Evaluation of microRNA Expression in Patients with Herpes Zoster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction
2.3. Analysis of microRNA (miRNA) Profiles by TaqMan Low Density Array (TLDA)
2.4. Confirming and Quantifying Candidate miRNAs through Real-Time qRT-PCR
2.5. Analysis of Target Genes
2.6. Luciferase Assay
2.7. Statistical Analysis
3. Results
3.1. Clinical Characteristics of the Participants
3.2. Using TLDA to Analyze the Expression Profiles of Serum miRNAs
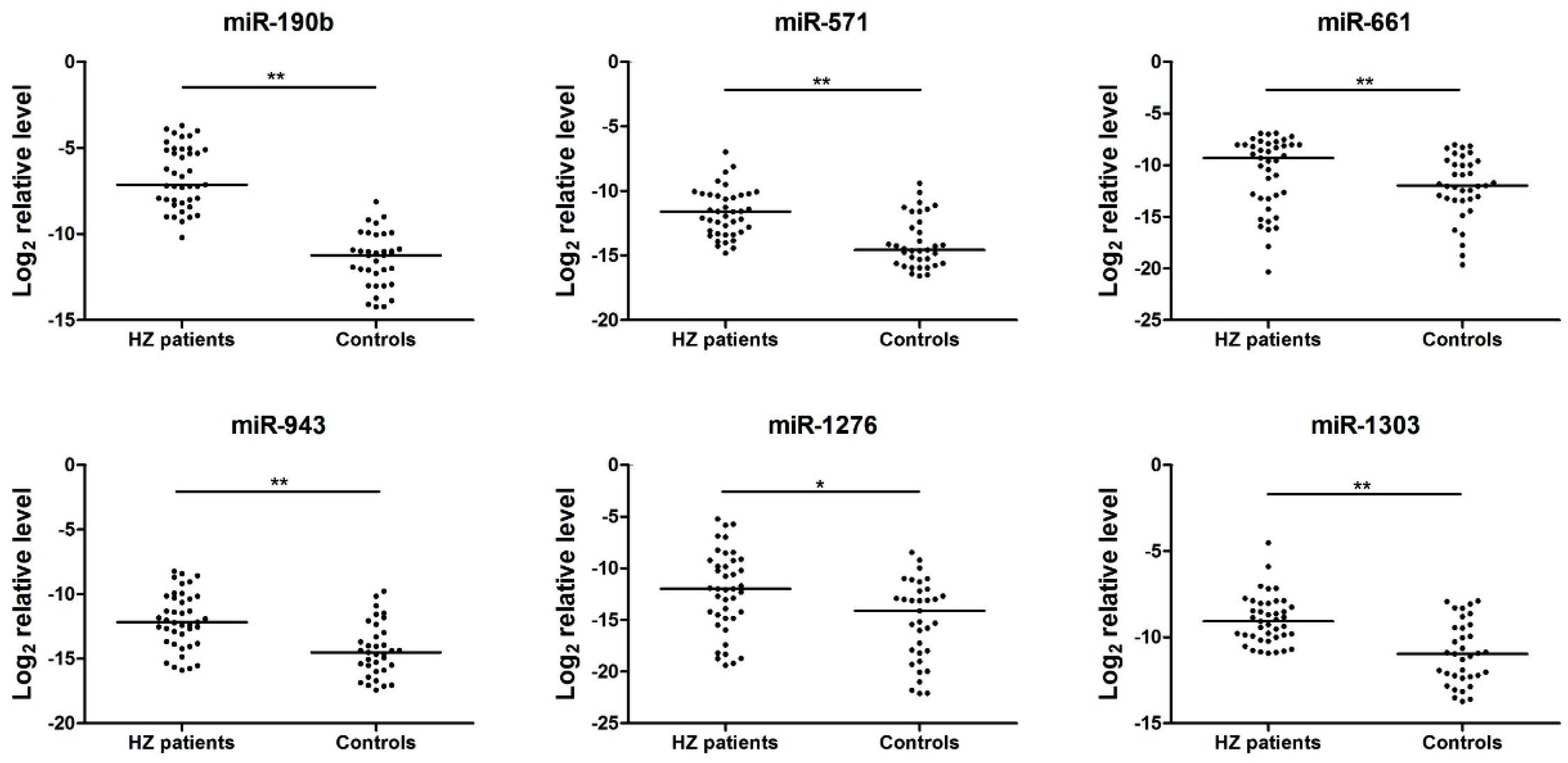
3.3. Analysis of the Serum miRNAs Expression Profiles in the Herpes Zoster (HZ) Group by quantitative reverse transcription PCR (qRT-PCR)
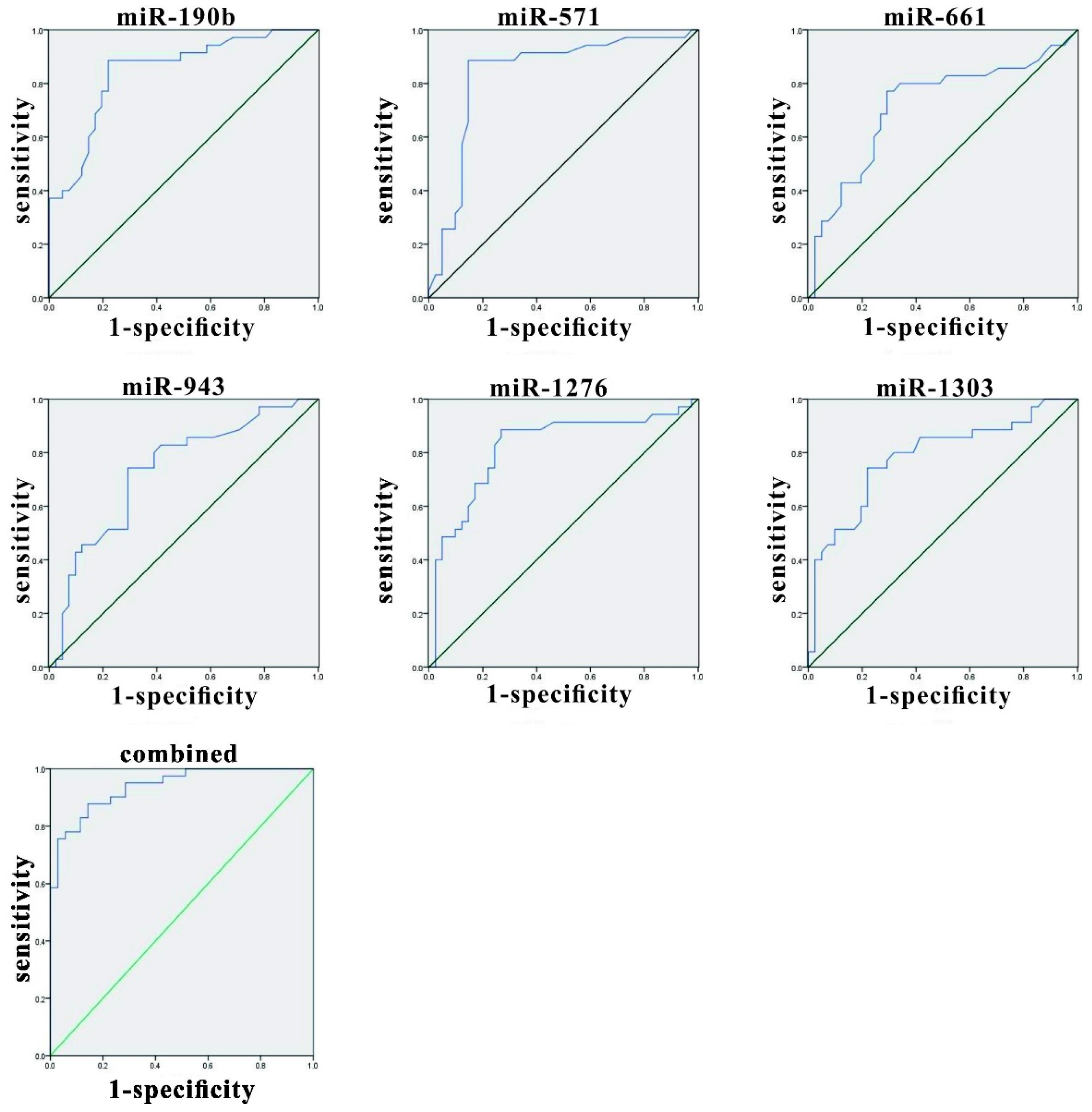
3.4. Evaluation of miRNAs in Herpes Zoster (HZ) and Analysis of Variables Using Receiver Operating Characteristic (ROC) Curves
3.5. Target Gene Prediction
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kang, C.I.; Choi, C.M.; Park, T.S.; Lee, D.J.; Oh, M.D.; Choe, K.W. Incidence of herpes zoster and seroprevalence of varicella-zoster virus in young adults of South Korea. Int. J. Infect. Dis. 2008, 12, 245–247. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.W. Herpes zoster and postherpetic neuralgia. Expert Rev. Vaccines 2010, 9 (Suppl. S3), 21–26. [Google Scholar] [CrossRef] [PubMed]
- Sampathkumar, P.; Drage, L.A.; Martin, D.P. Herpes zoster (shingles) and postherpetic neuralgia. Mayo Clin. Proc. 2009, 84, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Nalamachu, S.; Morley-Forster, P. Diagnosing and managing postherpetic neuralgia. Drugs Aging 2012, 29, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Zajkowska, A.; Garkowski, A.; Swierzbinska, R.; Kulakowska, A.; Krol, M.E.; Ptaszynska-Sarosiek, I.; Nowicka-Cieluszecka, A.; Pancewicz, S.; Czupryna, P.; Moniuszko, A.; et al. Evaluation of Chosen Cytokine Levels among Patients with Herpes Zoster as Ability to Provide Immune Response. PLoS ONE 2016, 11, e0150301. [Google Scholar] [CrossRef] [PubMed]
