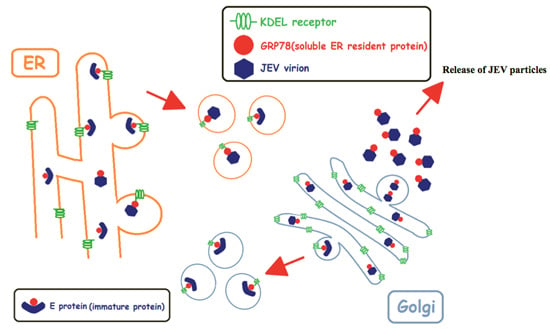
A KDEL Retrieval System for ER-Golgi Transport of Japanese Encephalitis Viral Particles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus and Cell Culture
2.2. Viral Plaque Assay
2.3. Immunoprecipitation, Western Blotting and Antisera
2.4. Immunofluorescent Staining
2.5. Sucrose Density Gradient Analysis
2.6. shGRP78 Knocked down Stable Cells and siRNA Knocking down of KDELR Proteins
2.7. Construction of the GRP78 with C-Terminal Flag-Tagged Expression Plasmids
2.8. Transmission Electron Microscopy
3. Results
3.1. JEV Infection Activates the Expression and Secretion of ER Luminal Proteins
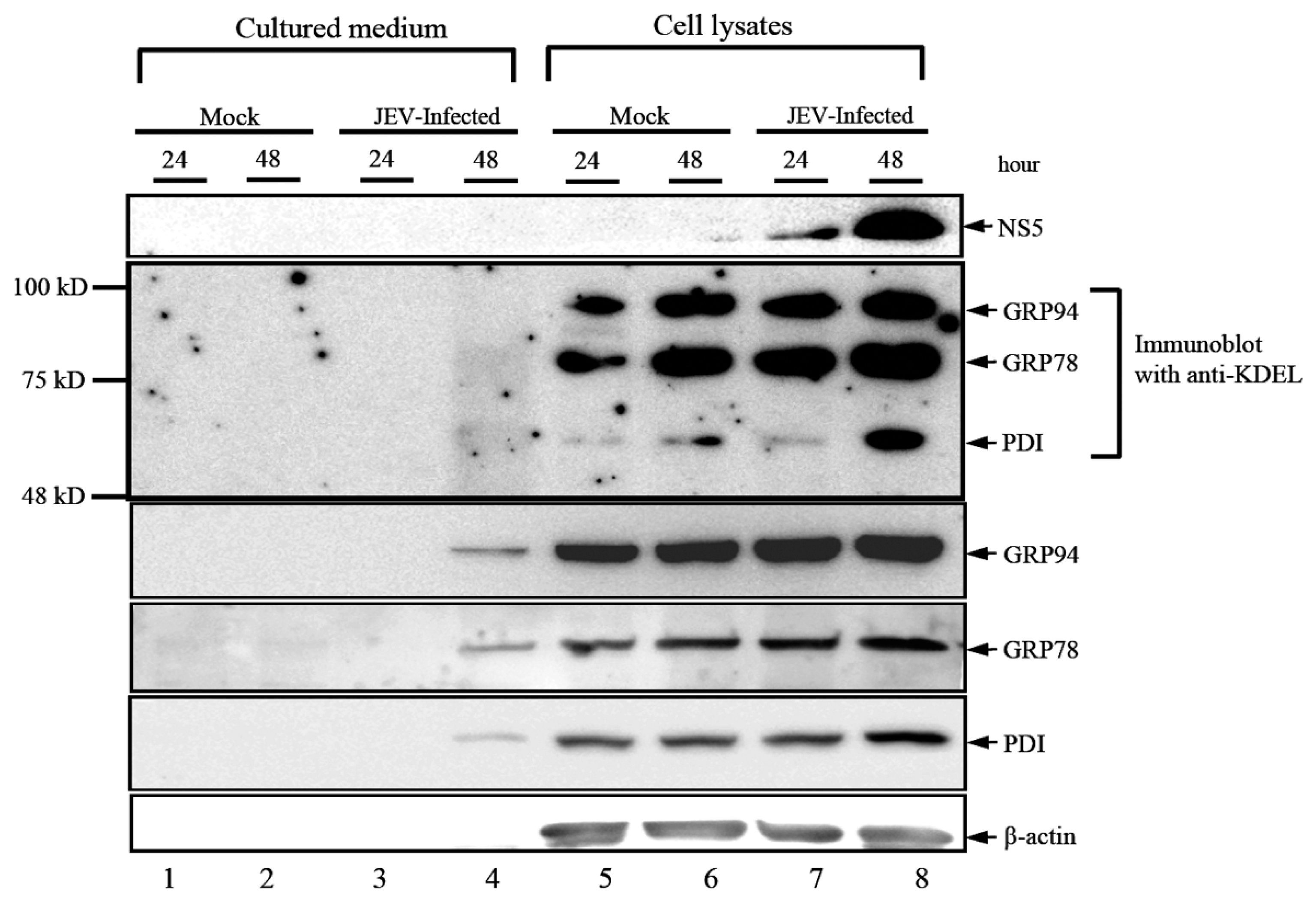
3.2. KDEL-Motif Is Not Essential for GRP78 Function in the Viral Protein Production
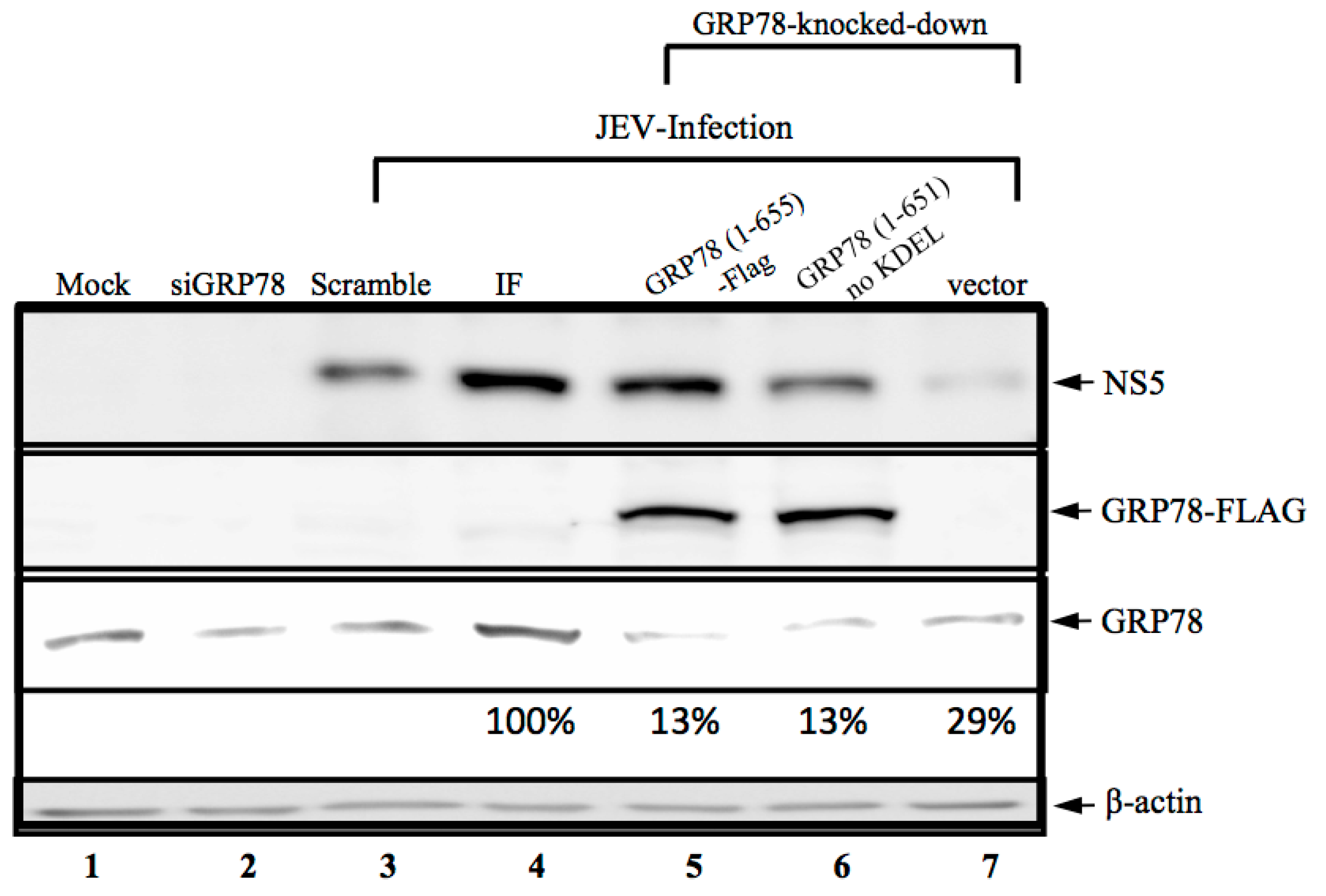
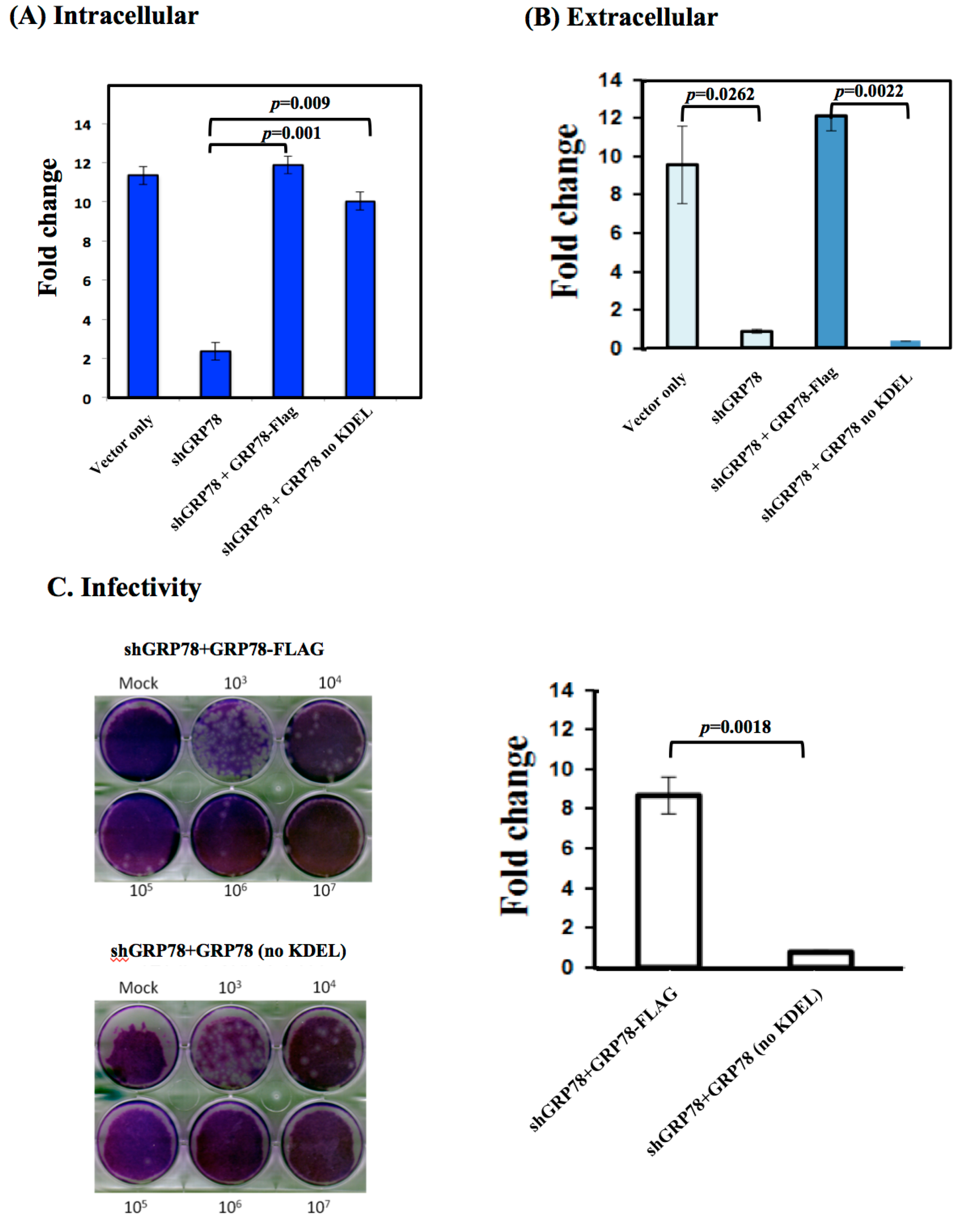
3.3. KDEL-Motif Is Required for GRP78 Function in the Release of JE Viral Virions
3.4. KDELR Protein Co-Fractionation with JE Viral Enveloped Proteins

3.5. KDELR Protein is Engaged in the Subcellular Localization of JE Viral Particles in the Golgi

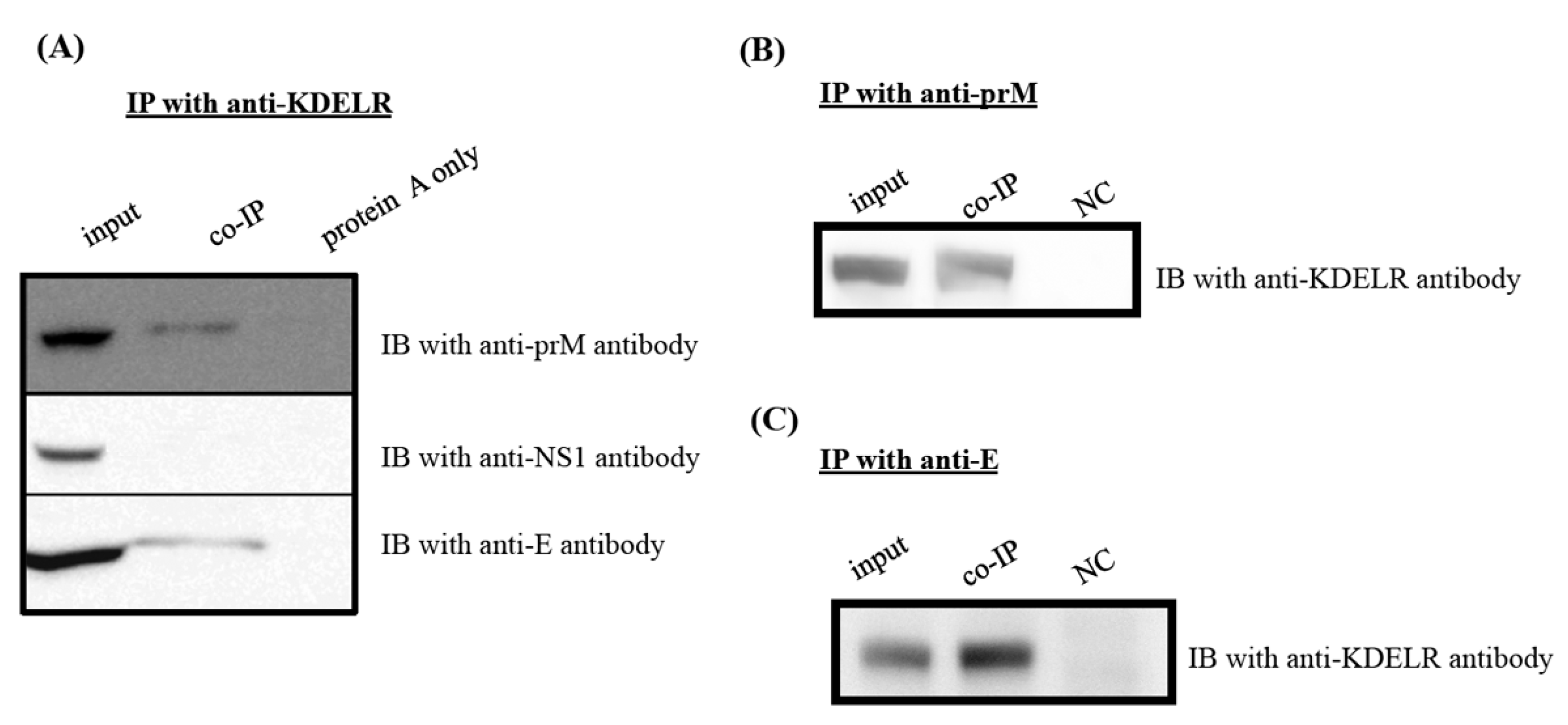
