CRISPR-Cas9 as a Powerful Tool for Efficient Creation of Oncolytic Viruses
Abstract
:1. Introduction
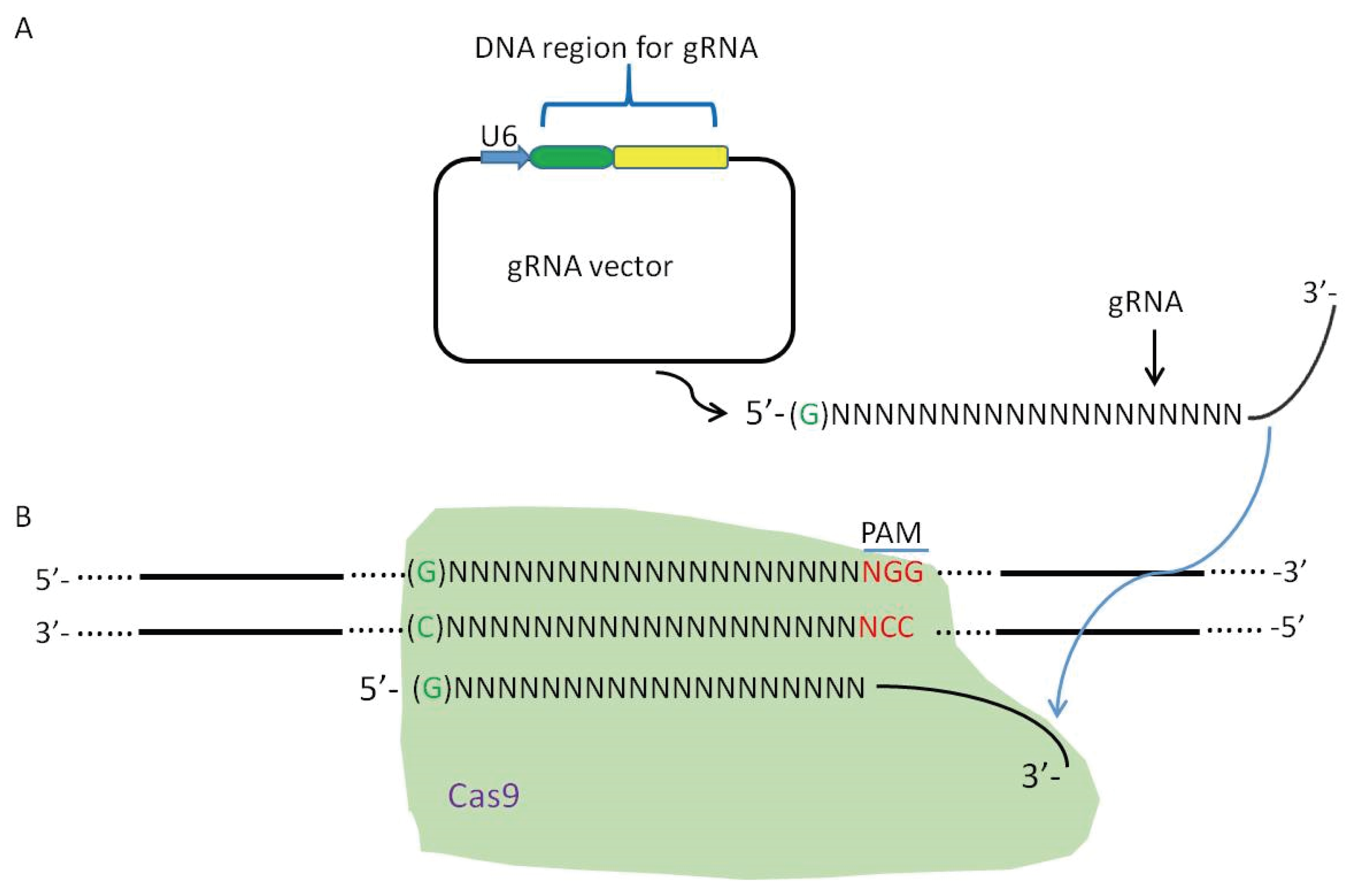
2. Viral Genome Editing Using the CRISPR-Cas9 System
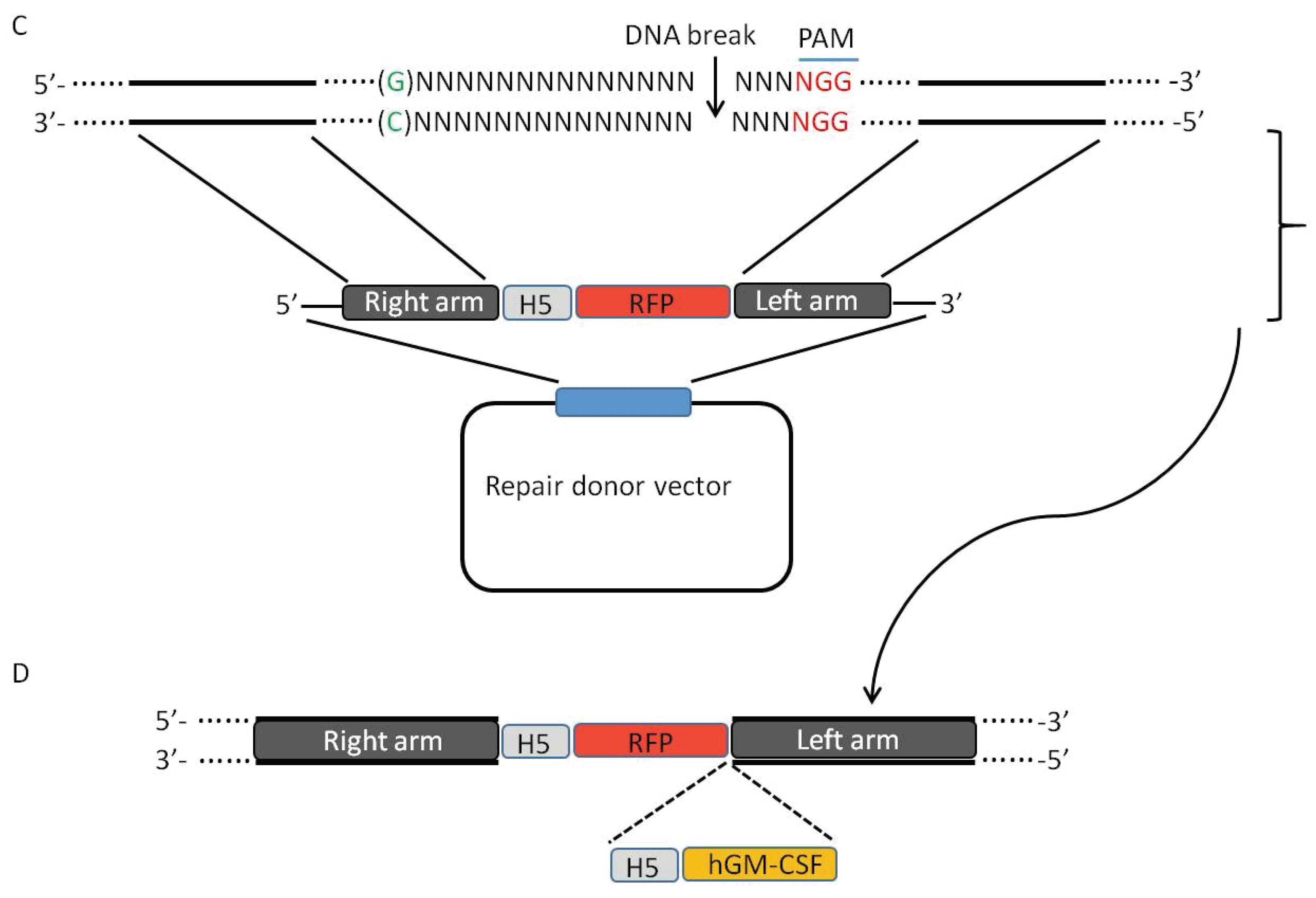
3. CRISPR-Cas9 System Induces High Efficiency of Homologous Recombination
4. Off-Target Effects by CRISPR-Cas9
5. Using CRISPR-Cas9 System to Create Mutant Oncolytic VV
6. Summary
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Russell, S.J.; Peng, K.W. Viruses as anticancer drugs. Trends Pharmacol. Sci. 2007, 28, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Senzer, N.N.; Kaufman, H.L.; Amatruda, T.; Nemunaitis, M.; Reid, T.; Daniels, G.; Gonzalez, R.; Glaspy, J.; Whitman, E.; Harrington, K.; et al. Phase II clinical trial of a granulocyte-macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J. Clin. Oncol. 2009, 27, 5763–5771. [Google Scholar] [CrossRef] [PubMed]
- Au, G.G.; Lindberg, A.M.; Barry, R.D.; Shafren, D.R. Oncolysis of vascular malignant human melanoma tumors by Coxsackievirus A21. Int. J. Oncol. 2005, 26, 1471–1476. [Google Scholar] [CrossRef] [PubMed]
- Martuza, R.L.; Malick, A.; Markert, J.M.; Ruffner, K.L.; Coen, D.M. Experimental therapy of human glioma by means of a genetically engineered virus mutant. Science 1991, 252, 854–856. [Google Scholar] [CrossRef] [PubMed]
- Park, B.H.; Hwang, T.; Liu, T.C.; Sze, D.Y.; Kim, J.S.; Kwon, H.C.; Oh, S.Y.; Han, S.Y.; Toon, J.H.; Hong, S.H.; et al. Use of a targeted oncolytic poxvirus, JX-594, in patients with refractory primary or metastatic liver cancer: A phase I trial. Lancet Oncol. 2008, 9, 533–542. [Google Scholar] [CrossRef]
- Harrington, K.J.; Hingorani, M.; Tanay, M.A.; Hickey, J.; Bhide, S.A.; Clarke, P.M.; Renouf, L.C.; Thway, K.; Sibtain, A.; Mcneish, I.A.; et al. Phase I/II study of oncolytic HSV GM-CSF in combination with radiotherapy and cisplatin in untreated stage III/IV squamous cell cancer of the head and neck. Clin. Cancer Res. 2010, 16, 4005–4015. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.; Breitbach, C.J.; Moon, A.; Kim, C.W.; Patt, R.; Kim, M.K.; Lee, Y.K.; Oh, S.Y.; Woo, H.Y.; Parato, K.; et al. Sequential therapy with JX-594, a targeted oncolytic poxvirus, followed by sorafenib in hepatocellular carcinoma: Preclinical and clinical demonstration of combination efficacy. Mol. Ther. 2011, 19, 1170–1179. [Google Scholar] [CrossRef] [PubMed]
