spectrometric Analysis
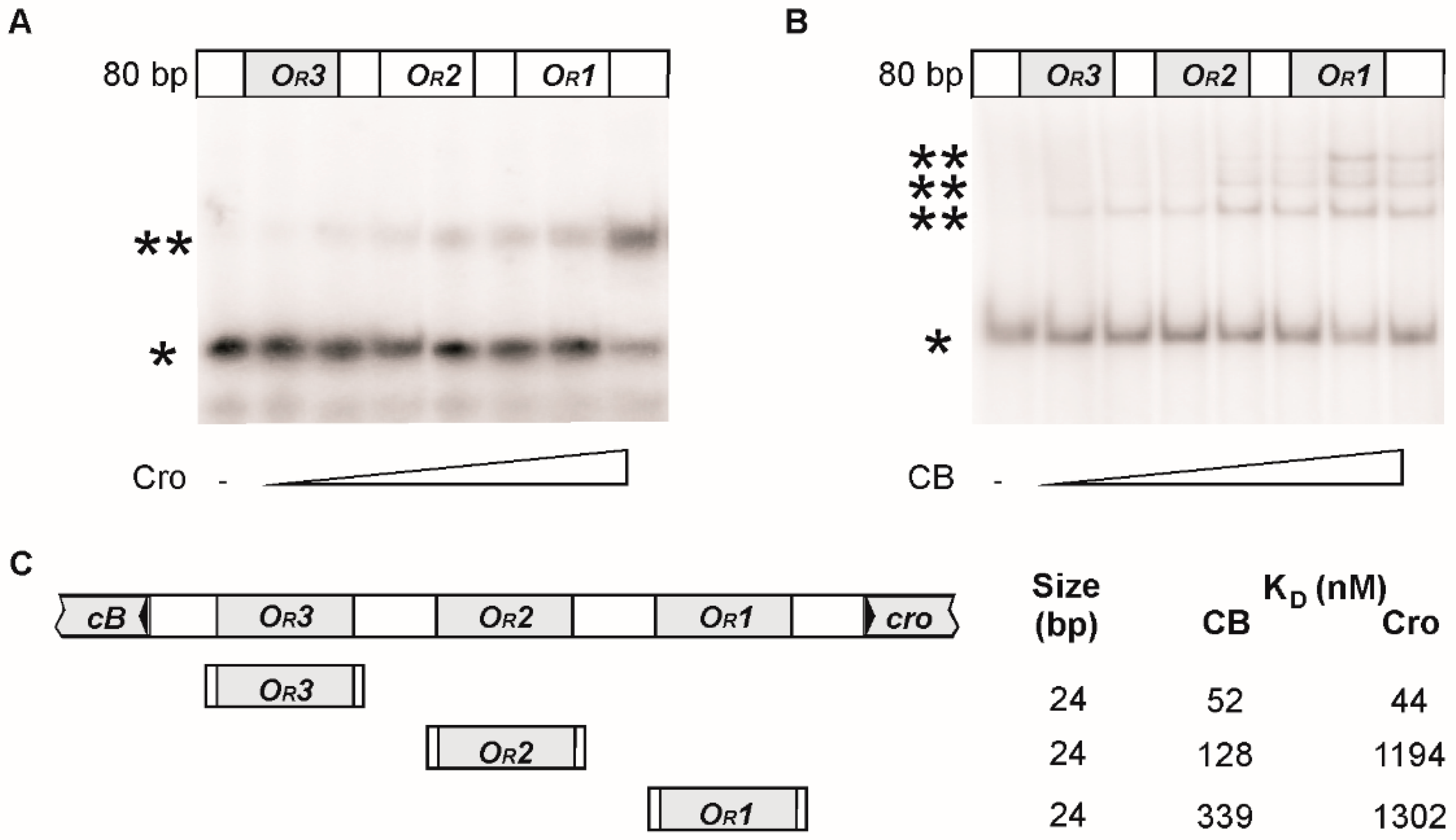
2.4. Electrophoretic Mobility Shift Assays (EMSA)
2.5. In Silico Analyses
2.6. Analysis of Promoter Activity
3. Results and Discussion
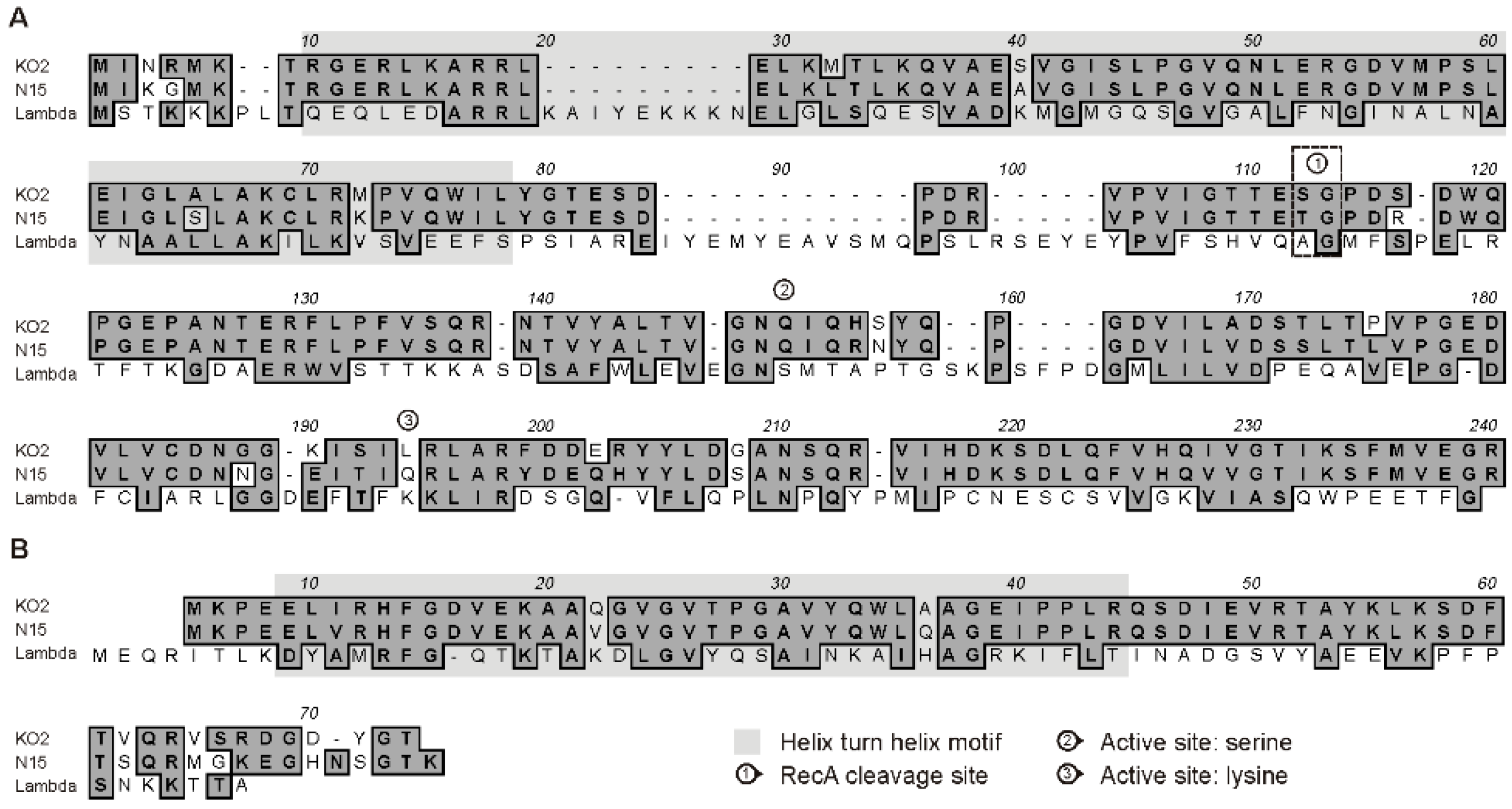
3.1. ϕKO2 and N15 Possess Closely Related immB Regions
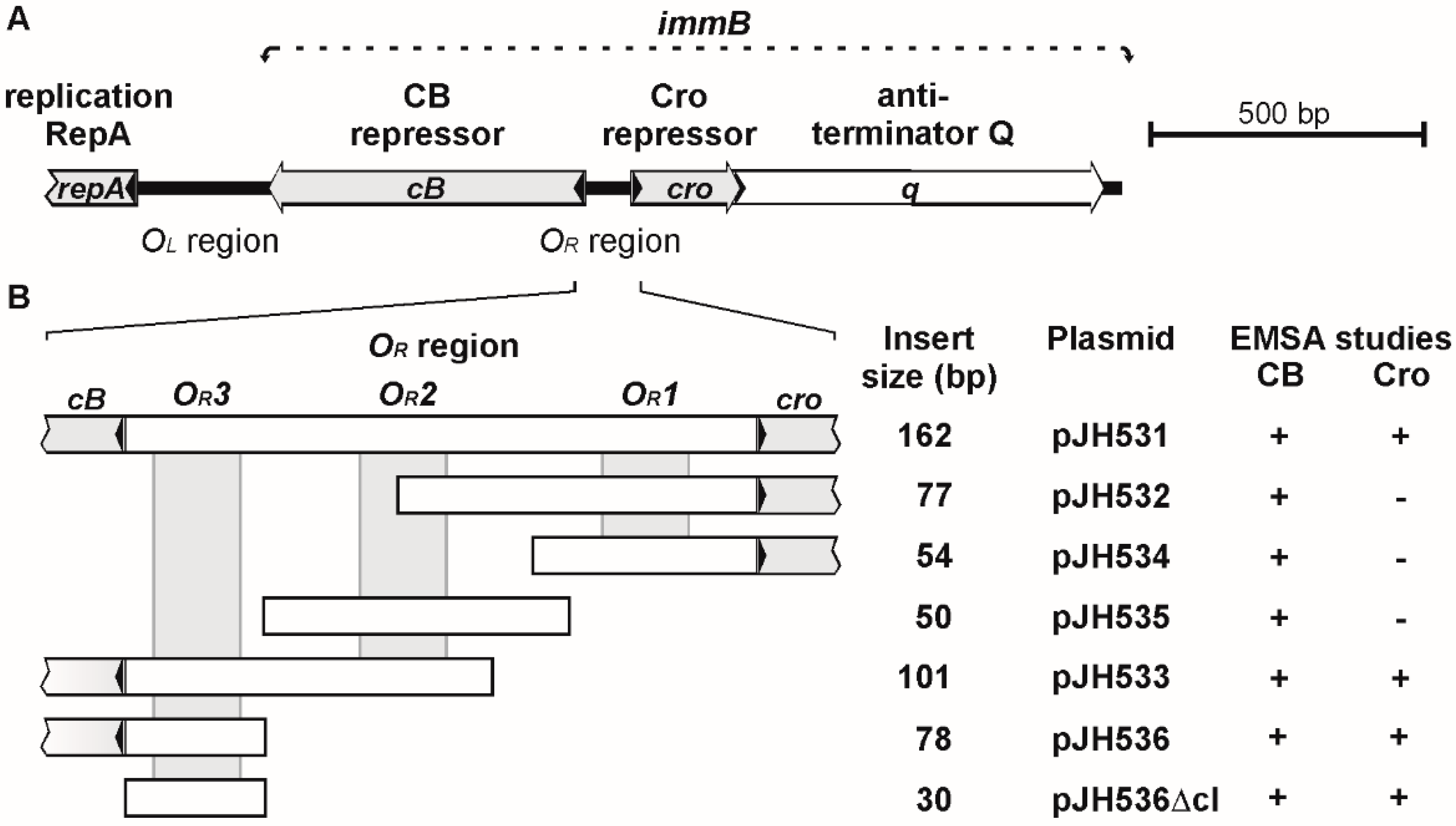
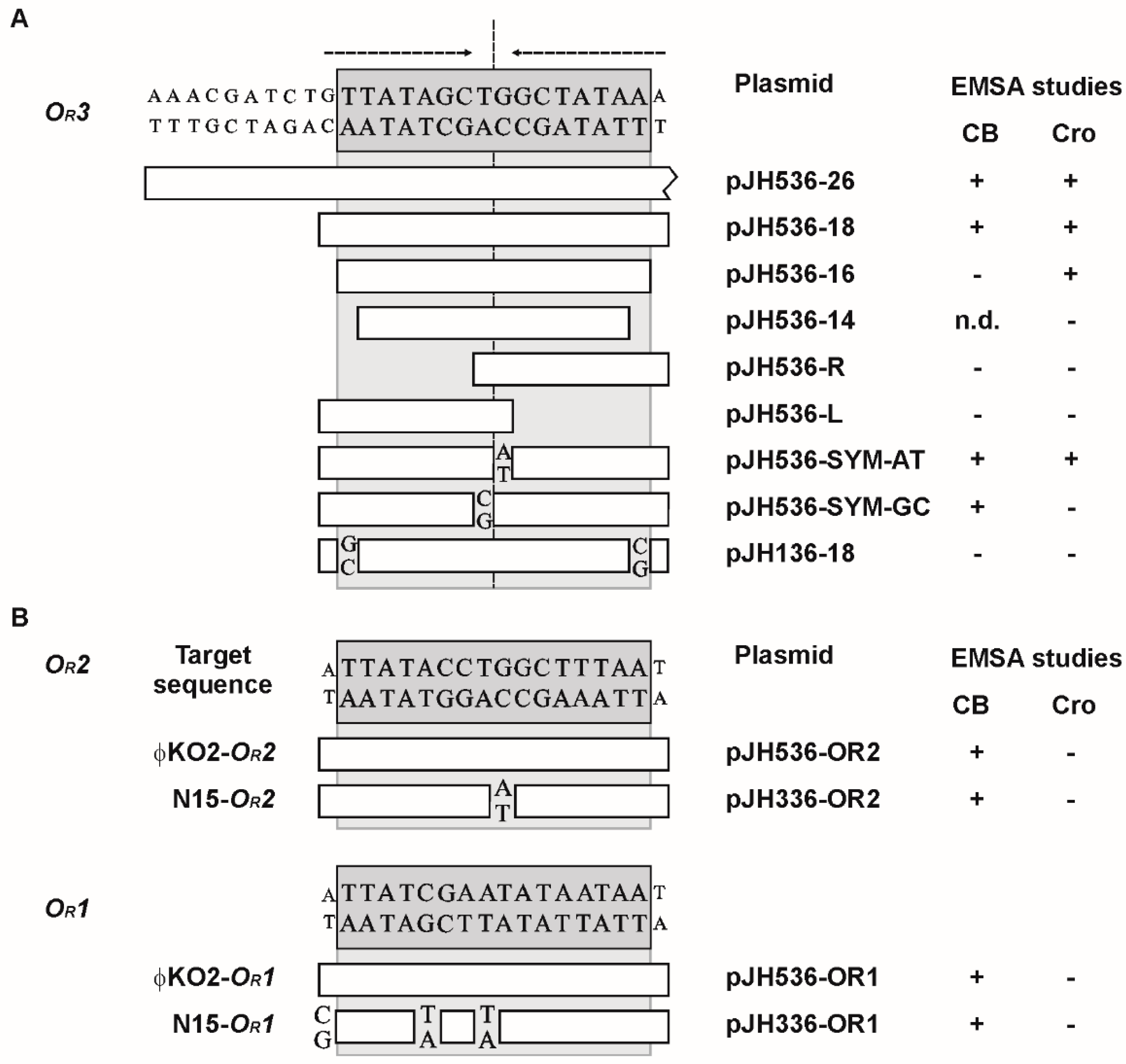
3.2. CB and Cro Do Not Bind to the Same Operators
3.3. Mutational Analysis of the Target Sites
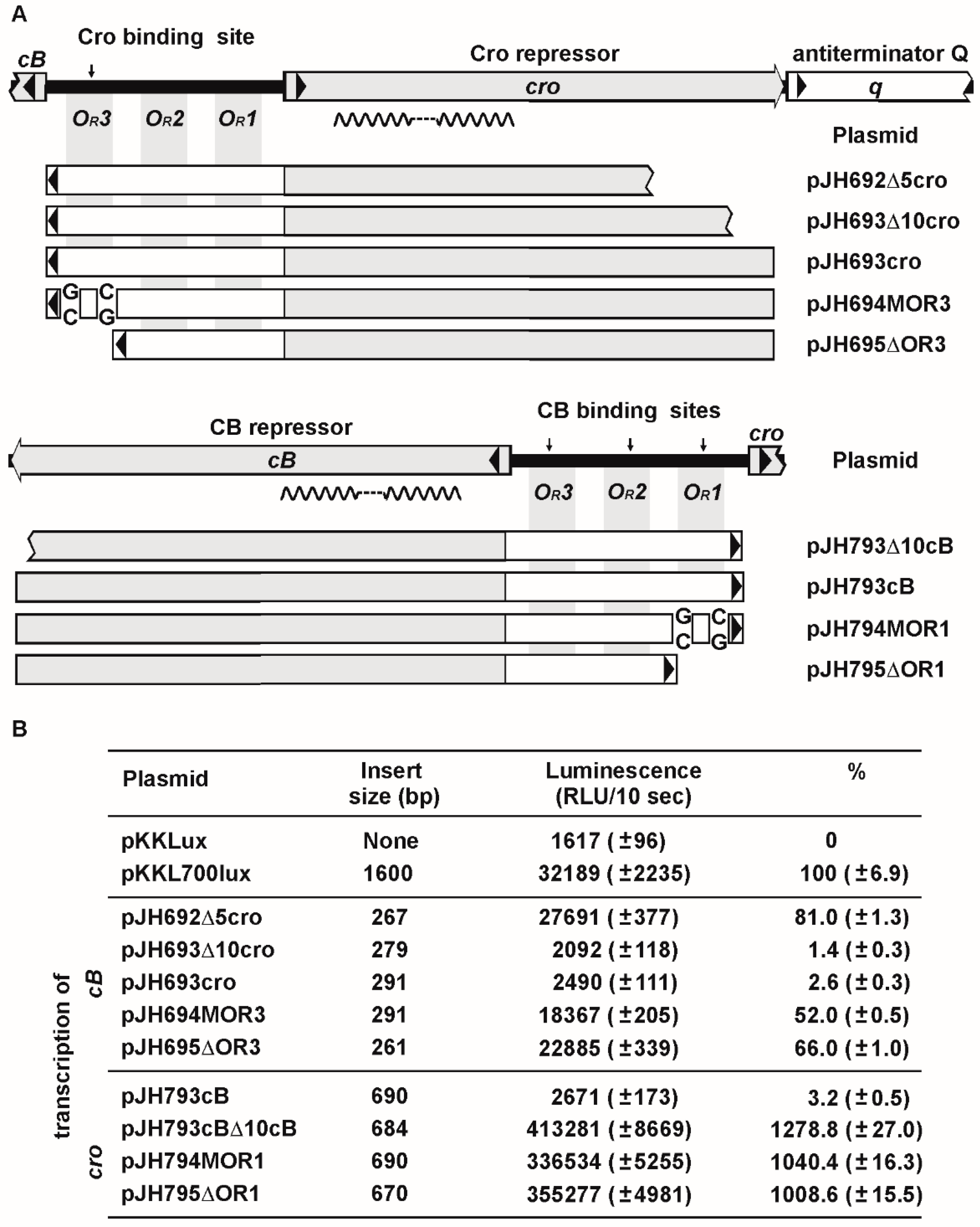
3.4. ϕKO2 Plasmid Prophage Replication Is Controlled by CB But Not by Cro
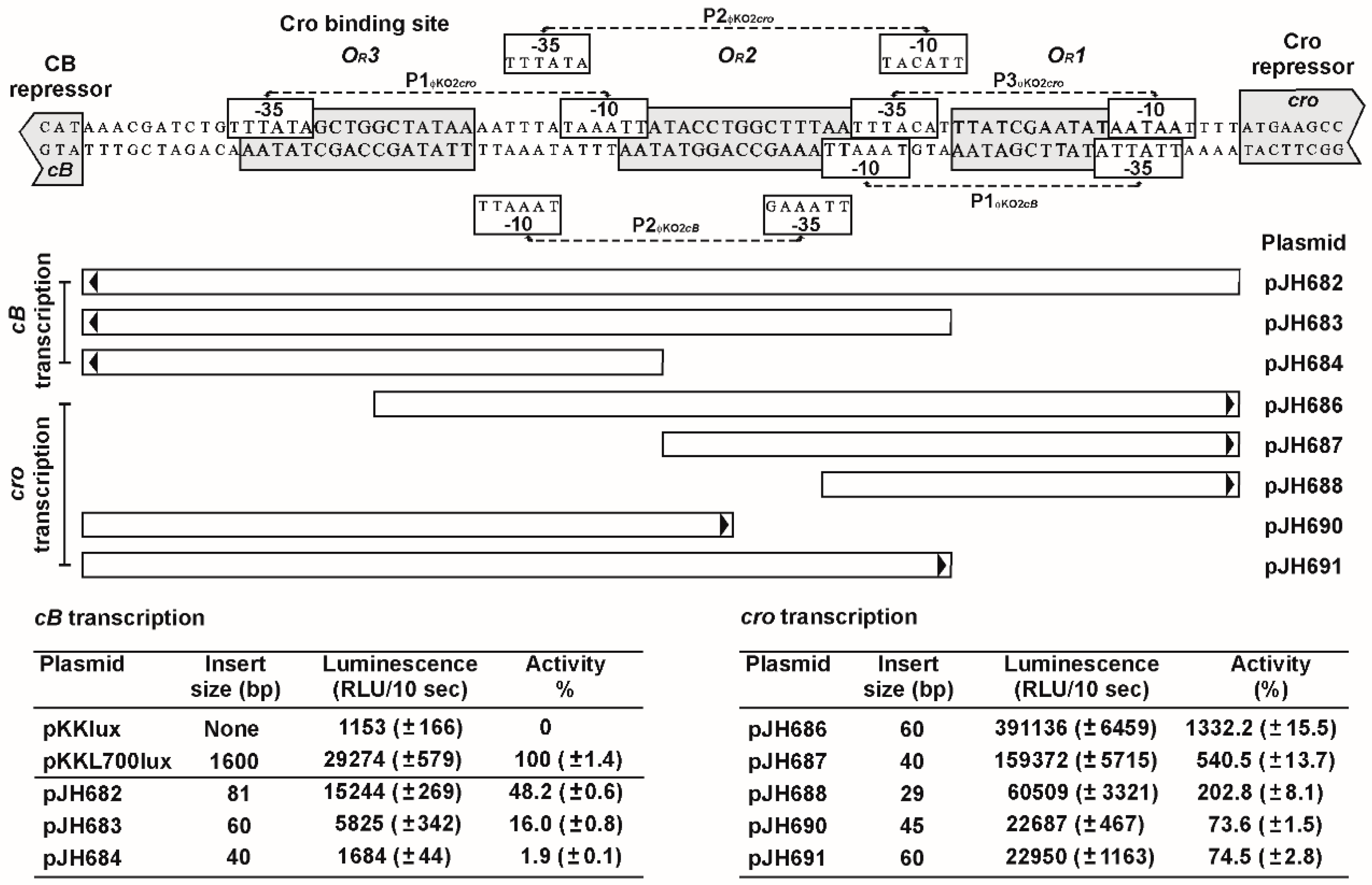
3.5. The OR Region Harbors Several Very Strong cro Promoters
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Casjens, S.R.; Gilcrease, E.B.; Huang, W.M.; Bunny, K.L.; Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Hatfull, G.F.; Hendrix, R.W. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 2004, 186, 1818–1832. [Google Scholar] [CrossRef] [PubMed]
