Badnaviruses: The Current Global Scenario
Abstract
:1. Introduction
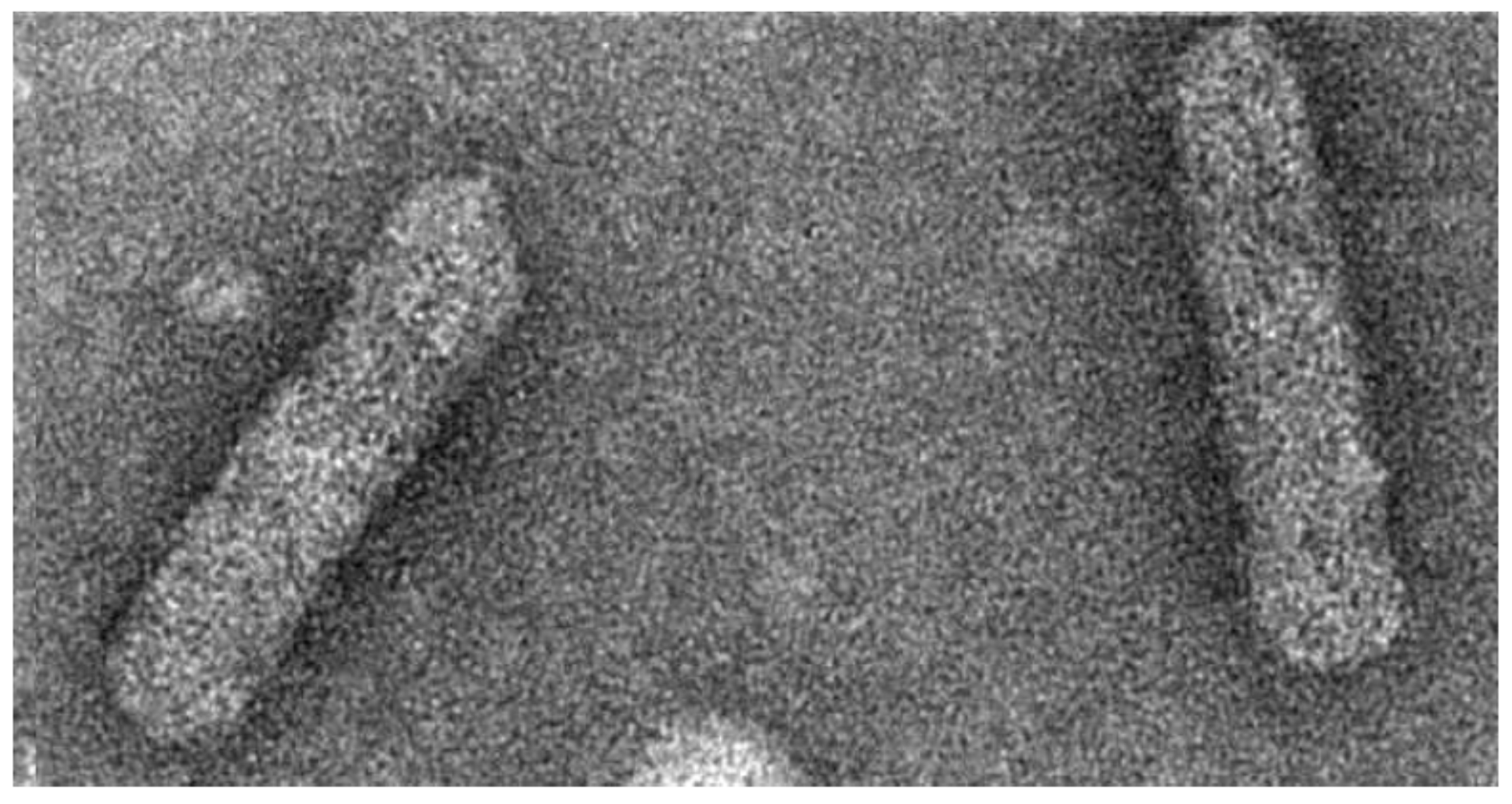
2. Symptomatology, Host Range, and Transmission
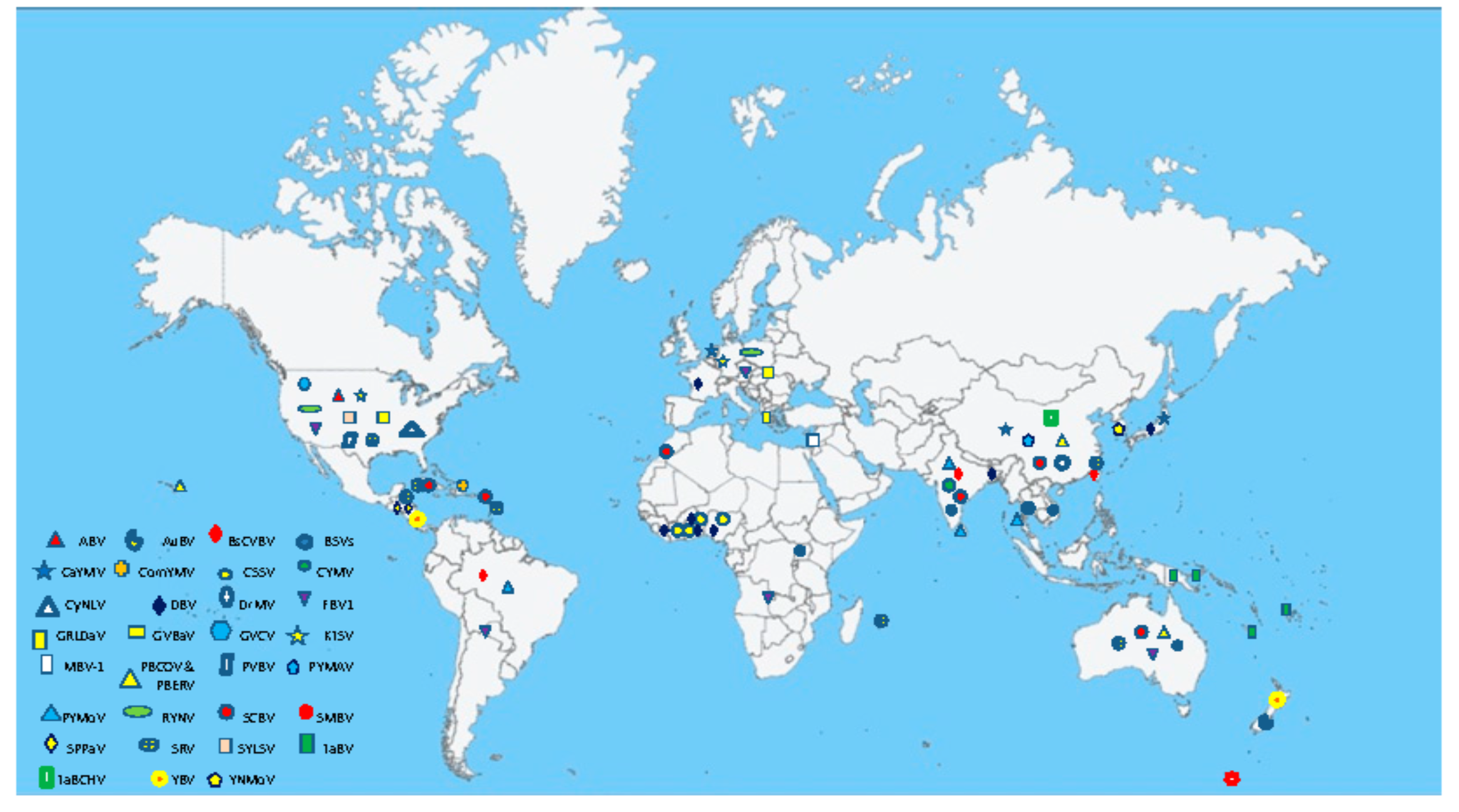
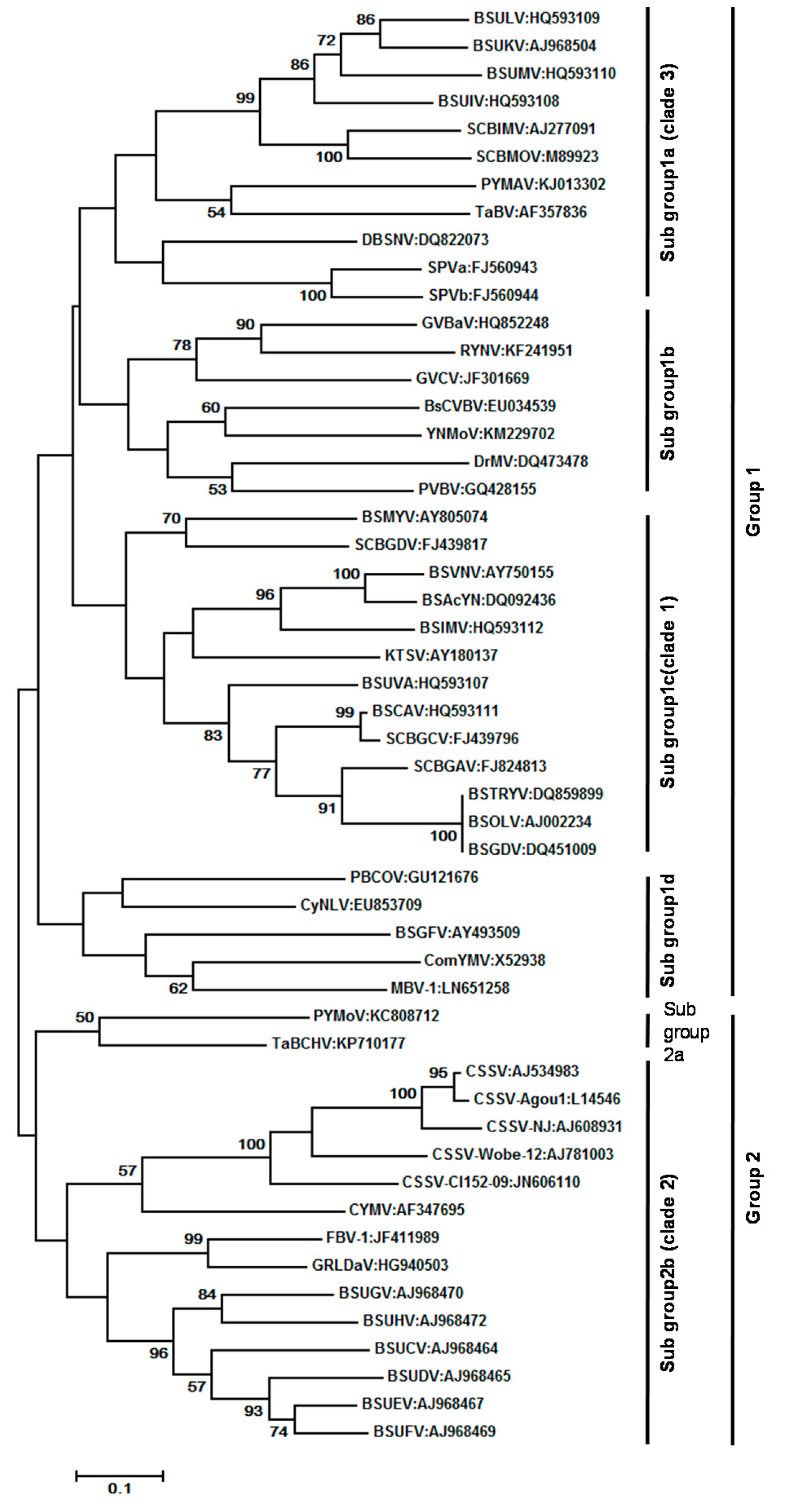
3. Geographical Distribution
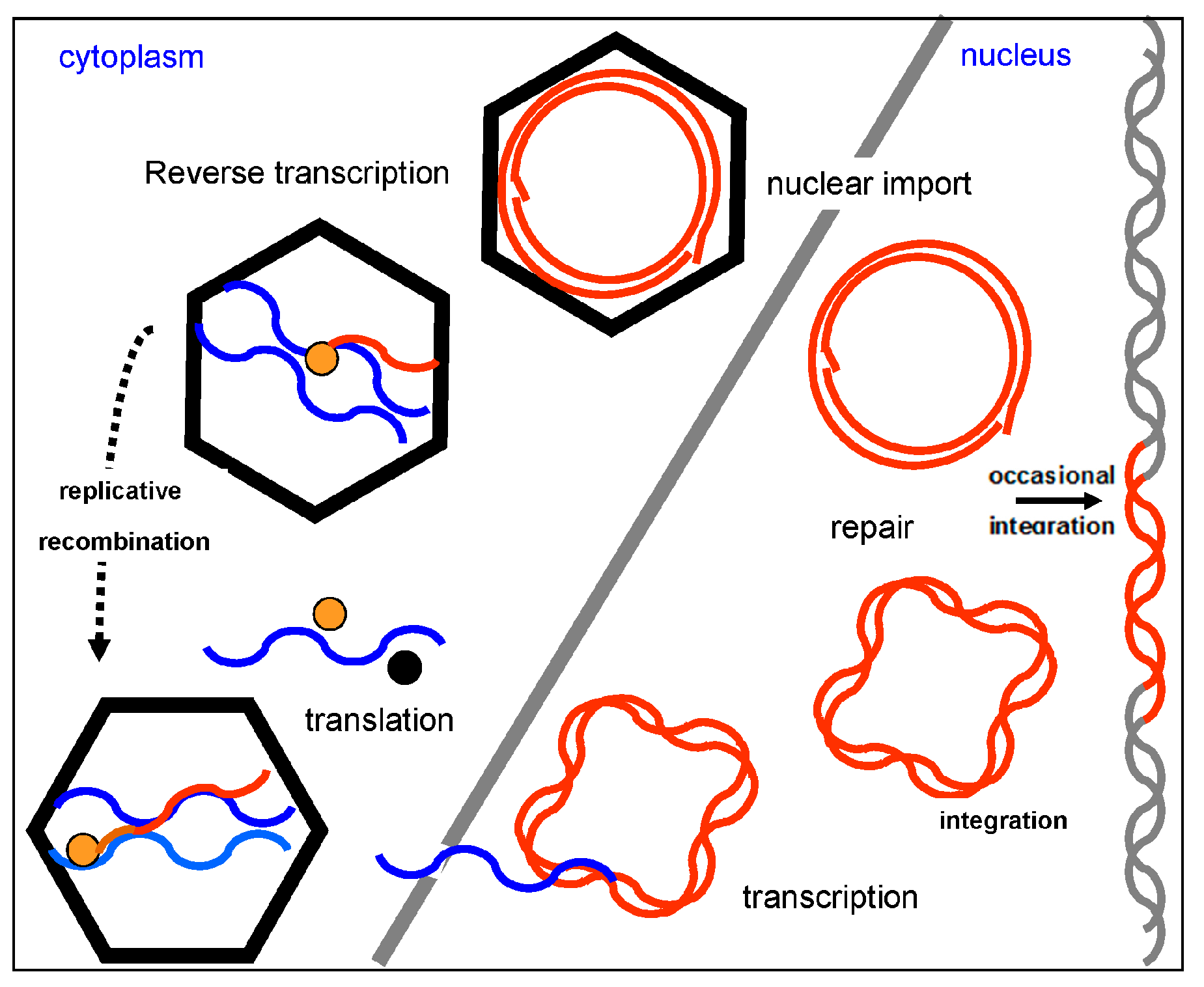
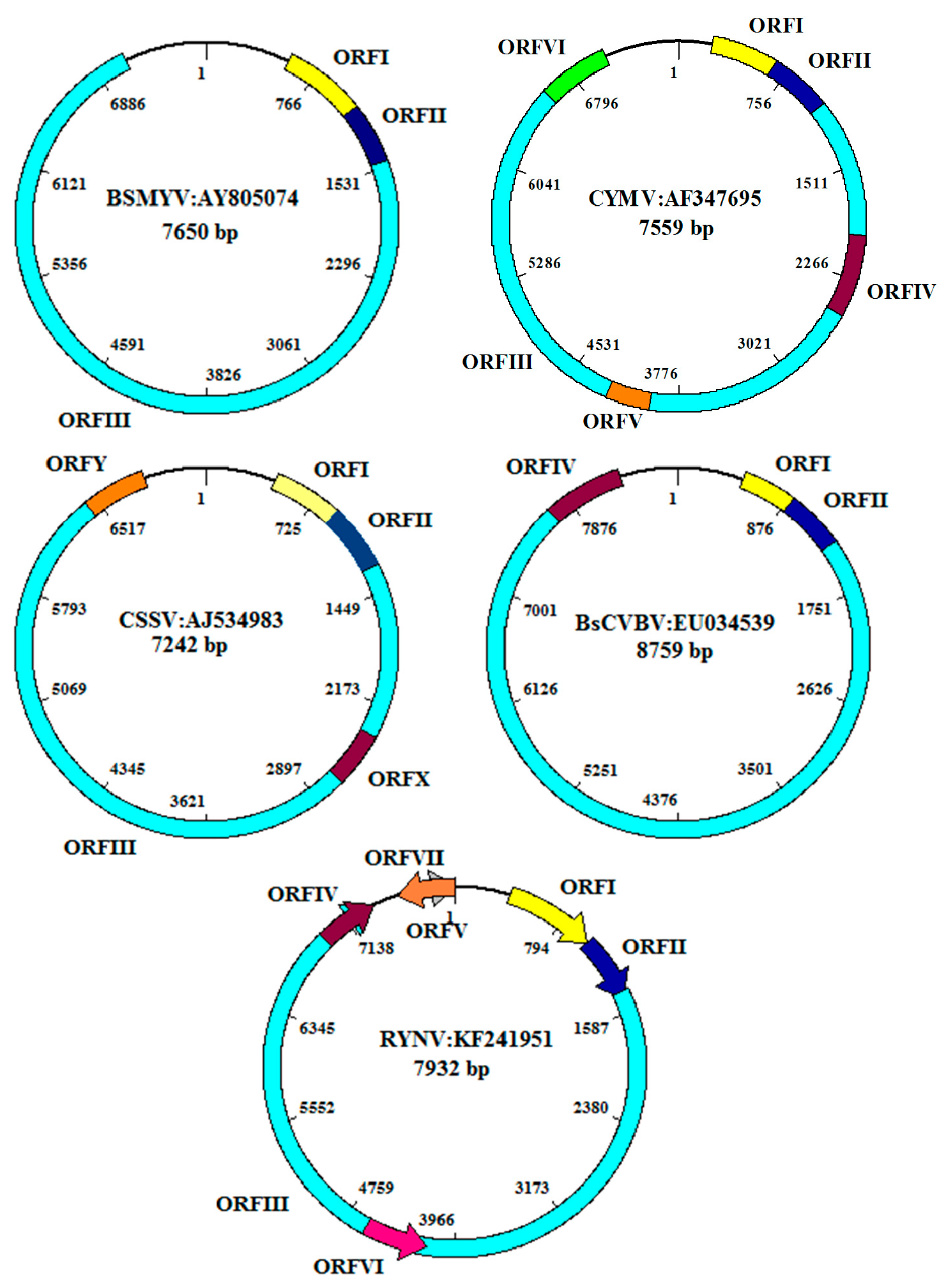
4. Genome Organization and Replication Cycle
5. Nuclear Integration of Badnaviruses (Endogenous Badnaviruses)
6. Diagnosis and Cure of Badnaviruses
7. Virus Characterization
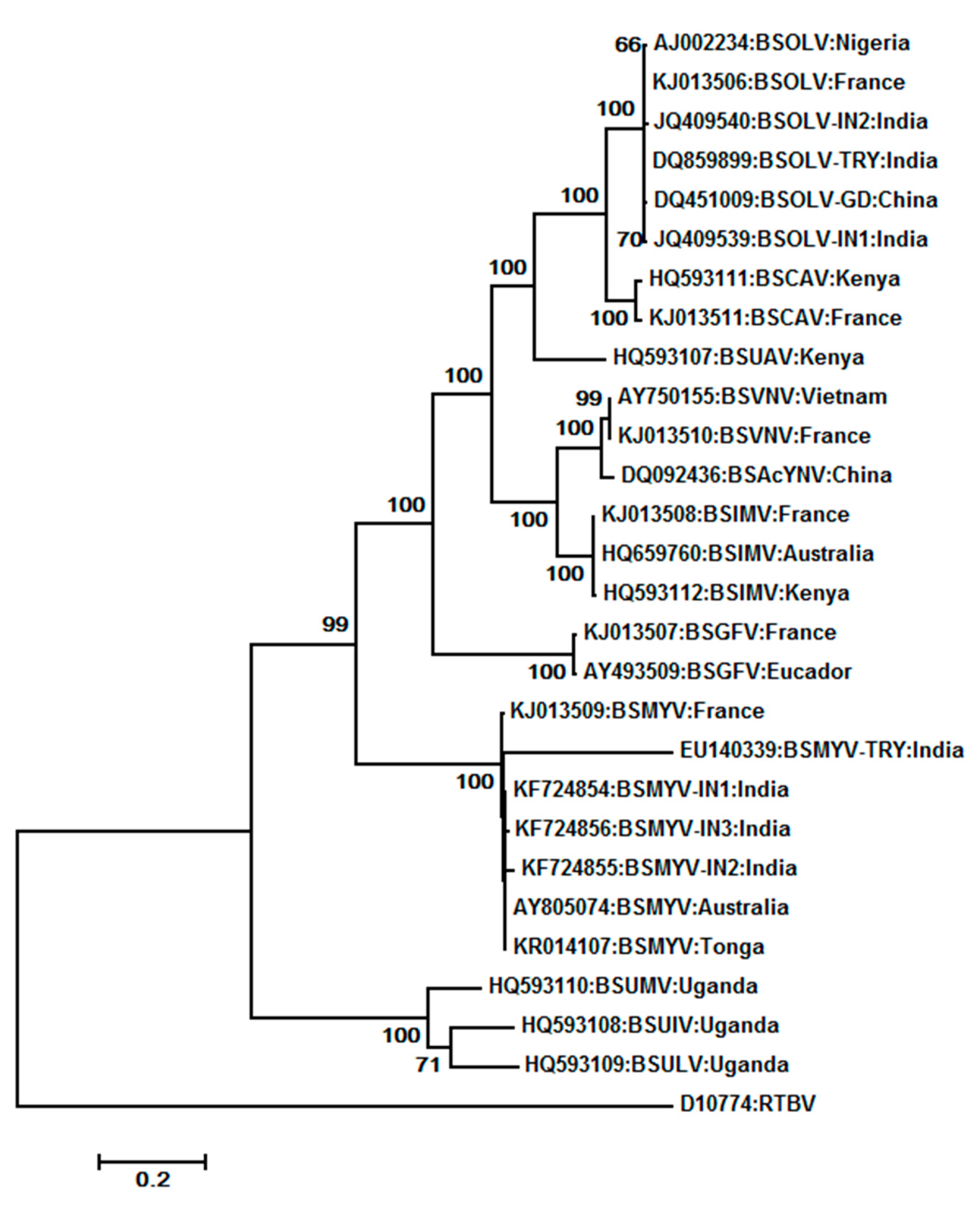
7.1. Banana streak virus (BSV) (BSGFV, BSIMV BSMYV, BSOLV, BSUAV, BSUIV, BSULV, BSUMV, and BSVNV)
7.2. Bougainvillea spectabilis chlorotic vein-banding virus (BsCVBV)
7.3. Cocoa swollen shoot virus (CSSV)
7.4. Canna yellow mottle virus (CaYMV)
7.5. Citrus yellow mosaic virus (CYMV)
7.6. Commelina yellow mottle virus (ComYMV)
7.7. Dioscorea bacilliform virus (DBV) (DBALV and DBSNV)
7.8. Fig badnavirus 1 (FBV-1)
7.9. Gooseberry vein banding associated virus (GVBaV)
7.10. Grapevine vein clearing virus (GVCV)
7.11. Kalanchoë top-spotting virus (KTSV)
7.12. Pagoda yellow mosaic associated virus (PYMAV)
7.13. Pineapple bacilliform comosus virus (PBCOV) and Pineapple bacilliform erectifolius virus (PBERV)
7.14. Piper yellow mottle virus (PYMoV)
7.15. Rubus yellow net virus (RYNV)
7.16. Schefflera ringspot virus (SRV)
7.17. Spiraea yellow leaf spot virus (SLSV)
7.18. Sugarcane bacilliform virus (SCBV) (SCBIMV and SCBMOV)
7.19. Sweet potato pakakuy virus (Sweet potato badnavirus a + Sweet potato badnavirus b)
7.20. Taro bacilliform virus (TaBV)
8. Putative Additional Species