- Oxman, M.N.; Levin, M.J.; Johnson, G.R.; Schmader, K.E.; Straus, S.E.; Gelb, L.D.; Arbeit, R.D.; Simberkoff, M.S.; Gershon, A.A.; Davis, L.E.; et al. A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N. Engl. J. Med. 2005, 352, 2271–2284. [Google Scholar] [CrossRef] [PubMed]
- Irmak, M.K.; Erdem, U.; Kubar, A. Antiviral activity of salivary microRNAs for ophthalmic herpes zoster. Theor. Biol. Med. Model. 2012, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Ristori, E.; Lopez-Ramirez, M.A.; Narayanan, A.; Hill-Teran, G.; Moro, A.; Calvo, C.F.; Thomas, J.L.; Nicoli, S. A Dicer-miR-107 Interaction Regulates Biogenesis of Specific miRNAs Crucial for Neurogenesis. Dev. Cell 2015, 32, 546–560. [Google Scholar] [CrossRef] [PubMed]
- Broderick, J.A.; Zamore, P.D. MicroRNA therapeutics. Gene Ther. 2011, 18, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Hennessey, P.T.; Sanford, T.; Choudhary, A.; Mydlarz, W.W.; Brown, D.; Adai, A.T.; Ochs, M.F.; Ahrendt, S.A.; Mambo, E.; Califano, J.A. Serum microRNA biomarkers for detection of non-small cell lung cancer. PLoS ONE 2012, 7, e32307. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, M.; Lin, G.; Sun, S.; Li, X.; Qi, J.; Li, J. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS ONE 2012, 7, e47003. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.; Bajracharya, S.D.; Yuen, P.S.; Zhou, H.; Star, R.A.; Illei, G.G.; Alevizos, I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.W.; Lin, L.Z.; Hsu, S.D.; Hsu, J.B.; Huang, H.D. ViTa: Prediction of host microRNAs targets on viruses. Nucleic Acids Res. 2007, 35, D381–D385. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhu, Z.; Shi, Z.; Ge, Y.; Zhao, K.; Zhou, M.; Cui, L. Dysregulated microRNA expression in serum of non-vaccinated children with varicella. Viruses 2014, 6, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, R.; Jeang, K.T. The roles of microRNAs in mammalian virus infection. Biochim. Biophys. Acta 2008, 1779, 706–711. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Xu, S.; Liu, X.; Zhang, Q.; Xu, X.; Liu, M.; Hua, M.; Li, N.; Yao, H.; Cao, X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat. Immunol. 2011, 12, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Cizeron-Clairac, G.; Lallemand, F.; Vacher, S.; Lidereau, R.; Bieche, I.; Callens, C. MiR-190b, the highest up-regulated miRNA in ERalpha-positive compared to ERalpha-negative breast tumors, a new biomarker in breast cancers? BMC Cancer 2015, 15, 499. [Google Scholar] [CrossRef] [PubMed]
- Au, K.Y.; Pong, J.C.; Ling, W.L.; Li, J.C. MiR-1303 Regulates Mycobacteria Induced Autophagy by Targeting Atg2B. PLoS ONE 2016, 11, e0146770. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hwang, J.S.; Lee, B.; Hong, J.; Lee, S. Newly Identified Cancer-Associated Role of Human Neuronal Growth Regulator 1 (NEGR1). J. Cancer 2014, 5, 598–608. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, Q.; Zhan, Y.; Chen, X.; Yu, Q.; Zhang, J.; Wang, Y.; Xu, X.J.; Zhu, L. Transcriptome sequencing uncovers a three-long noncoding RNA signature in predicting breast cancer survival. Sci. Rep. 2016, 6, 27931. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Torben, W.; Kenway, C.S.; Schiro, F.R.; Mohan, M. Longitudinal Examination of the Intestinal Lamina Propria Cellular Compartment of Simian Immunodeficiency Virus-Infected Rhesus Macaques Provides Broader and Deeper Insights into the Link between Aberrant MicroRNA Expression and Persistent Immune Activation. J. Virol. 2016, 90, 5003–5019. [Google Scholar] [PubMed]
- Salimi, Z.; Sadeghi, S.; Tabatabaeian, H.; Ghaedi, K.; Fazilati, M. rs11895168 C allele and the increased risk of breast cancer in Isfahan population. Breast 2016, 28, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.C.; Liang, Z.D.; Pei, L. Propofol-induced rno-miR-665 targets BCL2L1 and influences apoptosis in rodent developing hippocampal astrocytes. Neurotoxicology 2015, 51, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Kamat, V.; Paluru, P.; Myint, M.; French, D.L.; Gadue, P.; Diamond, S.L. MicroRNA