- Hertwig, S.; Klein, I.; Schmidt, V.; Beck, S.; Hammerl, J.A.; Appel, B. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 2003, 331, 605–622. [Google Scholar] [CrossRef]
- Ravin, V.; Ravin, N.; Casjens, S.; Ford, M.E.; Hatfull, G.F.; Hendrix, R.W. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 2000, 299, 53–73. [Google Scholar] [CrossRef] [PubMed]
- Alanis Villa, A.; Kropinski, A.M.; Abbasifar, R.; Griffiths, M.W. Complete genome sequence of Vibrio parahaemolyticus bacteriophage vB_VpaM_MAR. J. Virol. 2012, 86, 13138–13149. [Google Scholar] [CrossRef] [PubMed]
- Lan, S.F.; Huang, C.H.; Chang, C.H.; Liao, W.C.; Lin, I.H.; Jian, W.N.; Wu, Y.G.; Chen, S.Y.; Wong, H.C. Characterization of a new plasmid-like prophage in a pandemic Vibrio parahaemolyticus O3:K6 strain. Appl. Environ. Microbiol. 2009, 75, 2659–2667. [Google Scholar] [CrossRef] [PubMed]
- Mobberley, J.M.; Authement, R.N.; Segall, A.M.; Paul, J.H. The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J. Virol. 2008, 82, 6618–6630. [Google Scholar] [CrossRef] [PubMed]
- Zabala, B.; Hammerl, J.A.; Espejo, R.T.; Hertwig, S. The linear plasmid prophage Vp58.5 of Vibrio parahaemolyticus is closely related to the integrating phage VHML and constitutes a new incompatibility group of telomere phages. J. Virol. 2009, 83, 9313–9320. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Jäckel, C.; Funk, E.; Pinnau, S.; Mache, C.; Hertwig, S. The diverse genetic switch of enterobacterial and marine telomere phages. Bacteriophage 2016, 6, e1148805. [Google Scholar] [CrossRef]
