Capsule-Targeting Depolymerase, Derived from Klebsiella KP36 Phage, as a Tool for the Development of Anti-Virulent Strategy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phage, Bacterial Strains, and Culture Conditions
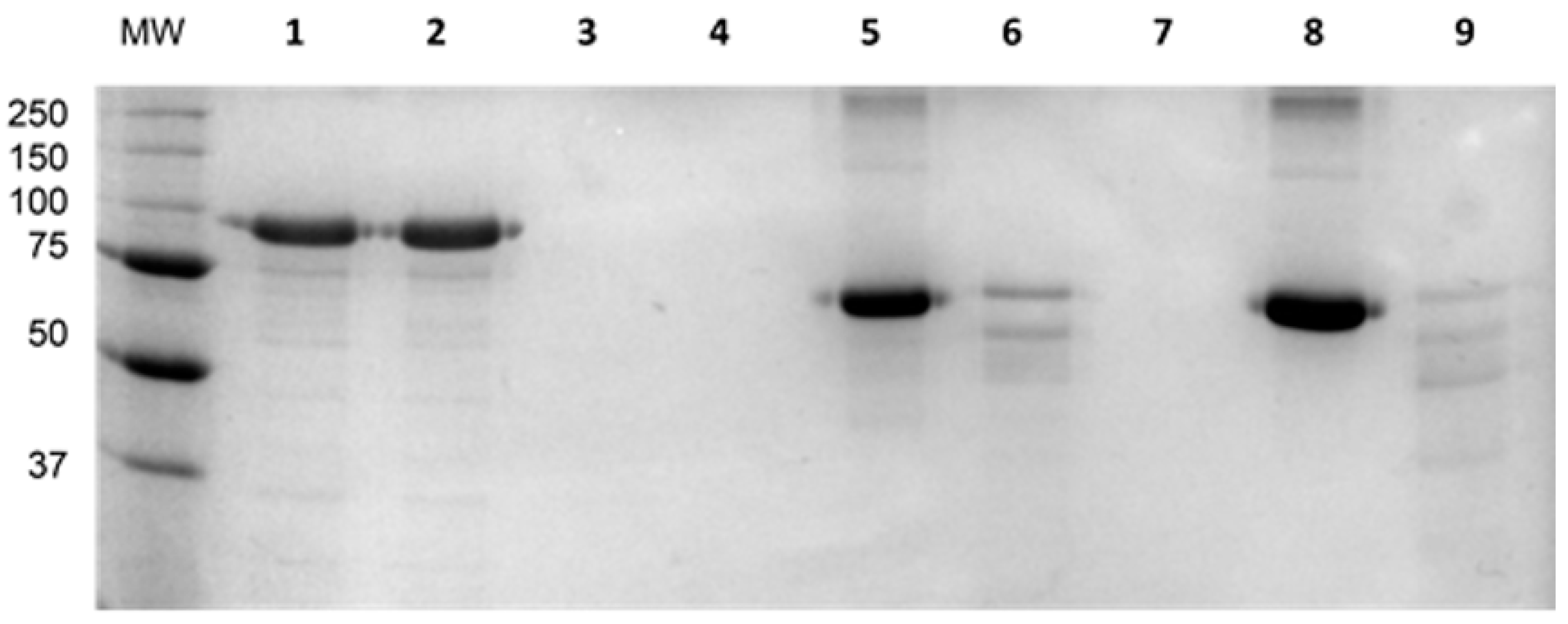
2.2. Cloning, Expression, and Purification
2.3. Protein Analysis Assays
2.4. Extraction and Purification of Exopolysaccharide (EPS)
2.5. Functional Assays
2.6. Galleria Mellonella Larvae Infection Model
2.7. Antimicrobial Susceptibility Testing
2.8. Light Scattering Experiments
2.9. Circular Dichroism (CD) Studies
3. Results
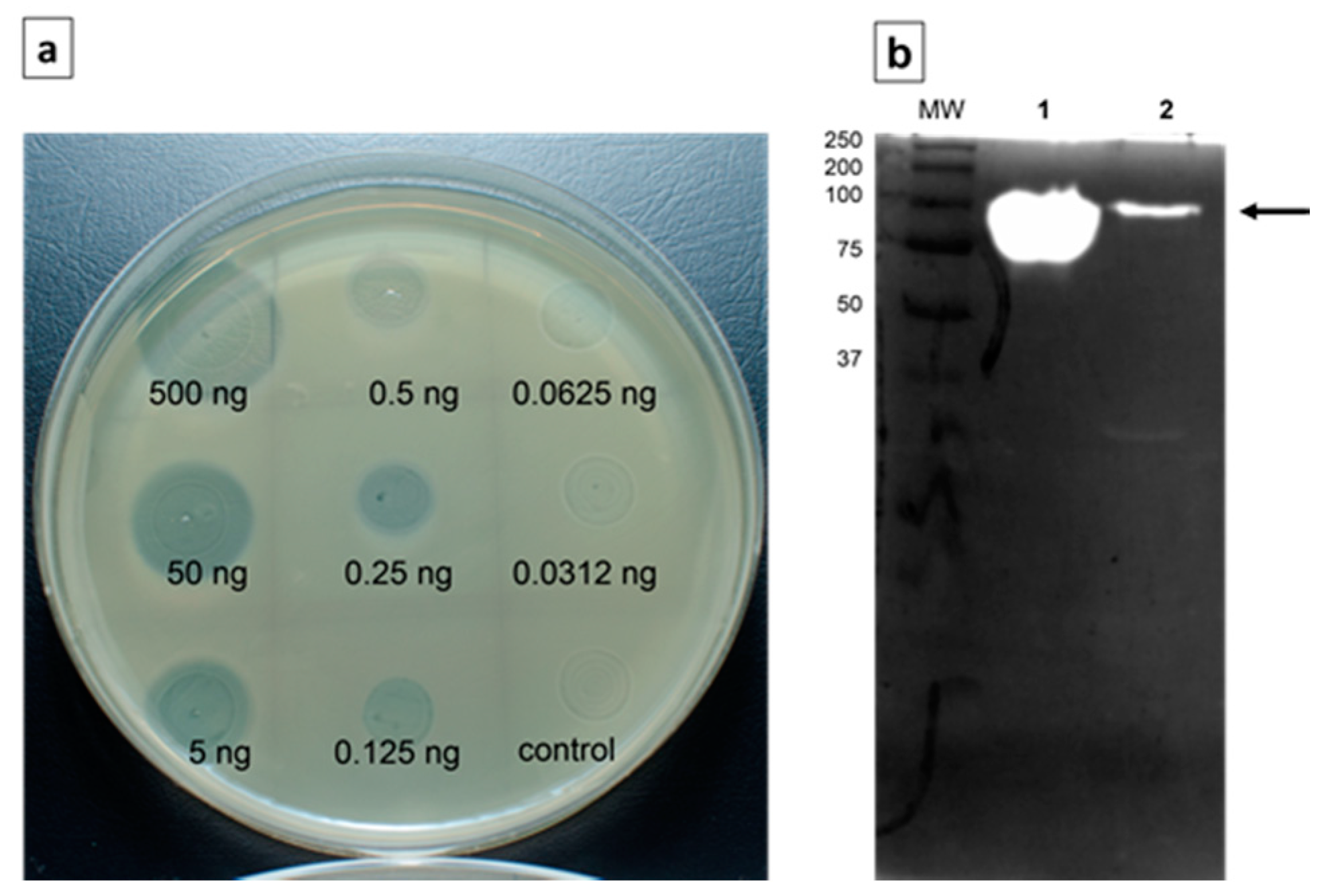
3.1. Identification of Phage KP36 gp50 as a Putative EPS Depolymerase
3.2. Recombinant depolymerase enzyme encoded by Klebsiella phage KP36 (depoKP36) Degrades Bacterial EPS and Shows a Narrow Spectrum Activity Against Specific K. pneumoniae Strains
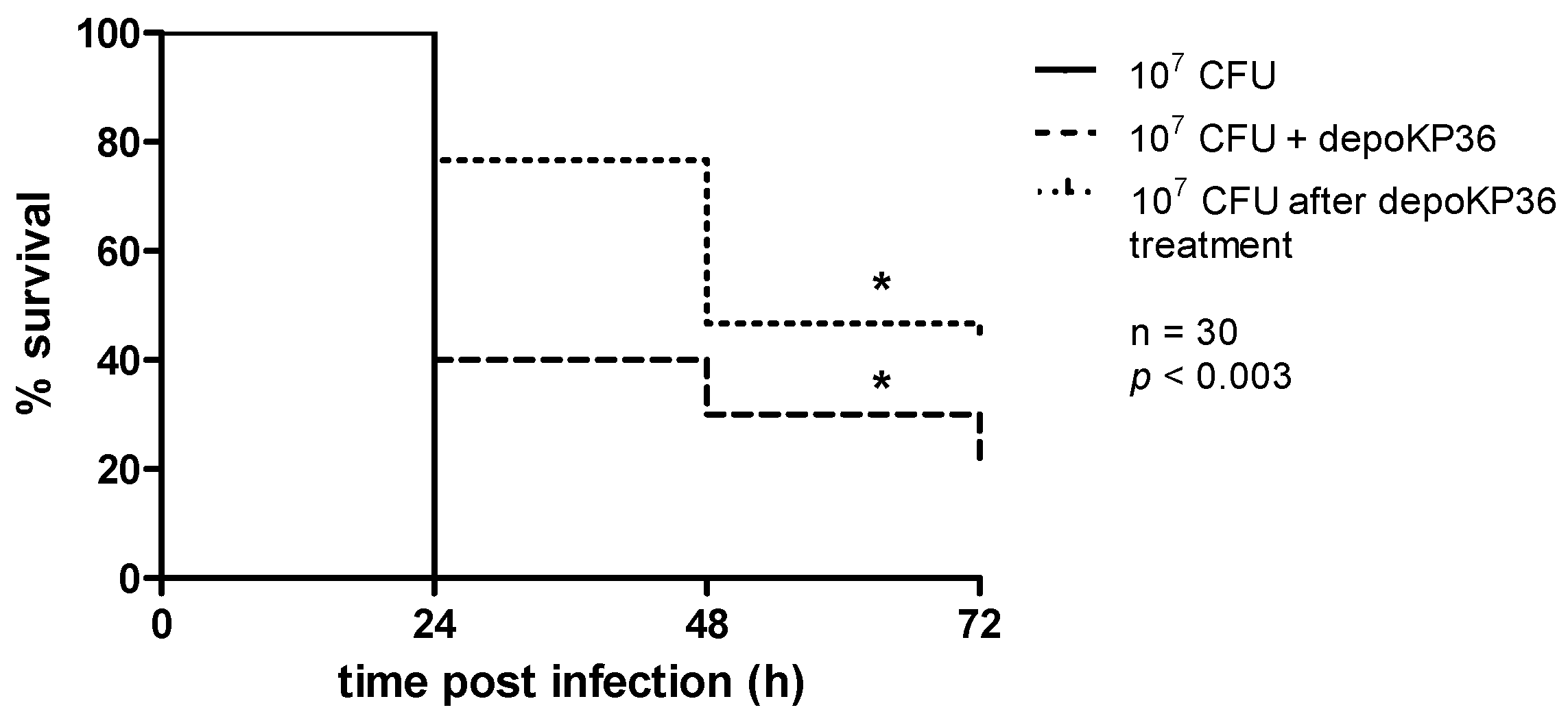
3.3. Anti-Virulence Efficacy of depoKP36 in the G. mellonella Infection Model
3.4. DepoKP36 Do Not Affect the Action of Antibiotics Against K. pneumoniae Strains
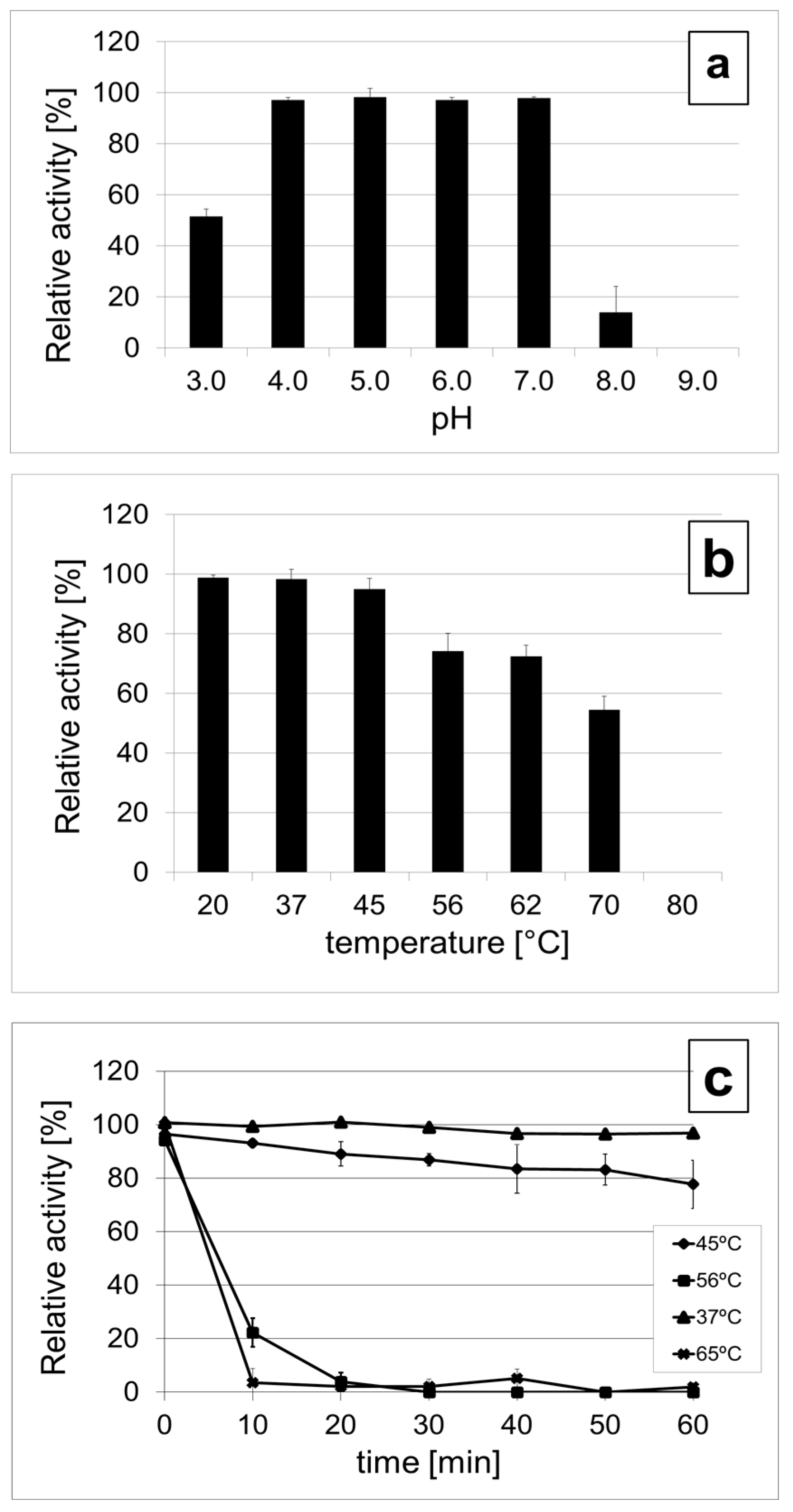
3.5. DepoKP36 Remains Stable at Moderately Acidic Conditions and Is Mesophilic
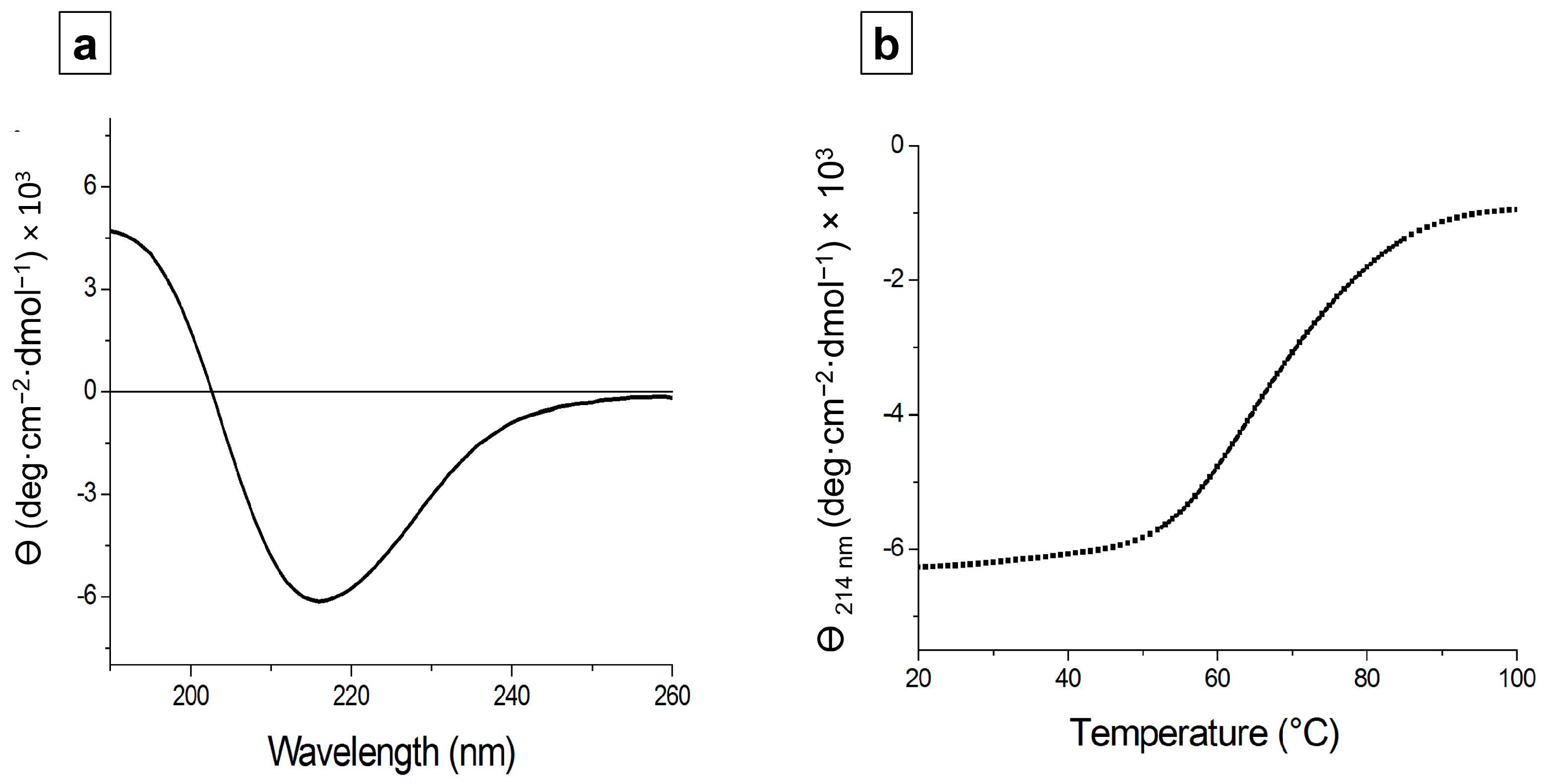
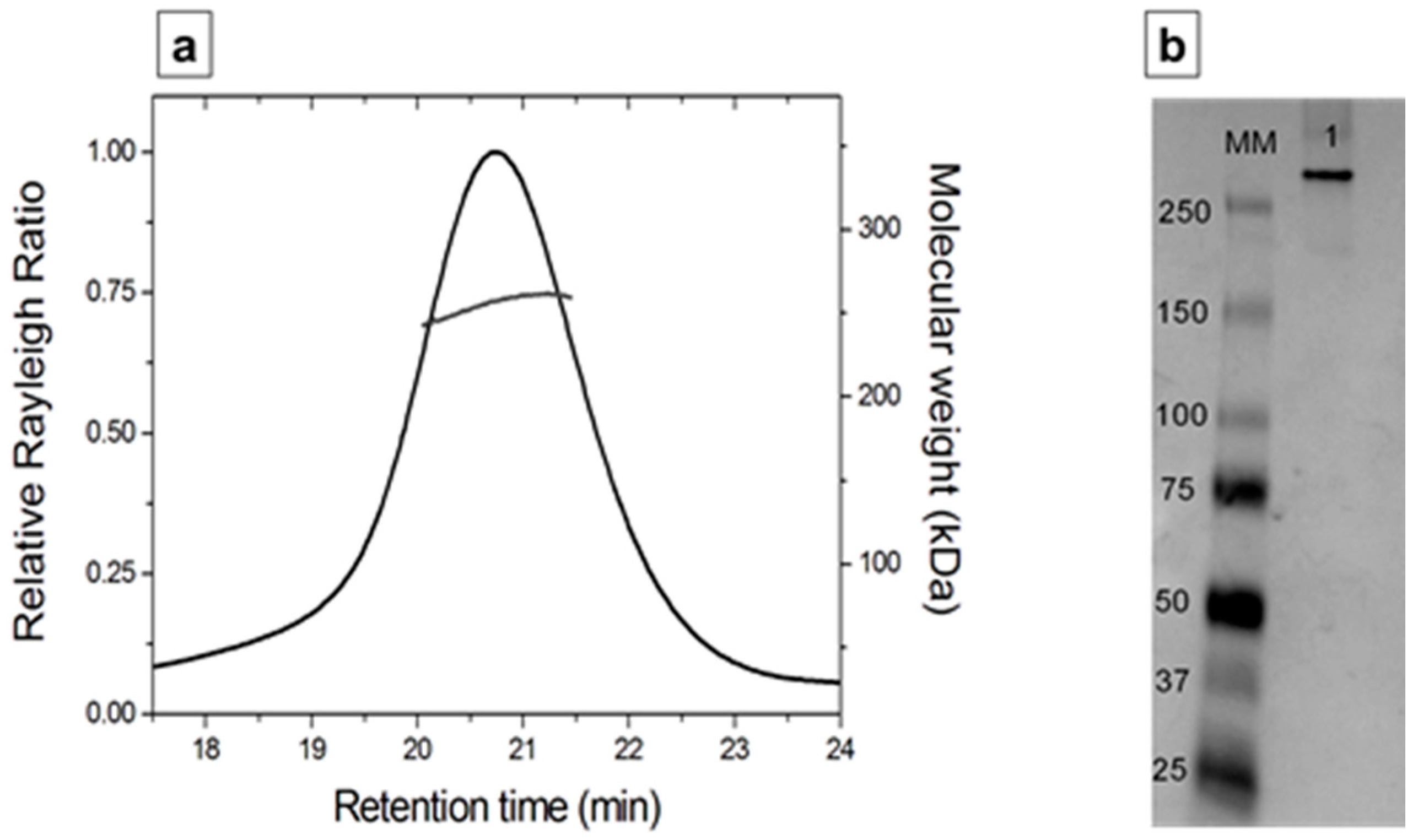
3.6. Structural Characterization of depoKP36 in Solution
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [PubMed]
- Keynan, Y.; Rubinstein, E. The changing face of Klebsiella pneumoniae infections in the community. Int. J. Antimicrob. Agents 2007, 30, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Coque, T.M.; Baquero, F.; Cantón, R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008, 13, 5437–5453. [Google Scholar]
- Munoz-Price, L.S.; Poirel, L.; Bonomo, R.A.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K.; et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef]