- Frew, S.E.; Sammut, S.M.; Shore, A.F.; Ramjist, J.K.; Al-Bader, S.; Rezaie, R.; Daar, A.S.; Singer, P.A. Chinese health biotech and the billion-patient market. Nat. Biotechnol. 2008, 26, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Garber, K. China approves world’s first oncolytic virus therapy for cancer treatment. J. Natl. Cancer Inst. 2006, 98, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, M.J.; Maguire, H.C.; Lattime, E.C. Intralesional vaccinia/GM-CSF recombinant virus in the treatment of metastatic melanoma. Adv. Exp. Med. Biol. 2000, 465, 391–400. [Google Scholar] [PubMed]
- Melcher, A.; Parato, K.; Rooney, C.M.; Bell, J.C. Thunder and lightning: Immunotherapy and oncolytic viruses collide. Mol. Ther. 2011, 19, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- He, T.C.; Zhou, S.; da Costa, L.T.; Yu, J.; Kinzler, K.W.; Vogelstein, B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 1998, 95, 2509–2514. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.H.; Jiang, G.; Gangeswaran, R.; Wang, P.; Wang, J.; Yuan, M.; Wang, H.; Bhakta, V.; Müller, H.; Lemoine, N.R.; et al. Modification of the early gene enhancer-promoter improves the oncolytic potency of adenovirus 11. Mol. Ther. 2012, 20, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Ruzsics, Z.; Lemnitzer, F.; Thirion, C. Engineering adenovirus genome by bacterial artificial chromosome (BAC) technology. Methods Mol. Biol. 2014, 1089, 143–158. [Google Scholar] [PubMed]
- Hokanson, C.A.; Dora, E.; Donahue, B.A.; Rivkin, M.; Finer, M.; Mendez, M.J. Hybrid yeast-bacteria cloning system used to capture and modify adenoviral and nonviral genomes. Hum. Gene Ther. 2003, 14, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Nakano, E.; Panicali, D.; Paoletti, E. Molecular genetics of vaccinia virus: Demonstration of marker rescue. Proc. Natl. Acad. Sci. USA 1982, 79, 1593–1596. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Zhang, W.; Wang, J.; Al Yaghchi, C.; Ahmed, J.; Chard, L.; Lemoine, N.R.; Wang, Y. Efficiently editing the vaccinia virus genome by using the CRISPR-Cas9 system. J. Virol. 