8.1. Ambrosia asymptomatic virus 2 and Ambrosia asymptomatic virus 4
8.2. Aucuba bacilliform virus (AuBV)
8.3. Cycad necrotic leafspot virus (CyLNV)
8.4. Dracaena mottle virus (DrMV)
8.5. Grapevine Roditis leaf discoloration-associated virus (GRLDaV)
8.6. Mulberry badnavirus 1 (MBV-1)
8.7. Red clover bacilliform virus (RCBV)
8.8. Stilbocarpa mosaic bacilliform virus (SMBV)
8.9. Yacon necrotic mottle virus (YNMoV)
8.10. Yucca bacilliform virus (YBV)
9. Conclusions and Future Research Needs
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jakowitsch, J.; Mette, M.F.; van der Winden, J.; Matzke, M.A.; Matzke, A.J.M. Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proc. Natl. Acad. Sci. USA 1999, 96, 13241–13246. [Google Scholar] [PubMed]
- LaFleur, D.A.; Lockhart, B.E.L.; Olszewski, N.E. Portions of the banana streak badnavirus genome are integrated in the genome of its host Musa sp. Phytopathology 1996, 86, S100. [Google Scholar]
- Richert-Poggeler, K.R.; Shepherd, R.J. Petunia vein clearing virus: A plant pararetrovirus with the core sequence of an integrase function. Virology 1997, 236, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Kunii, M.; Kanda, M.; Nagano, H.; Uyeda, I.; Kishima, Y.; Sano, Y. Reconstruction of putative DNA virus from endogenous rice tungro bacilliform virus-like sequences in the rice genome: Implications for integration and evolution. BMC Genom. 2004, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.N.; Harper, G.; Heslop-Harrison, J.S. Characterization of pararetrovirus-like sequences in the genome of potato (Solanum tuberosum). Cytogenet. Genome Res. 2005, 110, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Staginnus, C.; Richert-Poggeler, K.R. Endogenous pararetroviruses: Two-faced travelers in the plant genome. Trends Plant Sci. 2006, 11, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Geering, A.D.W.; Maumus, F.; Copetti, D.; Choisne, N.; Zwickl, D.J.; Zytnicki, M.; McTaggart, A.R.; Scalabrin, S.; Vezzulli, S.; Wing, R.A.; et al. Endogenous florendoviruses are major components of plant genomes and hallmarks of virus evolution. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Chabannes, M.; Baurens, F.-C.; Duroy, P.-O.; Bocs, S.; Vernerey, M.S.; Rodier-Goud, M.; Barbe, V.; Gayral, P.; Iskra-Caruana, M.-L. Three infectious viral species lying in wait in the banana genome. J. Virol. 2013, 87, 8624–8637. [Google Scholar] [CrossRef] [PubMed]
- Borah, B.K.; Sharma, S.; Kant, R.; Anthony-Johnson, A.M.; Saigopal, D.V.R.; Dasgupta, I. Bacilliform DNA-containing plant viruses in the tropics: Commonalities within a genetically diverse group. Mol. Plant Pathol. 2013, 14, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, B.E.L.; Olsweski, N.E. Plant virus badnavirus group. In The Encyclopedia of Virology; Webster, R.G., Granoff, A., Eds.; Academic Press: New York, NY, USA, 1994; Volume 1, pp. 139–143. [Google Scholar]
- Medberry, S.L.; Lockhart, B.E.; Olszewski, N.E. Properties of Commelina yellow mottle virus’s complete DNA sequence, genomic discontinuities and transcript suggest that it is a pararetrovirus. Nucleic Acids Res. 1990, 18, 5505–5513. [Google Scholar] [CrossRef] [PubMed]
- The Springer Index of Viruses Online. Available online: http://oesys.springer.de/viruses/database/login.shtm (accessed on 24 February 2016).