- Robilotti, E.; Deresinski, S. Carbapenemase-producing Klebsiella pneumoniae. F1000Prime Rep. 2014, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Fang, H.C.; Yang, H.C.; Lin, T.L.; Hsieh, P.F.; Tsai, F.C.; Keynan, Y.; Wang, J.T. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J. Clin. Microbiol. 2008, 46, 2231–2240. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Lin, T.L.; Chen, Y.H.; Hsu, C.R.; Hsieh, P.F.; Wu, M.C.; Wang, J.T. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS ONE 2013, 8, e80670. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.R.; Lin, T.L.; Pan, Y.J.; Hsieh, P.F.; Wang, J.T. Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLoS ONE 2013, 8, e70092. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, Y.; Liu, C.; Chen, Z.; Zhou, D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014, 9, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother. 1996, 40, 2517–2522. [Google Scholar] [PubMed]
- Samson, J.E.; Magadán, A.H.; Sabri, M.; Moineau, S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Microbiol. 2013, 11, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H.; Park, B.H. An enzyme produced by a phage host-cell system. II. The properties of the polysaccharide depolymerase. Virology 1956, 2, 719–736. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B. Bacteriophages and phage-derived proteins-application approaches. Curr. Med. Chem. 2015, 22, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Bessler, W.; Freund-Mölbert, E.; Knüfermann, H.; Rudolph, C.; Thurow, H.; Stirm, S. A bacteriophage-induced depolymerase active on Klebsiella K11 capsular polysaccharide. Virology 1973, 56, 134–151. [Google Scholar] [CrossRef]
- Niemann, H.; Kwiatkowski, B.; Westphal, U.; Stirm, S. Klebsiella serotypes 25 capsular polysaccharide: Primary structure and depolymerization by a bacteriophage-borne glycanase. J. Bacteriol. 1977, 130, 366–374. [Google Scholar] [PubMed]