2015, 89, 5176–5179. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, H.; Ino, Y.; Kuroda, T.; Martuza, R.L.; Todo, T. Triple gene-deleted oncolytic herpes simplex virus vector double-armed with interleukin 18 and soluble B7-1 constructed by bacterial artificial chromosome-mediated system. Cancer Res. 2005, 65, 10663–10668. [Google Scholar] [CrossRef] [PubMed]
- Agarwalla, P.K.; Aghi, M.K. Oncolytic herpes simplex virus engineering and preparation. Methods Mol. Biol. 2012, 797, 1–19. [Google Scholar] [PubMed]
- Mocarski, E.S.; Post, L.E.; Roizman, B. Molecular engineering of the herpes simplex virus genome: Insertion of a second L-S junction into the genome causes additional genome inversions. Cell 1980, 22, 243–255. [Google Scholar] [CrossRef]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; Embden, J.D.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Van der Oost, J.; Jore, M.M.; Westra, E.R.; Lundgren, M.; Brouns, S.J. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009, 34, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2013, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.; Sun, C.; Gao, L.; Zhu, D.; Xu, X.; Zhu, X.; Xiong, J.W.; Xi, J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013, 23, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.J.; Cummings, A.M.; Nguyen, J.N.; Hamm, D.C.; Donohue, L.K.; Harrison, M.M.; Wildonger, J.; O’Connor-Giles, K.M. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 2013, 194, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Friedland, A.E.; Tzur, Y.B.; Esvelt, K.M.; Colaiacovo, M.P.; Church, G.M.; Calarco, J.A. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 2013, 10, 741–743. [Google Scholar] [CrossRef] [PubMed]
- DiCarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Sun, L.; Gao, D.; Ding, C.; Li, Z.; Li, Y.; Cun, W.; Li, Q. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS Pathog. 2014, 10, e1004090. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Gao, X.; Chard, L.S.; Ali, Z.; Ahmed, J.; Li, Y.; Liu, P.; Lemoine, N.R.; Wang, Y. A marker-free system for highly efficient construction of vaccinia virus vectors using CRISPR Cas9. Mol. Ther. Methods Clin. Dev. 2015, 2, 15035. [Google Scholar] [CrossRef] [PubMed]