- Hohn, T.; Richert-Poggeler, K.R.; Harper, G.; Schawarzacher, T.; Teo, C.; Teycheney, P.-Y.; Iskra-Caruana, M.-L.; Hull, R. Evolution of integrated plant viruses. In Plant Virus Evolution; Roosinck, M., Ed.; Academic Springer: Heidelberg, Germany, 2008; pp. 54–76. [Google Scholar]
- Staginnus, C.; Iskra-Caruana, M.; Lockhart, B.; Hohn, T.; Richert-Pöggeler, K.R. Suggestions for a nomenclature of endogenous pararetroviral sequences in plants. Arch. Virol. 2009, 154, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Côte, F.X.; Galzi, S.; Folliot, M.; Lamagnère, Y.; Teycheney, P.-Y.; Iskra-Caruana, M.-L. Micropropagation by tissue culture triggers differential expression of infectious endogenous Banana streak virus sequences (eBSV) present in the B genome of natural and synthetic interspecific banana plantains. Mol. Plant Pathol. 2010, 11, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Dallot, S.; Accuna, P.; Rivera, C.; Ramirez, P.; Cote, F.; Lockhart, B.E.L.; Caruana, M.-L. Evidence that the proliferation stage of micropropagation procedure is determinant in the expression of Banana streak virus integrated into the genome of the FHIA21 hybrid (Musa AAAB). Arch. Virol. 2001, 146, 2179–2190. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, F.; Carreel, F.; Jenny, C.; Lockhart, B.E.L.; Iskra-Caruana, M.-L. Identification of genetic markers linked to banana streak disease expression in inter-specific Musa hybrids. Theor. Appl. Genet. 2003, 106, 594–598. [Google Scholar] [PubMed]
- Hearon, S.S.; Locke, J.C. Graft, pollen, and seed transmission of an agent associated with top spotting in Kalanchoë blossfeldiana. Plant Dis. 1984, 68, 347–350. [Google Scholar] [CrossRef]
- Quainoo, A.K.; Wetten, A.C.; Allainguillaume, J. Transmission of cocoa swollen shoot virus by seeds. J. Virol. Methods 2008, 150, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Hareesh, P.S.; Bhat, A.I. Seed transmission of Piper yellow mottle virus in black pepper (Piper nigrum L.). J. Plant. Crops 2010, 38, 62–65. [Google Scholar]
- Deeshma, K.P.; Bhat, A.I. Further evidence of true seed transmission of Piper yellow mottle virus in black pepper (Piper nigrum L.). J. Plant. Crops 2014, 42, 289–293. [Google Scholar]
- Devitt, L.; Ebenebe, A.; Gregory, H.; Harding, R.; Hunter, D.; Macanawai, A. Investigations into the seed and mealybug transmission of Taro bacilliform virus. Aust. Plant Pathol. 2005, 34, 73–76. [Google Scholar]
- Hohn, T.; Rothnie, H. Plant pararetroviruses: Replication and expression. Curr. Opin. Virol. 2013, 3, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Hull, R. Matthew’s Plant Virology, 4th ed.; Academic Press: London, UK, 2002; p. 1001. [Google Scholar]
- Sanfaçon, H.; Hohn, T. Proximity to the promoter inhibits recognition of cauliflower mosaic virus polyadenylation signal. Nature 1990, 346, 8–84. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.P.; Lockhart, B.E.; Olszewski, N.E. The ORF I and II Proteins of Commelina yellow mottle virus are virion-associated. Virology 1996, 223, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Jacquot, E.; Hagen, L.S.; Jacquemond, M.; Yot, P. The open reading frame 2 product of cacao swollen shoot badnavirus is a nucleic acid-binding protein. Virology 1996, 225, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Hagen, L.S.; Jacquemond, M.; Lepingle, A.; Lot, H.; Tepfer, M. Nucleotide sequence and genomic organization of Cacao swollen shoot virus. Virology 1993, 196, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Hohn, T.; Fütterer, J. The proteins and functions of plant pararetroviruses: Knowns and unknowns. Crit. Rev. Plant Sci. 1997, 16, 133–161. [Google Scholar] [CrossRef]
- Hany, U.; Adams, I.P.; Glover, R.; Bhat, A.I.; Boonham, N. The complete genome sequence of Piper yellow mottle virus (PYMoV). Arch. Virol. 2014, 159, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kwak, H.R.; Lee, Y.K.; Kim, M.K.; Choi, H.S.; Seo, J.K. Complete genome sequence of Yacon necrotic mottle virus, a novel putative member of the genus Badnavirus. Arch. Virol. 2015, 160, 1139–1142. [Google Scholar] [CrossRef] [PubMed]
- Maliogka, V.I.; Olmos, A.; Pappi, P.G.; Lotos, L.; Efthimiou, K.; Grammatikaki, G.; Candresse, T.; Katis, N.I.; Avgeli, A.D. A novel grapevine badnavirus is associated with the Roditis leaf discoloration disease. Virus Res. 2015, 203, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Seal, S.; Muller, E. Molecular analysis of a full-length sequence of a new yam badnavirus from Dioscorea sansibarensis. Arch. Virol. 2007, 152, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Chuang, W.H.; Wang, I.C. Sequencing and Molecular Analysis of the Viral Genome of Bougainvillea spectabilis chlorotic vein-banding virus from Bougainvillea spectabilis. Available online: http://www.ncbi.nlm.nih.gov/nuccore/NC_011592 (accessed on 18 January 2016).
- Yang, I.C.; Hafner, G.J.; Dale, J.L.; Harding, R.M. Genomic characterisation of taro bacilliform virus. Arch. Virol. 2003, 148, 937–949. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Wu, X.; Wang, A.; Wu, X. Characterization of complete genome and small RNA profile of Pagoda yellow mosaic associated virus, a novel Badnavirus in China. Virus Res. 2014, 188, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Borah, B.K.; Johnson, A.M.A.; Sai-Gopal, D.V.R.; Dasgupta, I. Sequencing and computational analysis of complete genome sequences of Citrus yellow mosaic badnavirus from acid lime and pummelo. Virus Genes 2009, 39, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Hartung, J.S. Cloning and sequence analysis of an infectious clone of Citrus yellow mosaic virus that can infect sweet orange via Agrobacterium-mediated inoculation. J. Gen. Virol. 2001, 82, 2549–2558. [Google Scholar] [CrossRef] [PubMed]
- Kazmi, S.A.; Yang, Z.; Hong, N.; Wang, G.; Wang, Y. Characterization by small RNA sequencing of Taro Bacilliform CH Virus (TaBCHV), a novel Badnavirus. PLoS ONE 2015, 10, e0134147. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Gao, S.; Huang, Y.; Ji, C.; Wang, D.; Ma, Y.; Fang, R.; Chen, X. Complete genomic sequence of Dracaena mottle virus, a distinct badnavirus. Virus Genes 2007, 35, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Kalischuk, M.L.; Fusaro, A.F.; Waterhouse, P.M.; Pappu, H.R.; Kawchuk, L.M. Complete genomic sequence of a Rubus yellow net virus isolate and detection of genome-wide pararetrovirus-derived small RNAs. Virus Res. 2013, 178, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Fütterer, J.; Kiss-László, Z.; Hohn, T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell 1993, 73, 789–802. [Google Scholar] [CrossRef]
- Ryabova, L.A.; Hohn, T. Ribosome shunting in cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes Dev. 2000, 14, 817–829. [Google Scholar] [PubMed]
- Pooggin, M.M.; Fütterer, J.; Skryabin, K.G.; Hohn, T. A short open reading frame terminating in front of a stable hairpin is the conserved feature in pregenomic RNA leaders of plant pararetroviruses. J. Gen. Virol. 1999, 80, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Fütterer, J.; Rothnie, H.M.; Hohn, T.; Potrykus, I. Rice tungro bacilliform virus open reading frames II and III are translated from polycistronic pregenomic RNA by leaky scanning. J. Virol. 1997, 71, 7984–7989. [Google Scholar] [PubMed]
- Iskra-Caruana, M.; Duroy, P.O.; Chabannes, M.; Muller, E. The common evolutionary history of badnaviruses and banana. Infect. Genet. Evol. 2014, 21, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Hull, R.; Lockhart, B.E.N.; Olsweski, N. Viral sequences integrated in to plant genomes. Ann. Rev. Phytopathol. 2002, 40, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Pahalawatta, V.; Druffel, K.; Pappu, H. A new and distinct species in the genus Caulimovirus exists as an endogenous plant pararetroviral sequence in its host, Dahlia variabilis. Virology 2008, 376, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Gayral, P.; Noa-Carrazana, J.-C.; Lescot, M.; Lheureux, F.; Lockhart, B.E.L.; Matsumoto, T.; Piffanelli, P.; Iskra-Caruana, M.-L. A single Banana streak virus integration event in the banana genome as the origin of infectious endogenous pararetrovirus. J. Virol. 2008, 82, 6697–6710. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, B.E.L.; Menke, J.; Dahal, G.; Olszewski, N.E. Characterization and genomic analysis of tobacco vein clearing virus, a plant pararetrovirus that is transmitted vertically and related to sequences integrated in the host genome. J. Gen. Virol. 2000, 81, 1579–1585. [Google Scholar] [CrossRef] [PubMed]