- Rieger-Hug, D.; Stirm, S. Comparative study of host capsule depolymerase associated with Klebsiella bacteriophages. Virology 1981, 113, 363–378. [Google Scholar] [CrossRef]
- Lin, T.L.; Hsieh, P.F.; Huang, Y.T.; Lee, W.C.; Tsai, Y.T.; Su, P.A.; Pan, Y.J.; Hsu, C.R.; Wu, M.C.; Wang, J.T. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: Implication in typing and treatment. J. Infect. Dis. 2014, 210, 1734–1744. [Google Scholar] [CrossRef] [PubMed]
- Dutton, G.G.; DiFabio, J.L.; Leek, D.M.; Merrifield, E.H.; Nunn, J.R.; Stephen, A.M. Preparation of oligosaccharides by the action of bacteriophage-borne enzymes on Klebsiella capsular polysaccharides. Carbohydr. Res. 1981, 97, 127–138. [Google Scholar] [CrossRef]
- Mou, H.; Wang, J.; Jiang, X.; Liu, Z. Preparation and properties bacteriophage-borne enzyme degrading bacterial exopolysaccharide. High Technol. Lett. 2008, 14, 210–215. [Google Scholar]
- Pan, Y.J.; Lin, T.L.; Lin, Y.T.; Su, P.A.; Chen, C.T.; Hsieh, P.F.; Hsu, C.R.; Chen, C.C.; Hsieh, Y.C.; Wang, J.T. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob. Agents Chemother. 2015, 59, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Kęsik-Szeloch, A.; Drulis-Kawa, Z.; Weber-Dąbrowska, B.; Kassner, J.; Majkowska-Skrobek, G.; Augustyniak, D.; Lusiak-Szelachowska, M.; Zaczek, M.; Górski, A.; Kropinski, A.M. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol. J. 2013, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.D.; McAllister, T.A.; Nash, J.H.; Kropinski, A.M.; Stanford, K. Four E. coli O157:H7 phages: A new bacteriophage genus and taxonomic classification of T1-like phages. PLoS ONE 2014, 10, e0142287. [Google Scholar] [CrossRef] [PubMed]