- Coen, D.M.; Schaffer, P.A. Two distinct loci confer resistance to acycloguanosine in herpes simplex virus type 1. Proc. Natl. Acad. Sci. USA 1980, 77, 2265–2269. [Google Scholar] [CrossRef] [PubMed]
- Sarisky, R.T.; Quail, M.R.; Clark, P.E.; Nguyen, T.T.; Halsey, W.S.; Wittrock, R.J.; O’Leary, B.J.; Van Horn, M.M.; Sathe, G.M.; Van Horn, S.; et al. Characterization of herpes simplex viruses selected in culture for resistance to penciclovir or acyclovir. J. Virol. 2001, 75, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, T.; Kohyama, M.; Hirayasu, K.; Arase, H. Engineering large viral DNA genomes using the CRISPR-Cas9 system. Microbiol. Immunol. 2014, 58, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Saito, I.; Oya, Y.; Yamamoto, K.; Yuasa, T.; Shimojo, H. Construction of nondefective adenovirus type 5 bearing a 2.8-kilobase hepatitis B virus DNA near the right end of its genome. J. Virol. 1985, 54, 711–719. [Google Scholar] [PubMed]
- Becker, T.C.; Noel, R.J.; Coats, W.S.; Gomez-Foix, A.M.; Alam, T.; Gerard, R.D.; Newgard, C.B. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 1994, 43 Pt A, 161–189. [Google Scholar] [PubMed]
- Mittal, S.K.; McDermott, M.R.; Johnson, D.C.; Prevec, L.; Graham, F.L. Monitoring foreign gene expression by a human adenovirus-based vector using the firefly luciferase gene as a reporter. Virus Res. 1993, 28, 67–90. [Google Scholar] [CrossRef]
- Stratford-Perricaudet, L.D.; Makeh, I.; Perricaudet, M.; Briand, P. Widespread long-term gene transfer to mouse skeletal muscles and heart. J. Clin. Investig. 1992, 90, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, M.J.; Maguire, H.C., Jr.; Eisenlohr, L.C.; Laughlin, C.E.; Monken, C.E.; McCue, P.A.; Kovatich, A.J.; Lattime, E.C. Intratumoral recombinant GM-CSF-encoding virus as genetherapy in patients with cutaneous melanoma. Cancer Gene Ther. 1998, 6, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.D.; Jasin, M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 2001, 29, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015, 33, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Zhang, W.; Zhang, J.; Zhou, J.; Wang, J.; Chen, L.; Wang, L.; Hodgekins, A.; Iyer, V.; Huang, X.; et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 2014, 11, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014, 32, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014, 32, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Wyvekens, N.; Topkar, V.V.; Khayter, C.; Joung, J.K.; Tsai, S.Q. Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum. Gene Ther. 2015, 26, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Panicali, D.; Davis, S.W.; Weinberg, R.L.; Paoletti, E. Construction of live vaccines by using genetically engineered poxviruses: Biological activity of recombinant vaccinia virus expressing influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 1983, 80, 5364–5368. [Google Scholar] [CrossRef] [PubMed]
- Wiktor, T.J.; Macfarlan, R.I.; Reagan, K.J.; Dietzschold, B.; Curtis, P.J.; Wunner, W.H.; Kieny, M.P.; Lathe, R.; Lecocq, J.P.; Mackett, M.; et al. Protection from rabies by a vaccinia virus recombinant containing the rabies virus glycoprotein gene. Proc. Natl. Acad. Sci. USA 1984, 81, 7194–7198. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.S.; Bartlett, D.L. Vaccinia as a vector for gene delivery. Expert Opin. Biol. Ther. 2004, 4, 901–917. [Google Scholar] [CrossRef] [PubMed]
- Kwak, H.; Horig, H.; Kaufman, H.L. Poxviruses as vectors for cancer immunotherapy. Curr. Opin. Drug Discov. Dev. 2003, 6, 161–168. [Google Scholar]
- Tysome, J.R.; Briat, A.; Alusi, G.; Cao, F.; Gao, D.; Yu, J.; Wang, P.; Yang, S.; Dong, Z.; Wang, S.; et al. Lister strain of vaccinia virus armed with endostatin-angiostatin fusion gene as a novel therapeutic agent for human pancreatic cancer. Gene Ther. 2009, 16, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Kirn, D.H.; Wang, Y.; Liang, W.; Contag, C.H.; Thorne, S.H. Enhancing poxvirus oncolytic effects through increased spread and immune evasion. Cancer Res. 2008, 68, 2071–2075. [Google Scholar] [CrossRef] [PubMed]