3.6. KDELR Protein Interacts with Viral Structural Proteins, but Not with Nonstructural NS1 Protein
3.7. Impaired Secretory Trafficking of JEV Particles in the KDELR Knocked down Cells

4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lindenbach, B.D.; Rice, C.M. Flaviviridae: The viruses and their replication. In Fields Virology, 4th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott William & Wilkins: Philadephia, PA, USA, 2001; Volume 1, pp. 991–1042. [Google Scholar]
- Lad, V.J.; Gupta, A.K. Inhibition of Japanese encephalitis virus maturation and transport in PS cells to cell surface by brefeldin A. Acta Virol. 2002, 46, 187–190. [Google Scholar] [PubMed]
- Mukhopadhyay, S.; Kuhn, R.J.; Rossmann, M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005, 3, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Jackson, W.T. Poliovirus-induced changes in cellular membranes throughout infection. Curr. Opin. Virol. 2014, 9, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Neveu, G.; Barouch-Bentov, R.; Ziv-Av, A.; Gerber, D.; Jacob, Y.; Einav, S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 2012, 8, e1002845. [Google Scholar] [CrossRef] [PubMed]
- Hsu, N.Y.; Ilnytska, O.; Belov, G.; Santiana, M.; Chen, Y.H.; Takvorian, P.M.; Pau, C.; van der Schaar, H.; Kaushik-Basu, N.; Balla, T.; et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 2010, 141, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lee, S.Y.; Beznoussenko, G.V.; Peters, P.J.; Yang, J.S.; Gilbert, H.Y.; Brass, A.L.; Elledge, S.J.; Isaacs, S.N.; Moss, B.; et al. A role for the host coatomer and KDEL receptor in early vaccinia biogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Capitani, M.; Sallese, M. The KDEL receptor: New functions for an old protein. FEBS Lett. 2009, 583, 3863–3871. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.P.; Chang, C.M.; Hung, C.Y.; Tsai, M.C.; Schuyler, S.C.; Wang, R.Y. Japanese encephalitis virus co-opts the ER-stress response protein GRP78 for viral infectivity. Virol. J. 2011, 8. [Google Scholar] [CrossRef]
- Hung, C.Y.; Tsai, M.C.; Wu, Y.P.; Wang, R.Y. Identification of heat-shock protein 90 β in Japanese encephalitis virus-induced secretion proteins. J. Gen. Virol. 2011, 92, 2803–2809. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chuang, C.K.; Chen, W.J. In situ reverse-transcription loop-mediated isothermal amplification (in situ RT-LAMP) for detection of Japanese encephalitis viral RNA in host cells. J. Clin. Virol. 2009, 46, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Lietzen, N.; Ohman, T.; Rintahaka, J.; Julkunen, I.; Aittokallio, T.; Matikainen, S.; Nyman, T.A. Quantitative subcellular proteome and secretome profiling of influenza a virus-infected human primary macrophages. PLoS Pathog. 2011, 7, e1001340. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.; Kuo, R.L.; Ma, W.C.; Huang, H.I.; Yu, J.S.; Yen, S.M.; Huang, C.R.; Shih, S.R. Heat shock protein-90-β facilitates enterovirus 71 viral particles assembly. Virology 2013, 443, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Higa, L.M.; Caruso, M.B.; Canellas, F.; Soares, M.R.; Oliveira-Carvalho, A.L.; Chapeaurouge, D.A.; Almeida, P.M.; Perales, J.; Zingali, R.B.; Da Poian, A.T. Secretome of HepG2 cells infected with dengue virus: Implications for pathogenesis. Biochim. Biophys. Acta 2008, 1784, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, R.; Ni, M.; Gill, P.; Lee, A.S. Cell surface relocalization of the endoplasmic reticulum chaperone and unfolded protein response regulator GRP78/BiP. J. Biol. Chem. 2010, 285, 15065–15075. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.; Pelham, H.R. A C-terminal signal prevents secretion of luminal ER proteins. Cell 1987, 48, 899–907. [Google Scholar] [CrossRef]
- Ni, M.; Zhang, Y.; Lee, A.S. Beyond the endoplasmic reticulum: Atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem. J. 2011, 434, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Aoe, T.; Lee, A.J.; van Donselaar, E.; Peters, P.J.; Hsu, V.W. Modulation of intracellular transport by transported proteins: Insight from regulation of COPI-mediated transport. Proc. Natl. Acad. Sci. USA 1998, 95, 1624–1629. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Grandadam, M.; Kwok, K.; Lagache, T.; Siu, Y.L.; Zhang, J.S.; Sayteng, K.; Kudelko, M.; Qin, C.F.; Olivo-Marin, J.C.; et al. KDEL receptors assist dengue virus exit from the endoplasmic reticulum. Cell Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Bruzzone, R.; Wang, P.G. New tricks for KDEL receptors. Oncotarget 2015, 6, 30425–30426. [Google Scholar] [PubMed]
- Wang, R.; Li, K. Host factors in the replication of positive-strand RNA viruses. Chang Gung Med. J. 2012, 35, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Rauschert, N.; Brandlein, S.; Holzinger, E.; Hensel, F.; Muller-Hermelink, H.K.; Vollmers, H.P. A new tumor-specific variant of GRP78 as target for antibody-based therapy. Lab. Investig. 2008, 88, 375–386. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.Y.; Wu, Y.-J.; Chen, H.-S.; Chen, C.-J. A KDEL Retrieval System for ER-Golgi Transport of Japanese Encephalitis Viral Particles. Viruses 2016, 8, 44. https://doi.org/10.3390/v8020044
Wang RY, Wu Y-J, Chen H-S, Chen C-J. A KDEL Retrieval System for ER-Golgi Transport of Japanese Encephalitis Viral Particles. Viruses. 2016; 8(2):44. https://doi.org/10.3390/v8020044
Chicago/Turabian StyleWang, Robert YL, Yu-Jen Wu, Han-Shan Chen, and Chih-Jung Chen. 2016. "A KDEL Retrieval System for ER-Golgi Transport of Japanese Encephalitis Viral Particles" Viruses 8, no. 2: 44. https://doi.org/10.3390/v8020044
APA StyleWang, R. Y., Wu, Y.-J., Chen, H.-S., & Chen, C.-J. (2016). A KDEL Retrieval System for ER-Golgi Transport of Japanese Encephalitis Viral Particles. Viruses, 8(2), 44. https://doi.org/10.3390/v8020044