- Brisse, S.; Passet, V.; Björk Haugaard, A.; Babosan, A.; Kassis-Chikhani, N.; Struve, C.; Decrée, D. Wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J. Clin. Microbiol. 2013, 51, 4073–4078. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond.) 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Clifton, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Bales, P.M.; Renke, E.M.; May, S.L.; Shen, Y.; Nelson, D.C. Purification and characterization of biofilm-associated EPS exopolysaccharides from ESKAPE organisms and other pathogens. PLoS ONE 2013, 8, e67950. [Google Scholar] [CrossRef] [PubMed]
- Bellemann, P.; Bereswill, S.; Berger, S.; Geider, K. Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. Int. J. Biol. Macromol. 1994, 16, 290–296. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, Approved Standard—Ninth Edition; CLSI document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed]
- Buchan, D.W.A.; Minneci, F.; Nugent, T.C.O.; Bryson, K.; Jones, D.T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013, 41, W340–W348. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.; Mayans, O.; Pickersgill, R. Structure and evolution of parallel beta-helix proteins. J. Struct. Biol. 1998, 122, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Mitraki, A.; Miller, S.; van Raaij, M.J. Review: Conformation and folding of novel beta-structural elements in viral fiber proteins: The triple beta-spiral and triple beta-helix. J. Struct. Biol. 2002, 137, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Insua, J.L.; Llobet, E.; Moranta, D.; Pérez-Gutiérrez, C.; Tomás, A.; Garmendia, J.; Bengoechea, J.A. Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect. Immun. 2013, 81, 3552–3565. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Harjai, K.; Chhibber, S. Structural changes induced by a lytic bacteriophage make ciprofloxacin effective against older biofilm of Klebsiella pneumoniae. Biofouling 2010, 26, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Kassa, T.; Chhibber, S. Thermal treatment of the bacteriophage lysate of Klebsiella pneumoniae B5055 as a step for