- Puhlmann, M.; Gnant, M.; Brown, C.K.; Alexander, H.R.; Bartlett, D.L. Thymidine kinase-deleted vaccinia virus expressing purine nucleoside phosphorylase as a vector for tumor-directed gene therapy. Hum. Gene Ther. 1999, 10, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Puhlmann, M.; Brown, C.K.; Gnant, M.; Huang, J.; Libutti, S.K.; Alexander, H.R.; Bartlett, D.L. Vaccinia as a vector for tumor-directed gene therapy: Biodistribution of a thymidine kinase-deleted mutant. Cancer Gene Ther. 2000, 7, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Chard, L.S.; Maniati, E.; Wang, P.; Zhang, Z.; Gao, D.; Wang, J.; Cao, F.; Ahmed, J.; El Khouri, M.; Hughes, J.; et al. A vaccinia virus armed with interleukin-10 is a promising therapeutic agent for treatment of murine pancreatic cancer. Clin. Cancer Res. 2015, 21, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.; Wang, P.; Alusi, G.; Shi, H.; Chu, Y.; Wang, J.; Bhakta, V.; McNeish, I.; McCart, A.; Lemoine, N.R.; et al. Lister strain vaccinia virus with thymidine kinase gene deletion is a tractable platform for development of a new generation of oncolytic virus. Gene Ther. 2015, 22, 476–484. [Google Scholar] [CrossRef] [PubMed]
- McCart, J.A.; Ward, J.M.; Lee, J.; Hu, Y.; Alexander, H.R.; Libutti, S.K.; Moss, B.; Bartlett, D.L. Systemic cancer therapy with a tumor-selective vaccinia virus mutant lacking thymidine kinase and vaccinia growth factor genes. Cancer Res. 2001, 61, 8751–8757. [Google Scholar] [PubMed]
- Ma, H.; Wu, Y.; Dang, Y.; Choi, J.G.; Zhang, J.; Wu, H. Pol III Promoters to Express Small RNAs: Delineation of Transcription Initiation. Mol. Ther. Nucleic Acids 2014, 3, e161. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, M.; Webb, E.; Lemoine, N.R.; Wang, Y. CRISPR-Cas9 as a Powerful Tool for Efficient Creation of Oncolytic Viruses. Viruses 2016, 8, 72. https://doi.org/10.3390/v8030072
Yuan M, Webb E, Lemoine NR, Wang Y. CRISPR-Cas9 as a Powerful Tool for Efficient Creation of Oncolytic Viruses. Viruses. 2016; 8(3):72. https://doi.org/10.3390/v8030072
Chicago/Turabian StyleYuan, Ming, Eika Webb, Nicholas Robert Lemoine, and Yaohe Wang. 2016. "CRISPR-Cas9 as a Powerful Tool for Efficient Creation of Oncolytic Viruses" Viruses 8, no. 3: 72. https://doi.org/10.3390/v8030072
APA StyleYuan, M., Webb, E., Lemoine, N. R., & Wang, Y. (2016). CRISPR-Cas9 as a Powerful Tool for Efficient Creation of Oncolytic Viruses. Viruses, 8(3), 72. https://doi.org/10.3390/v8030072