screen of human embryonic stem cell differentiation reveals miR-105 as an enhancer of megakaryopoiesis from adult CD34+ cells. Stem Cells 2014, 32, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Rawlings-Goss, R.A.; Campbell, M.C.; Tishkoff, S.A. Global population-specific variation in miRNA associated with cancer risk and clinical biomarkers. BMC Med. Genom. 2014, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wan, S.; Yang, T.; Niu, D.; Zhang, A.; Yang, C.; Cai, J.; Wu, J.; Song, J.; Zhang, C.Y.; et al. Increased serum microRNAs are closely associated with the presence of microvascular complications in type 2 diabetes mellitus. Sci. Rep. 2016, 6, 20032. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | Herpes Zoster (HZ) Group (n = 41) | Healthy Control Group (n = 35) | |
|---|---|---|---|
| Mean age in years (range) | 58.3 (26–85) | 55.9 (22–79) | |
| Sex (percentage) | |||
| Male | 23 (56.1%) | 16 (45.7%) | |
| Female | 18 (43.9%) | 19 (54.3%) | |
| chi square test | p = 0.367 | ||
| HZ localization (percentage) | |||
| C2-L1 | 27 (65.9%) | 0 | |
| Facial nerve dermatomes | 7 (17.1%) | 0 | |
| Ophthalmic nerve dermatomes | 4 (9.7%) | 0 | |
| Ramsay Hunt syndrome | 3 (7.3%) | 0 | |
| miRNA | ΔCtHZ | ΔCtcontrol | ΔΔCt |
|---|---|---|---|
| miR-1243 | 7.86978 | 23.06114 | −15.1914 |
| miR-190b | 7.273803 | 21.02214 | −13.7483 |
| miR-520d-3p | 8.989834 | 22.06215 | −13.0723 |
| miR-627 | 8.4600315 | 21.14534 | −12.6853 |
| miR-541 | 10.102917 | 22.06213 | −11.9592 |
| miR-571 | 12.504569 | 23.02614 | −10.5216 |
| miR-1238 | 12.89762 | 23.16214 | −10.2645 |
| miR-1303 | 11.215716 | 21.06214 | −9.84642 |
| miR-943 | 13.273769 | 23.06799 | −9.79422 |
| miR-1225-3p | 13.785332 | 23.40662 | −9.62129 |
| miR-1276 | 14.727001 | 23.58135 | −8.85435 |
| miR-661 | 11.416535 | 20.06211 | −8.64558 |
| miR-605 | 12.586351 | 21.06514 | −8.47879 |
| miR-1 | 15.524136 | 21.14534 | −5.6212 |
| miR-1233 | 6.993761 | 12.05311 | −5.05935 |