the purification of capsular depolymerase enzyme. J. Virol. Methods 2012, 179, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Chhibber, S.; Nag, D.; Bansal, S. Inhibiting biofilm formation by Klebsiella pneumoniae B5055 using an iron antagonizing molecule and a bacteriophage. BMC Microbiol. 2013, 13, 174. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, G.; Mo, Z.; Chai, Z.; Shang, A.; Mou, H. Properties of Klebsiella phage P13 and associated exopolysaccharide depolymerase. J. Ocean Univ. Chin. 2014, 13, 163–168. [Google Scholar] [CrossRef]
- Mushtaq, N.; Redpath, M.B.; Luzio, J.P.; Taylor, P.W. Treatment of experimental E. coli infection with recombinant bacteriophage-derived capsule depolymerase. J. Antimicrob. Chemother. 2005, 56, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Harjai, K.; Chhibber, S. Depolymerase improves gentamicin efficacy during Klebsiella pneumoniae induced murine infection. BMC Infect. Dis. 2014, 14, 456. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, N.; Redpath, M.B.; Luzio, J.P.; Taylor, P.W. Prevention and cure of systemic E. coli K1 infection by modification of the bacterial phenotype. Antimicrob. Agents Chemother. 2004, 48, 1503–1508. [Google Scholar] [CrossRef] [PubMed]
- Shang, A.; Liu, Y.; Wang, J.; Mo, Z.; Li, G.; Mou, H. Complete nucleotide sequence of Klebsiella phage P13 and prediction of an EPS depolymerase gene. Virus Genes 2015, 50, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Hoyles, L.; Murphy, J.; Neve, H.; Heller, K.J.; Turton, J.F.; Mahony, J.; Sanderson, J.D.; Hudspith, B.; Gibson, G.R.; McCartney, A.L.; et al. Klebsiella pneumoniae subsp. pneumoniae-bacteriophage combination from the caecal effluent of a healthy woman. Peer J. 2015, 3, e1061. [Google Scholar] [CrossRef] [PubMed]