- Richert-Pöggeler, K.R.; Noreen, F.; Schwarzacher, T.; Harper, G.; Hohn, T. Induction of infectious petunia vein clearing (pararetro) virus from endogenous provirus in petunia. EMBO J. 2003, 22, 4836–4845. [Google Scholar] [CrossRef] [PubMed]
- Chabannes, M.; Iskra-Caruana, M.-L. Endogenous pararetroviruses—A reservoir of virus infection in plants. Curr. Opin. Virol. 2013, 3, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Iskra-Caruana, M.-L.; Chabannes, M.; Durou, P.-O. A possible scenario for the evolution of Banana streak virus in banana. Virus Res. 2014, 186, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Geering, A.D.W.; Olszewski, N.E.O.; Dahal, G.; Thomas, J.E. Analysis of the distribution and structure of integrated Banana streak virus DNA in a range of Musa cultivars. Mol. Plant Pathol. 2001, 2, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Laney, A.G.; Hassan, M.; Tzanetakis, I.E. An integrated badnavirus is prevalent in fig germplasm. Phytopathology 2012, 102, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Seal, S.; Turaki, A.; Muller, E.; Kumar, P.L.; Kenyon, L.; Filloux, D.; Galzi, S.; Lopez-Montes, A.; Iskra-Caruana, M.-L. The prevalence of badnaviruses in West African yams (Dioscorea cayenensis-rotundata) and evidence of endogenous pararetrovirus sequences in their genomes. Virus Res. 2014, 186, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.C.; Hafner, G.J.; Revill, P.A.; Dale, J.L.; Harding, R.M. Sequence diversity of South Pacific isolates of Taro bacilliform virus and the development of a PCR-based diagnostic test. Arch. Virol. 2003, 148, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nicotaisen, M.; Olszewski, N.E.; Lockhart, B.E. Sequencing, improved detection, and a novel form of Kalanchoë top-spotting virus. Plant Dis. 2005, 89, 298–302. [Google Scholar] [CrossRef]
- Mette, M.F.; Kanno, T.; Aufsatz, W.; Jakowitsch, J.; van der Winden, J.; Matzke, M.A.; Matzke, A.J.M. Endogenous viral sequences and their potential contribution to heritable virus resistance in plants. EMBO J. 2002, 21, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, B.E.L. Banana Streak Badnavirus Infection in Musa: Epidemiology, Diagnosis, and Control; ASPAC Food & Fertilizer Technology Center: Taipei, Taiwan, 1995; Volume 143, p. 11. [Google Scholar]
- Lockhart, B.E.L. Purification and serology of a bacilliform virus associated with banana streak disease. Phytopathology 1986, 76, 995–999. [Google Scholar] [CrossRef]
- Selvarajan, R.; Balasubramanian, V.; Gayathrie, T. Highly efficient immunodiagnosis of episomal Banana streak MY virus (BSMYV) using polyclonal antibodies raised against recombinant viral associated protein. J. Phytpathol. 2016. [Google Scholar] [CrossRef]
- Braithwaite, K.S.; Egeskov, N.M.; Smith, G.R. Detection of Sugarcane bacilliform virus using the polymerase chain reaction. Plant Dis. 1995, 79, 792–796. [Google Scholar] [CrossRef]
- Harper, G.; Dahal, G.; Thottappilly, G.; Hull, R. Detection of episomal banana streak badnavirus by IC-PCR. J. Virol. Methods 1999, 79, 1–8. [Google Scholar] [CrossRef]
- Jones, T.A.; McGavin, W.J.; Geering, A.D.W.; Lockhart, B.E.L. A new badnavirus in Ribes species, its detection by PCR, and its close association with Gooseberry vein banding disease. Plant Dis. 2001, 85, 417–422. [Google Scholar] [CrossRef]
- Muller, E.; Jacquot, E.; Yot, P. Early detection of cacao swollen shoot virus using the polymerase chain reaction. J. Virol. Methods 2001, 93, 15–22. [Google Scholar] [CrossRef]
- Thomson, K.G.; Dietzgen, R.G.; Thomas, J.E.; Teakle, D.S. Detection of pineapple bacilliform virus using the polymerase chain reaction. Ann. Appl. Biol. 1996, 129, 57–69. [Google Scholar] [CrossRef]
- Anthony-Johnson, A.M.; Dasgupta, I.; Sai-Gopal, D.V.R. Development of loop-mediated isothermal amplification and SYBR green real-time PCR methods for the detection of Citrus yellow mosaic badnavirus in citrus species. J. Virol. Methods 2014, 203, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.I.; Siljo, A.; Deeshma, K.P. Rapid detection of Piper yellow mottle virus and Cucumber mosaic virus infecting black pepper (Piper nigrum) by loop-mediated isothermal amplification (LAMP). J. Virol. Methods 2013, 193, 190–196. [Google Scholar]
- Bhat, A.I.; Siljo, A. Detection of viruses infecting black pepper by SYBR Green-based real-time PCR assay. J. Plant Pathol. 2014, 96, 105–109. [Google Scholar]
- Geering, A.D.W.; Mc Michael, L.A.; Dietzgen, R.G.; Thomas, J.E. Genetic diversity among banana streak virus isolates from Australia. Phytopathology 2000, 90, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Le Provost, G.; Iskra-Caruana, M.L.; Acina, I.; Teycheney, P.Y. Improved detection of episomal banana streak viruses by multiplex immunocapture PCR. J. Virol. Methods 2006, 137, 7–13. [Google Scholar] [CrossRef] [PubMed]
- James, A.P.; Geijskes, R.J.; Dale, J.L.; Harding, R.M. Development of a novel rolling-circle amplification technique to detect Banana streak virus which also discriminates between integrated and episomal virus sequences. Plant Dis. 2011, 95, 57–62. [Google Scholar] [CrossRef]
- Blanco, L.; Bernad, A.; Lazaro, J.M.; Martin, G.; Gar-mendia, C.; Salas, M. Highly efficient DNA synthesis by the phage Phi29 DNA polymerase. J. Biol. Chem. 1989, 264, 8935–8940. [Google Scholar] [PubMed]
- Dean, F.; Nelson, J.; Giesler, T.; Lasken, R. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply primed rolling circle amplification. Genome Res. 2001, 11, 1095–1099. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Müller, H.; Rector, A.; van Ranst, M.; Stevens, H. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol. 2009, 17, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Wambulwa, M.C.; Wachira, F.N.; Karanja, L.S.; Muturi, S.M. Rolling circle amplification is more sensitive than PCR and serology-based methods in detection of Banana streak virus in Musa germplasm. Am. J. Plant Sci. 2012, 3, 1581–1587. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Virus Taxonomy: 2014 Release. Available online: http://www.ictvonline.org/virusTaxonomy.asp (accessed on 28 February 2016).
- King, A.M.Q.; Adams, M.J.; Lefkowitz, E.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Academic Press: San Diego, CA, USA, 2012; p. 1463. [Google Scholar]
- Kumar, P.L.; Selvarajan, R.; Iskra-Caruana, M.-L.; Chabannes, M.; Hanna, R. Biology, etiology, and control of virus diseases of banana and plantain. Adv. Virus Res. 2015, 91, 229–269. [Google Scholar] [PubMed]
- Lockhart, B.E.L.; Jones, D.R. Banana streak mosaic. In Diseases of Banana, Abaca and Enset; Jones, D.R., Ed.; CAB International: Wallingford, UK, 2000; pp. 263–274. [Google Scholar]
- Dahal, G.; Hughes, J.D.A.; Thottappilly, G.; Lockhart, B.E.L. Effect of temperature on symptom expression and reliability of banana streak badnavirus detection in naturally infected plantain and banana (Musa spp.). Plant Dis. 1998, 82, 16–21. [Google Scholar] [CrossRef]
- Dahal, G.; Ortiz, R.; Tenkouano, A.; Hughes, J.D.A.; Thottappilly, G.; Vuylsteke, D.; Lockhart, B.E.L. Relationship between natural occurrence of Banana streak badnavirus and symptom expression, relative concentration of viral antigen, and yield characteristics of some micropropagated Musa spp. Plant Pathol. 2000, 49, 68–79. [Google Scholar] [CrossRef]
- Selvarajan, R.; (ICAR-National Research Centre for Banana, Tiruchirapalli, Tamil Nadu, India). Personal Communication, 2015.