- Lobocka, M.B.; Svarchevsky, A.N.; Rybchin, V.N.; Yarmolinsky, M.B. Characterization of the primary immunity region of the Escherichia coli linear plasmid prophage N15. J. Bacteriol. 1996, 178, 2902–2910. [Google Scholar] [PubMed]
- Hammerl, J.A.; Roschanski, N.; Lurz, R.; Johne, R.; Lanka, E.; Hertwig, S. The molecular switch of telomere phages: High binding specificity of the PY54 Cro lytic repressor to a single operator site. Viruses 2015, 7, 2771–2793. [Google Scholar] [CrossRef] [PubMed]
- Dubrava, M.S.; Ingram, W.M.; Roberts, S.A.; Weichsel, A.; Montfort, W.R.; Cordes, M.H. N15 Cro and lambda Cro: Orthologous DNA-binding domains with completely different but equally effective homodimer interfaces. Protein Sci. 2008, 17, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Klein, I.; Appel, B.; Hertwig, S. Interplay between the temperate phages PY54 and N15, linear plasmid prophages with covalently closed ends. J. Bacteriol. 2007, 189, 8366–8370. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.M.; Lefevre, K.R.; Cordes, M.H. Sequence correlations between Cro recognition helices and cognate O(R) consensus half-sites suggest conserved rules of protein-DNA recognition. J. Mol. Biol. 2005, 350, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.F.; Russel, D.W. Molecular Cloning: A Laboratory Manual (3-Volume Set), 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [PubMed]
- Bertani, G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 2004, 186, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Slilaty, S.N.; Little, J.W. Lysine-156 and serine-119 are required for LexA repressor cleavage: A possible mechanism. Proc. Natl. Acad. Sci. USA 1987, 84, 3987–3991. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Jäckel, C.; Reetz, J.; Beck, S.; Alter, T.; Lurz, R.; Baretto, C.; Brüssow, H.; Hertwig, S. Campylobacter jejuni group III phage CP81 contains many T4-like genes without belonging to the T4-type phage group: Implications for the evolution of T4 phages. J. Virol. 2011, 85, 8597–8605. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Covarrubias, L.; Bolivar, F. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene 1982, 17, 79–89. [Google Scholar] [CrossRef]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Jacob, D.; Lewin, A.; Meister, B.; Appel, B. Plant-specific promoter sequences carry elements that are recognised by the eubacterial transcription machinery. Transgenic Res. 2002, 11, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Dodd, I.B.; Shearwin, K.E.; Perkins, A.J.; Burr, T.; Hochschild, A.; Egan, J.B. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004, 18, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Darling, P.J.; Holt, J.M.; Ackers, G.K. Coupled energetics of lambda cro repressor self-assembly and site-specific DNA operator binding II: Cooperative interactions of cro dimers. J. Mol. Biol. 2000, 302, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Darling, P.J.; Holt, J.M.; Ackers, G.K. Coupled energetics of lambda cro repressor self-assembly and site-specific DNA operator binding I: Analysis of cro dimerization from nanomolar to micromolar concentrations. Biochemistry 2000, 39, 11500–11507. [Google Scholar] [CrossRef] [PubMed]
- Jana, R.; Hazbun, T.R.; Mollah, A.K.; Mossing, M.C. A folded monomeric intermediate in the formation of lambda Cro dimer-DNA complexes. J. Mol. Biol. 1997, 273, 402–416. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammerl, J.A.; Jäckel, C.; Lanka, E.; Roschanski, N.; Hertwig, S. Binding Specificities of the Telomere Phage ϕKO2 Prophage Repressor CB and Lytic Repressor Cro. Viruses 2016, 8, 213. https://doi.org/10.3390/v8080213
Hammerl JA, Jäckel C, Lanka E, Roschanski N, Hertwig S. Binding Specificities of the Telomere Phage ϕKO2 Prophage Repressor CB and Lytic Repressor Cro. Viruses. 2016; 8(8):213. https://doi.org/10.3390/v8080213
Chicago/Turabian StyleHammerl, Jens Andre, Claudia Jäckel, Erich Lanka, Nicole Roschanski, and Stefan Hertwig. 2016. "Binding Specificities of the Telomere Phage ϕKO2 Prophage Repressor CB and Lytic Repressor Cro" Viruses 8, no. 8: 213. https://doi.org/10.3390/v8080213
APA StyleHammerl, J. A., Jäckel, C., Lanka, E., Roschanski, N., & Hertwig, S. (2016). Binding Specificities of the Telomere Phage ϕKO2 Prophage Repressor CB and Lytic Repressor Cro. Viruses, 8(8), 213. https://doi.org/10.3390/v8080213