- Drulis-Kawa, Z.; Mackiewicz, P.; Kęsik-Szeloch, A.; Maciaszczyk-Dziubinska, E.; Weber-Dąbrowska, B.; Dorotkiewicz-Jach, A.; Augustyniak, D.; Majkowska-Skrobek, G.; Bocer, T.; Empel, J.; et al. Isolation and characterization of KP34-a novel φKMV-like bacteriophage for Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2011, 90, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.E.; Pourhossein, M.; Waterhouse, A.; Hudson, T.; Goldrick, M.; Derrick, J.P.; Roberts, I.S. The K5 lyase KflA combines a viral tail spike structure with a bacterial polysaccharide lyase mechanism. J. Biol. Chem. 2010, 285, 23963–23969. [Google Scholar] [CrossRef] [PubMed]
- Hooton, S.P.; Timms, A.R.; Rowsell, J.; Wilson, R.; Connerton, I.F. Salmonella typhimurium-specific bacteriophage ΦSH19 and the origins of species specificity in the Vi01-like phage family. Virol. J. 2010, 8, 498. [Google Scholar] [CrossRef] [PubMed]
- Joseleau, J.P.; Marais, M.F. Structure of the capsular polysaccharide of Klebsiella K-type 63. Carbohydr. Res. 1979, 77, 183–190. [Google Scholar] [CrossRef]
- Niemann, H.; Chakraborty, A.K.; Friebolin, H.; Stirm, S. Primary structure of the E. coli serotype K42 capsular polysaccharide and its serological identity with the Klebsiella K63 polysaccharide. J. Bacteriol. 1978, 133, 390–391. [Google Scholar] [PubMed]
- Simpson, D.J.; Sacher, J.C.; Szymanski, C.M. Exploring the interactions between bacteriophage-encoded glycan binding proteins and carbohydrates. Curr. Opin. Struct. Biol. 2015, 34, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Broeker, N.K.; Gohlke, U.; Muller, J.J.; Uetrecht, C.; Heinemann, U.; Seckler, R.; Barbirz, S. Single amino acid exchange in bacteriophage HK620 tailspike protein results in thousand-fold increase of its oligosaccharide affinity. Glycobiology 2013, 23, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Drexler, K.; Dannull, J.; Hindennach, I.; Mutschler, B.; Henning, U. Single mutations in a gene for a tail fiber component of an E. coli phage can cause an extension from a protein to a carbohydrate as a receptor. J. Mol. Biol. 1991, 219, 655–663. [Google Scholar] [CrossRef]