- Daniells, J.W.; Geering, A.D.W.; Bryde, N.J.; Thomas, J.E. The effect of Banana streak virus on the growth and yield of dessert bananas in tropical Australia. Ann. Appl. Biol. 2001, 139, 51–60. [Google Scholar] [CrossRef]
- Geering, A.D.; Parry, J.N.; Thomas, J.E. Complete genome sequence of a novel badnavirus, Banana streak IM virus. Arch. Virol. 2011, 156, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Geering, A.D.; Pooggin, M.M.; Olszewski, N.E.; Lockhart, B.E.; Thomas, J.E. Characterisation of Banana streak Mysore virus and evidence that it’s DNA is integrated in the B genome of cultivated Musa. Arch. Virol. 2005, 150, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Hart, D.; Moult, S.; Hull, R.; Geering, A.; Thomas, J. The diversity of Banana streak virus isolates in Uganda. Arch. Virol. 2005, 150, 2407–2420. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Hart, D.; Moult, S.; Hull, R.; Thresh, J.M.; Jones, R.A.C.; Kuehne, T. Banana streak virus is very diverse in Uganda. Virus Res. 2004, 100, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, F.; Laboureau, N.; Muller, E.; Lockhart, B.E.; Iskra-Caruana, M.L. Molecular characterization of banana streak acuminata Vietnam virus isolated from Musa acuminata siamea (banana cultivar). Arch. Virol. 2007, 152, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Hull, R. Cloning and sequence analysis of banana streak virus DNA. Virus Genes 1998, 17, 271–278. [Google Scholar] [CrossRef] [PubMed]
- James, A.P.; Geijskes, R.J.; Dale, J.L.; Harding, R.M. Molecular characterisation of six badnavirus species associated with leaf streak disease of banana in East Africa. Ann. Appl. Biol. 2011, 158, 346–353. [Google Scholar] [CrossRef]
- Geering, A.D.W.; Olszewski, N.E.; Harper, G.; Lockhart, B.E.L.; Hull, R.; Thomas, J.E. Banana contains a diverse array of endogenous badnaviruses. J. Gen. Virol. 2005, 86, 511–520. [Google Scholar] [CrossRef] [PubMed]
- D’Hont, A.; Denoeud, F.; Aury, J.-M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Gayral, P.; Iskra-Caruana, M.-L. Phylogeny of Banana streak virus reveals recent and repetitive endogenization in the genome of its banana host (Musa sp.). J. Mol. Evol. 2009, 69, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Osuji, J.O.; Heslop-Harrison, J.S.; Hull, R. Integration of banana streak badnavirus into the Musa genome: Molecular and cytogenetic evidence. Virology 1999, 255, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Ndowora, T.; Ganesh, D.; Daryl, L.F.; Harper, G.; Hull, R.; Neil, E.O.; Lockhart, B. Evidence that Badnavirus infection in Musa can originate from integrated Pararetroviral sequences. Virology 1999, 225, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Gayral, P.; Blondin, L.; Guidolin, O.; Carreel, F.; Hippolyte, I.; Perrier, X.; Iskra-Caruana, M.L. Evolution of endogenous sequences of Banana Streak Virus: What can we learn from banana (Musa sp.) evolution? J. Virol. 2010, 84, 7346–7359. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.B.; Kasdorf, G.G.F.; Nel, L.H.; Pietersen, G. Transmission of activated episomal Banana streak OL (badna) virus (BSOLV) to cv. Williams banana (Musa sp.) by three mealybug species. Plant Dis. 2008, 92, 1158–1163. [Google Scholar] [CrossRef]
- Delanoy, M.; Salmon, M.; Kummert, J.; Frison, E.; Lepoivre, P. Development of real-time PCR for the rapid detection of episomal Banana streak virus (BSV). Plant Dis. 2003, 87, 33–38. [Google Scholar] [CrossRef]
- Selvarajan, R.; Balasubramanian, V.; Kavitha, K.; Kavitha, K.S.; Sathiamoorthy, S.; Ahlawat, Y.S. Detection of Banana bunchy top virus and Banana streak Mysore virus by PCR: Impact of storing virus infected banana samples. Virus Dis. 2008, 19, 28–33. [Google Scholar]
- Selvarajan, R.; Balasubramanian, V.; Sheeba, M.M.; Raj Mohan, R.; Mustaffa, M.M. Virus-indexing technology for production of quality banana planting material: A boon to the tissue-culture industry and banana growers in India. Acta Hortic. 2011, 897, 463–469. [Google Scholar] [CrossRef]
- Selvarajan, R.; Sheeba, M.M.; Balasubramanian, V. Simultaneous detection of episomal Banana streak Mysore virus and Banana bunchy top virus using multiplex RT-PCR. Curr. Sci. 2011, 100, 31–34. [Google Scholar]
- Javer-Higginson, E.; Acina-Mambole, I.; González, J.E.; Font, C.; González, G.; Echemendía, A.L.; Muller, E.; Teycheney, P.-Y. Occurrence, prevalence and molecular diversity of banana streak viruses in Cuba. Eur. J. Plant Pathol. 2013, 138, 157–166. [Google Scholar] [CrossRef]
- Helliot, B.; Panis, B.; Poumay, Y.; Swennen, R.; Lepoivre, P.; Frison, E. Cryopreservation for the elimination of cucumber mosaic and banana streak viruses from banana (Musa spp.). Plant Cell Rep. 2002, 20, 1117–1122. [Google Scholar]
- Rivas, E.B.; Ligia, M.L.; Duarte, M.; Alexandre, A.V.; Flora, M.C.; Fernandes, R.H.; Chagas, C.M. A new Badnavirus species detected in Bougainvillea in Brazil. J. Gen. Plant Pathol. 2005, 71, 438–440. [Google Scholar] [CrossRef]
- Tsai, C.H.; Su, H.-J.; Liao, Y.C.; Hung, T.-H. First report of Bougainvillea spectabilis chlorotic vein-banding virus infecting Bougainvillea plants in Taiwan. Plant Dis. 2005, 89, 1363. [Google Scholar] [CrossRef]
- Baranwal, V.K.; Meenakshi, A.; Singh, J. First report of two distinct badnaviruses associated with Bougainvillea spectabilis in India. J. Gen. Plant Pathol. 2010, 76, 236–239. [Google Scholar] [CrossRef]
- Tsai, C.-H.; Su, H.-J.; Wu, M.-L.; Feng, Y.-C.; Hung, T.-H. Identification and detection of Bougainvillea spectabilis chlorotic vein-banding virus in different bougainvillea cultivars in Taiwan. Ann. Appl. Biol. 2008, 53, 187–193. [Google Scholar] [CrossRef]
- Thresh, M.J. The origin and epidemiology of some important plant virus diseases. Appl. Biol. 1980, 5, 1–65. [Google Scholar]
- Posnette, A.F. Virus diseases of cacao in West Africa VII: Virus transmission by different vector species. Ann. Appl. Biol. 1950, 37, 378–384. [Google Scholar] [CrossRef]
- Posnette, A.F. Virus diseases of cacao in West Africa. 1. Cacao viruses 1A, 1B, 1C and 1D. Ann. Appl. Biol. 1947, 34, 388–402. [Google Scholar] [CrossRef]
- Roivainen, O. Transmission of cocoa viruses by mealy bugs (Homoptera: Pseudococcidae). J. Sci. Agric. Soc. 1976, 48, 433–453. [Google Scholar]
- Ameyaw, G.A.; Wetten, A.; Dzahini-Obiatey, H.; Domfeh, O.; Allainguillaume, J. Investigation on Cacao swollen shoot virus (CSSV) pollen transmission through cross-pollination. Plant Pathol. 2013, 62, 421–427. [Google Scholar] [CrossRef]
- Oro, F.; Mississo, E.; Okassa, M.; Guilhaumon, C.; Fenouillet, C.; Cilas, C.; Muller, E. Geographical differentiation of the molecular diversity of Cacao swollen shoot virus in Togo. Arch. Virol. 2012, 157, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.; Sackey, S. Molecular variability analysis of five new complete cacao swollen shoot virus genomic sequences. Arch. Virol. 2005, 150, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Jacquot, E.; Hagen, L.S.; Michler, P.; Rohfritsch, O.; Stussi-Garaud, C.; Keller, M.; Jacquemond, M.; Yot, P. in situ localisation of cacao swollen shoot virus in agroinoculated Theobroma cacao. Arch. Virol. 1999, 144, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Quainoo, A.K.; Wetten, A.C.; Allainguillaume, J. The effectiveness of somatic embryogenesis in eliminating the cocoa swollen shoot virus from infected cocoa trees. J. Virol. Methods 2008, 149, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, B.E.L. Occurrence of Canna yellow mottle virus in North America. Acta Hort. 1988, 234, 69–72. [Google Scholar] [CrossRef]
- Pappu, H.R.; Druffel, K.B.; Eastwell, K.C. Canna yellow mottle virusin Canna spp. in Washington State. Plant Dis. 2008, 92, 1136. [Google Scholar] [CrossRef]
- Jianguo, S.; Nianwu, W.; Dongbin, Z.; Kehui, H.; Qiongxia, G. Detection of Canna yellow mottle virus by PCR. Plant Quar. 2008, 22, 287–289. (In Chinese) [Google Scholar]
- Marino, M.T.; Ragozzino, E.; Lockhart, B.E.L.; Miglino, R.; Alioto, D. First report of Canna yellow mottle virus (CaYMV) in Italy and in The Netherlands. Plant Pathol. 2008, 57, 394. [Google Scholar] [CrossRef]