| miR-99a | 10.33614 | 13.75413 | −3.41799 |
| miR-1825 | 14.144214 | 16.97566 | −2.83145 |
| miR-598 | 7.228752 | 9.272097 | −2.04335 |
| miRNA | AUC | SE | Asymptotic Significance | Asymptotic 95% CI | |
|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||
| miR-190b | 0.845 | 0.045 | <0.001 | 0.756 | 0.934 |
| miR-571 | 0.837 | 0.051 | <0.001 | 0.737 | 0.936 |
| miR-1276 | 0.716 | 0.062 | 0.001 | 0.595 | 0.837 |
| miR-1303 | 0.733 | 0.058 | 0.001 | 0.618 | 0.847 |
| miR-943 | 0.818 | 0.052 | <0.001 | 0.716 | 0.921 |
| miR-661 | 0.784 | 0.054 | <0.001 | 0.678 | 0.890 |
| Combined miRNAs | 0.939 | 0.025 | <0.001 | 0.890 | 0.987 |
| miRNAs | Go Term | Genes |
|---|---|---|
| miR-190b | Nervous system | NEUROD1, NLGN1, NEGR1, NRG3, NAV3 |
| Immune system | MTMR6, BNIP3L | |
| miR-571 | Nervous system | PAINP, NLGN3, NYAP2 |
| Immune system | FASLG, FCAMR, CASP8, IGSF6, IGLL5 | |
| miR-1303 | Nervous system | NEGR1, NXPE2 |
| Immune system | IGSF1, BAG2, CASP14, IGJ | |
| miR-943 | Nervous system | CDNF, NRP1 |
| Immune system | IL8, CASP10 | |
| miR-1276 | Nervous system | NMB |
| Immune system | BMP2, CASP9, BNIP2, ILDR1, BCL2L1 | |
| miR-661 | Nervous system | DRAXIN, IGF1, COL4A4, NRCAM, CDK5R1 |
| Immune system | TCTA, IL17REL, IL16, BOK, SIX4 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Huang, Y.; Zhang, Y.; He, N. Evaluation of microRNA Expression in Patients with Herpes Zoster. Viruses 2016, 8, 326. https://doi.org/10.3390/v8120326
Li X, Huang Y, Zhang Y, He N. Evaluation of microRNA Expression in Patients with Herpes Zoster. Viruses. 2016; 8(12):326. https://doi.org/10.3390/v8120326
Chicago/Turabian StyleLi, Xihan, Ying Huang, Yucheng Zhang, and Na He. 2016. "Evaluation of microRNA Expression in Patients with Herpes Zoster" Viruses 8, no. 12: 326. https://doi.org/10.3390/v8120326
APA StyleLi, X., Huang, Y., Zhang, Y., & He, N. (2016). Evaluation of microRNA Expression in Patients with Herpes Zoster. Viruses, 8(12), 326. https://doi.org/10.3390/v8120326