- Steinbacher, S.; Baxa, U.; Miller, S.; Weintraub, A.; Seckler, R.; Huber, R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc. Natl. Acad. Sci. USA 1996, 93, 10584–10588. [Google Scholar] [CrossRef] [PubMed]
- Muller, J.J.; Barbirz, S.; Heinle, K.; Freiberg, A.; Seckler, R.; Heinemann, U. An intersubunit active site between supercoiled parallel beta helices in the trimeric tailspike endorhamnosidase of Shigella flexneri phage Sf6. Structure 2008, 16, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Barbirz, S.; Muller, J.J.; Uetrecht, C.; Clark, A.J.; Heinemann, U.; Seckler, R. Crystal structure of E. coli phage HK620 tailspike: Podoviral tailspike endoglycosidase modules are evolutionarily related. Mol. Microbiol. 2008, 69, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Leiman, P.G.; Li, L.; Grimes, S.; Anderson, D.L.; Rossmann, M.G. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol. Cell 2009, 34, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.; Fiedler, C.; Grassl, R.; Biebl, M.; Rachel, R.; Hermo-Parrado, X.L.; Llamas-Saiz, A.L.; Seckler, R.; Miller, S.; van Raaij, M.J. Structure of the receptor-binding protein of bacteriophage det7: A podoviral tail spike in a myovirus. J. Virol. 2008, 82, 2265–2273. [Google Scholar] [CrossRef] [PubMed]
- Andres, D.; Roske, Y.; Doering, C.; Heinemann, U.; Seckler, R.; Barbirz, S. Tail morphology controls DNA release in two Salmonella phages with one lipopolysaccharide receptor recognition system. Mol. Microbiol. 2012, 83, 1244–1253. [Google Scholar] [CrossRef] [PubMed]