- Momol, M.T.; Lockhart, B.E.L.; Dankers, H.; Adkins, S. Canna yellow mottle virus detected in Canna in Florida. Plant Health Prog. 2004. [Google Scholar] [CrossRef]
- Kumari, A.; Kumar, S.; Raj, S.K.; Johri, J.K.; Nautiyal, C.S. First report of Canna yellow mottle virus associated with yellow vein mosaic disease of betel vine (Piper betel) in India. Plant Dis. 2015, 99, 1189. [Google Scholar] [CrossRef]
- Ahlawat, Y.S.; Pant, R.P.; Lockhart, B.E.L.; Srivastava, M.; Chakraborty, N.K.; Varma, A. Association of a badnavirus with citrus mosaic disease in India. Plant Dis. 1996, 80, 590–592. [Google Scholar] [CrossRef]
- Baranwal, V.K.; Majumder, S.; Ahlawat, Y.S.; Singh, R.P. Sodium sulphite yields improved DNA of higher stability for PCR detection of Citrus yellow mosaic virus from citrusleaves. J. Virol. Methods 2003, 112, 153–156. [Google Scholar] [CrossRef]
- Gopi, V.; Gopal, K.; Gourisankar, T.; Palanivel, S. Detection of citrus yellow mosaic virus by PCR and nucleic acid spot hybridisation using non-radioactive probes in commercial citrus species. Arch. Phytopathol. Plant Prot. 2010, 43, 892–899. [Google Scholar] [CrossRef]
- Anthony-Johnson, A.M.; Borah, B.K.; Sai Gopal, D.V.; Dasgupta, I. Analysis of full-length sequences of two Citrus yellow mosaic badnavirus isolates infecting Citrus jambhiri (Rough Lemon) and Citrus sinensis L. Osbeck (Sweet Orange) from a nursery in India. Virus Genes 2012, 45, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Tzafrir, I.; Ayala-Navarrete, L.; Lockhart, B.E.; Olszewski, N.E. The N-terminal portion of the 216-kD polyprotein of Commelina yellow mottle badnavirus is required for virus movement but not for replication. Virology 1997, 232, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.P.; Tzafrir, I.; Lockhart, B.E.; Olszewski, N.E. Tubules containing virions are present in plant tissues infected with Commelina yellow mottle badnavirus. J. Gen. Virol. 1998, 79, 925–929. [Google Scholar] [CrossRef] [PubMed]
- Van Lent, J.; Storms, M.; van der Meer, F.; Wellink, J.; Goldbach, R. Tubular structures involved in movement of cowpea mosaic virus are also formed in infected cowpea protoplast. J. Gen. Virol. 1991, 72, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Medberry, S.L.; Lockhart, B.E.L.; Olszewski, N.E. The Commelinayellow mottlevirus promoter is a strong promoter in vascular and reproductive tissues. Plant Cell 1992, 4, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Torbert, K.A.; Gopalraj, M.; Medberry, S.L.; Olszewski, N.E.; Somers, D.A. Expression of the Commelina yellow mottle virus promoter in transgenic oat. Plant Cell Rep. 1998, 17, 284–287. [Google Scholar] [CrossRef]
- Wu, B.; Pan, R.; Lu, R.; Tian, Y. Deletion analysis and functional studies of the promoter from commelina yellow mottle virus. Acta Microbiol. Sin. 1999, 39, 15–22. [Google Scholar]
- Matsuda, Y.; Liang, G.; Zhu, Y.; Ma, F.; Nelson, R.S.; Ding, B. The Commelina yellow mottle virus promoter drives companion-cell-specific gene expression in multiple organs of transgenic tobacco. Protoplasma 2002, 220, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.; Briddon, R.W.; Brunt, A.A.; Hull, R. The partial characterization of badnavirus infecting the Greater asiatic or water yam (Dioscorea alata). J. Phytopathol. 1999, 147, 265–269. [Google Scholar] [CrossRef]
- Briddon, R.W.; Phillips, S.; Brunt, A.; Hull, R. Analysis of the sequence of Dioscorea alata bacilliform virus; Comparison to other members of the badnavirus group. Virus Genes 1999, 18, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Eni, A.O.; Hughes, J.D.A.; Asiedu, R.; Rey, M.E.C. Sequence diversity among badnavirus isolates infecting yam (Dioscorea spp.) in Ghana, Togo, Benin and Nigeria. Arch. Virol. 2008, 153, 2263–2272. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, L.; Lebas, B.S.M.; Seal, S.E. Yams (Dioscorea spp.) from the South Pacific Islands contain many novel badnaviruses: Implications for international movement of yam germplasm. Arch. Virol. 2008, 153, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Bousalem, M.; Durand, O.; Scarcelli, N.; Lebas, B.S.M.; Kenyon, L.; Marchand, J.L.; Lefort, F.; Seal, S.E. Dilemmas caused by endogenous pararetroviruses regarding the taxonomy and diagnosis of yam (Dioscorea spp.). Arch. Virol. 2009, 145, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Umber, M.; Filloux, D.; Muller, E.; Laboureau, N.; Galzi, S.; Roumagnac, P.; Iskra-Caruana, M.-L.; Pavis, C.; Teycheney, P.-Y.; Seal, S. The genome of African yam (Dioscorea cayenensis-rotundata complex) hosts endogenous sequences from four distinct badnavirus species. Mol. Plant Pathol. 2014, 15, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Minafra, A.; Chiumenti, M.; Elbeaino, T.; Digiaro, M.; Bottalico, G.; Pantaleo, V.; Martelli, G.P. Occurrence of Fig badnavirus 1 in fig trees from different countries and in symptomless seedlings. J. Plant Pathol. 2012, 94, 94–105. [Google Scholar]
- Xu, D.; Mock, R.; Kinard, G.; Li, R. Molecular analysis of the complete genomic sequences of four isolates of Gooseberry vein banding associated virus. Virus Genes 2011, 43, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Singh, K.; Kaur, R.; Qiu, W. Association of a novel DNA virus with the grapevine vein-clearing and vine decline syndrome. Phytopathology 2011, 101, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, B.E.L.; Ferji, Z. Purification and mechanical transmission of Kalanchoë top spotting-associated virus. Acta Hortic. 1988, 234, 72–78. [Google Scholar] [CrossRef]
- Wakman, W.D.; Teakle, D.S.; Thomas, J.E.; Dietzgen, R.G. Presence of clostero-like virus and a bacilliform virus in pineapple plants in Queensland. Aust. J. Agric. Res. 1995, 46, 947–958. [Google Scholar] [CrossRef]
- Sether, D.M.; Hu, J.S. Yield impact and spread of Pineapple mealybug wilt associated virus-2 and mealybug wilt of pineapple in Hawaii. Plant Dis. 2002, 86, 867–874. [Google Scholar] [CrossRef]
- Gambley, C.F.; Geering, A.D.; Steele, V.; Thomas, J.E. Identification of viral and non-viral reverse transcribing elements in pineapple (Ananas comosus), including members of two new badnavirus species. Arch. Virol. 2008, 153, 1599–1604. [Google Scholar] [CrossRef] [PubMed]
- Liting, W.; Xiaolei, R.; Wenjin, S. Sequencing and analysis of the complete genomic sequence of pineapple bacilliform comosus virus. China Agric. Sci. 2010, 43, 1969–1976. [Google Scholar]
- Sether, D.M.; Melzer, M.J.; Borth, W.B.; Hu, J.S. Pineapple bacilliform CO virus: Diversity, detection, distribution, and transmission. Plant Dis. 2012, 96, 1798–1804. [Google Scholar] [CrossRef]
- Lockhart, B.E.L.; Kirtisak, K.A.; Jones, P.; Padmini, D.D.; Olszieewski, N.E.; Lockhart, N.; Nuarchan, D.; Sangalang, J. Identification of Piper yellow mottle virus, a mealy bug transmited badnavirus infecting Piper spp. in south East Asia. Eur. J. Plant Pathol. 1997, 103, 303–311. [Google Scholar] [CrossRef]
- Umadevi, P.; Bhat, A.I.; Krishamurthy, K.S.; Anandaraj, M. Influence of temperature on symptom expression and detection of host factors in Piper yellow mottle virus infected black pepper (Piper nigrum L.). Indian J. Exp. Biol. 2016, 54, 354–360. [Google Scholar] [PubMed]
- Bhat, A.I.; Devasahayam, S.; Sarma, Y.R.; Pant, R.P. Association of a Badnavirus in black pepper (Piper nigrum L.) transmitted by mealybug (Ferrisia virgata) in India. Curr. Sci. 2003, 84, 1547–1550. [Google Scholar]
- Hareesh, P.S.; Bhat, A.I. Detection and partial nucleotide sequence analysis of Piper yellow mottle virus infecting black pepper in India. Indian J. Virol. 2008, 19, 160–167. [Google Scholar]
- Siju, S.; Bhat, A.I.; Hareesh, P.S. Identification and characterization of a Badnavirus infecting betel vine and Indian long pepper. J. Plant Biochem. Biotechnol. 2008, 17, 73–76. [Google Scholar] [CrossRef]
- Bhat, A.I.; Sasi, S.; Revathy, K.A.; Deeshma, K.P.; Saji, K.V. Sequence diversity among badnavirus isolates infecting black pepper and related species in India. Virus Dis. 2014, 25, 402–407. [Google Scholar] [CrossRef] [PubMed]
- De Silva, D.P.; Jones, P.P.; Shaw, M.W. Identification and transmission of Piper yellow mottle virus and Cucumber mosaic virus infecting black pepper (Piper nigrum) in Sri Lanka. Plant Pathol. 2002, 51, 537–545. [Google Scholar] [CrossRef]
- Deeshma, K.P.; Bhat, A.I. Complete genome sequencing of Piper yellow mottle virus infecting black pepper, betelvine and Indian long pepper. Virus Genes 2015, 50, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.T.; McGavin, W.J.; Geering, A.D.W.; Lockhart, B.E.L. Identification of Rubus yellow net virus as a distinct badnavirus and its detection by PCR in Rubus species and in aphids. Ann. Appl. Biol. 2002, 141, 1–10. [Google Scholar] [CrossRef]
- Blevins, T.; Rajeswaran, R.; Aregger, M.; Borah, B.K.; Schepetilnikov, M.; Baerlocher, L.; Farinelli, L.; Meins, F., Jr.; Hohn, T.; Mikhail, M.; et al. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 2011, 39, 5003–5014. [Google Scholar] [CrossRef] [PubMed]
- Rajeswaran, R.; Golyaev, V.; Seguin, J.; Zvereva, A.S.; Farinelli, L.; Pooggin, M.M. Interactions of Rice tungro bacilliform pararetrovirus and its protein P4 with plant RNA-silencing machinery. Mol. Plant Microbe Interact. 2014, 27, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, B.E.L.; Olszewsk, N.E. Schefflera ringspot virus, a widely distributed mealybug-transmitted Badnavirus occurring in Schefflera and Aralia. Acta Hortic. 1996, 432, 196–202. [Google Scholar] [CrossRef]
- Lockhart, B.E.L.; Geering, A.D.W. Partial characterization of two aphid-transmitted viruses associated with yellow leafspot of spiraea. Acta Hortic. 2002, 568, 163–168. [Google Scholar] [CrossRef]
- Autrey, L.J.C.; Boolell, S.; Jones, P. Distribution of sugarcane bacilliform virus in various geographical regions. In Proceedings of the XXI Congress of the International Society of Sugar Cane Technologists, Bangkok, Thailand, 5–14 March 1992; Kasetsart University: Bangkok, Thailand, 1995; p. 657. [Google Scholar]
- Li, W.-F.; Huang, Y.-K.; Jiang, D.-M.; Zhang, Z.-X.; Zhang, B.-L.; Li, S.-F. Detection of Sugarcane bacilliform virus isolate and its influence on yield and quality of cane in Yunnan. Acta Phytopathol. Sin. 2010, 6, 651–654. (In Chinese) [Google Scholar]
- Lockhart, B.E.L.; Autrey, L.J.C. Occurrence in sugarcane of a bacilliform virus related serologically to banana streak virus. Plant Dis. 1988, 72, 230–233. [Google Scholar] [CrossRef]
- Singh, D.; Tewari, A.; Rao, G.; Karuppaiah, R.; Viswanathan, R.; Arya, M.; Baranwal, V. RT-PCR/PCR analysis detected mixed infection of DNA and RNA viruses infecting sugarcane crops in different states of India. Sugar Tech 2009, 11, 373–380. [Google Scholar] [CrossRef]
- Bouhida, M.; Lockhart, B.E.; Olszewski, N.E. An analysis of the complete sequence of a sugarcane bacilliform virus genome infectious to banana and rice. J. Gen. Virol. 1993, 74, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Geijskes, R.J.; Braithwaite, K.S.; Dale, J.L. Sequence analysis of an Australian isolate of sugarcane bacilliform badnavirus. Arch. Virol. 2002, 147, 2393–2404. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.; Dupuy, V.; Blondin, L.; Bauffe, F.; Daugrois, J.H.; Nathalie, L.; Iskra-Caruana, M.L. High molecular variability of sugarcane bacilliform viruses in Guadeloupe implying the existence of at least three new species. Virus Res. 2011, 160, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Karuppaiah, R.; Viswanathan, R.; Kumar, V.G. Genetic diversity of Sugarcane bacilliform virus isolates infecting Saccharum spp. in India. Virus Genes 2013, 46, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Geijskes, R.J.; Braithwaite, K.S.; Smith, G.R.; Dale, J.L.; Harding, R.M. Sugarcane bacilliform virus encapsidates genome concatamers and does not appear to integrate into the Saccharum officinarum genome. Arch. Virol. 2004, 149, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Xu, D.; Zhou, G. Evidence of sugarcane bacilliform virus DNA fragment integrated into the saccharum inter-specific hybrids genome. J. South China Agric. Univ. 2009, 30, 19–23. (In Chinese) [Google Scholar]
- Braithwaite, K.S.; Geijskes, R.J.; Smith, G.R. A variable region of the sugarcane bacilliform virus (SCBV) genome can be used to generate promoters for transgene expression in sugarcane. Plant Cell Rep. 2004, 23, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Schenk, P.M.; Sagi, L.; Remans, T.; Dietzgen, R.G.; Bernard, M.J.; Graham, M.W.; Manners, J.M. A promoter from sugarcane bacilliform badnavirus drives transgene expression in banana and other monocot and dicot plants. Plant Mol. Biol. 1999, 39, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Kreuze, J.F.; Perez, A.; Untiveros, M.; Quispe, D.; Fuentes, S.; Barker, I.; Simon, R. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: A generic method for diagnosis, discovery and sequencing of viruses. Virology 2009, 388, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kashif, M.; Pietilä, S.; Artola, K.; Jones, R.A.C.; Tugume, A.K.; Mäkinen, V.; Valkonen, J.P.T. Detection of viruses in sweetpotato from Honduras and Guatemala augmented by deep-sequencing of small-RNAs. Plant Dis. 2012, 96, 1430–1437. [Google Scholar] [CrossRef]
- Mbanzibwa, D.R.; Tugume, A.K.; Chiunga, E.; Mark, D.; Tairo, F.D. Small RNA deep sequencing-based detection and further evidence of DNA viruses infecting sweetpotato plants in Tanzania. Ann. Appl. Biol. 2014, 165, 329–339. [Google Scholar] [CrossRef]
- Yang, I.C.; Iommarini, J.P.; Becker, D.K.; Hafner, G.J.; Dale, J.L.; Harding, R.M. A promoter derived from taro bacilliform badnavirus drives strong expression in transgenic banana and tobacco plants. Plant Cell Rep. 2003, 21, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Melcher, U.; Muthukumar, V.; Wiley, G.B.; Min, B.E.; Palmer, M.W.; Verchot-Lubicz, J.; Ali, A.; Nelson, R.S.; Roe, B.A.; Thapa, V.; et al. Evidence for novel viruses by analysis of nucleic acids in virus-like particle fractions from Ambrosia psilostachya. J. Virol. Methods 2008, 152, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Lockhrat, B.; Fetzer, J.L.; Olszewski, N.E. Preliminary characterization of Cycad leaf necrosis virus, the first badnavirus identified in cycads. Phytopahtology 2006, 96, S70. [Google Scholar]
- Rumbos, I.C.; Avgelis, A.D. Roditis leaf discoloration—A new virus disease of grapevine: Symptomatology and transmission to indicators plants. J. Phytopathol. 1989, 125, 274–278. [Google Scholar] [CrossRef]
- Chiumenti, M.; Morelli, M.; Elbeaino, T.; Stavolone, L.; de Stradis, A.; Digiaro, M.; Minafra, A.; Martelli, G.P. Sequencing an unconventional virus genome: The mulberry badnavirus 1 case. J. Plant Pathol. 2014, 96, S4:36–S4:37. [Google Scholar]
- Elbeaino, T.; Chiumenti, M.; de Stradis, A.; Digiaro, M.; Minafra, A.; Martelli, G.P. Identification of a badnavirus infecting mulberry. J. Plant Pathol. 2013, 95, 207–210. [Google Scholar]
- Fránová, J.; Jakešová, H. First report of bacilliform badnavirus-like virus particles in red clover. J. Phytopathol. 2012, 160, 588–590. [Google Scholar] [CrossRef]
- Skotnicki, M.L.; Selkirk, P.M.; Kitajima, E.; McBride, T.P.; Shaw, J.; Mackenzie, A. The first subantarctic plant virus report: Stilbocarpa mosaic bacilliform badnavirus (SMBV) from Macquarie Island. Pol. Biol. 2003, 26, 1–7. [Google Scholar]
- Clover, C.R.G.; Pearson, M.N.; Elliott, D.R.; Tang, Z.; Smales, T.E.; Alexander, B.J.R. Amplification of badnavirus-like sequences from Yucca elephantipes. Aust. Plant Pathol. 2003, 32, 563–564. [Google Scholar] [CrossRef]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhat, A.I.; Hohn, T.; Selvarajan, R. Badnaviruses: The Current Global Scenario. Viruses 2016, 8, 177. https://doi.org/10.3390/v8060177
Bhat AI, Hohn T, Selvarajan R. Badnaviruses: The Current Global Scenario. Viruses. 2016; 8(6):177. https://doi.org/10.3390/v8060177
Chicago/Turabian StyleBhat, Alangar Ishwara, Thomas Hohn, and Ramasamy Selvarajan. 2016. "Badnaviruses: The Current Global Scenario" Viruses 8, no. 6: 177. https://doi.org/10.3390/v8060177
APA StyleBhat, A. I., Hohn, T., & Selvarajan, R. (2016). Badnaviruses: The Current Global Scenario. Viruses, 8(6), 177. https://doi.org/10.3390/v8060177