- Manning, M.; Colón, W. Structural basis of protein kinetic stability: Resistance to sodium dodecyl sulfate suggests a central role for rigidity and a bias toward beta-sheet structure. Biochemistry 2004, 43, 11248–11254. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Sasaoka, H.; Sasa, K.; Hirai, H.; Hachiya, K.; Moriyama, Y. Size and mobility of sodium dodecyl-sulfate bovine serum-albumin complex as studied by dynamic light-scattering and electrophoretic light-scattering. J. Coll. Int. Sci. 1992, 154, 385–392. [Google Scholar] [CrossRef]
- Ibel, K.; May, R.P.; Kirschner, K.; Szadkowski, H.; Mascher, E.; Lundahl, P. Protein-decorated micelle structure of sodium-dodecyl-sulfate-protein complexes as determined by neutron scattering. Eur. J. Biochem. 1990, 190, 311–318. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majkowska-Skrobek, G.; Łątka, A.; Berisio, R.; Maciejewska, B.; Squeglia, F.; Romano, M.; Lavigne, R.; Struve, C.; Drulis-Kawa, Z. Capsule-Targeting Depolymerase, Derived from Klebsiella KP36 Phage, as a Tool for the Development of Anti-Virulent Strategy. Viruses 2016, 8, 324. https://doi.org/10.3390/v8120324
Majkowska-Skrobek G, Łątka A, Berisio R, Maciejewska B, Squeglia F, Romano M, Lavigne R, Struve C, Drulis-Kawa Z. Capsule-Targeting Depolymerase, Derived from Klebsiella KP36 Phage, as a Tool for the Development of Anti-Virulent Strategy. Viruses. 2016; 8(12):324. https://doi.org/10.3390/v8120324
Chicago/Turabian StyleMajkowska-Skrobek, Grażyna, Agnieszka Łątka, Rita Berisio, Barbara Maciejewska, Flavia Squeglia, Maria Romano, Rob Lavigne, Carsten Struve, and Zuzanna Drulis-Kawa. 2016. "Capsule-Targeting Depolymerase, Derived from Klebsiella KP36 Phage, as a Tool for the Development of Anti-Virulent Strategy" Viruses 8, no. 12: 324. https://doi.org/10.3390/v8120324
APA StyleMajkowska-Skrobek, G., Łątka, A., Berisio, R., Maciejewska, B., Squeglia, F., Romano, M., Lavigne, R., Struve, C., & Drulis-Kawa, Z. (2016). Capsule-Targeting Depolymerase, Derived from Klebsiella KP36 Phage, as a Tool for the Development of Anti-Virulent Strategy. Viruses, 8(12), 324. https://doi.org/10.3390/v8120324







