Structural Maturation of HIV-1 Reverse Transcriptase—A Metamorphic Solution to Genomic Instability
Abstract
:1. Introduction
2. Structural Background
2.1. Nomenclature and Constructs Studied
2.2. Structural Features of the RT Heterodimer
2.3. The Alternate Folds of the Polymerase Domain
2.4. Structural Characterization of the p66 Monomer
3. The Metamorphic Transition of the Polymerase Domain
3.1. Monomer Domain Dissociation
3.2. Formation of New Interfaces in the Extended Conformation
4. Dimerization-Induced Structural Changes
4.1. Dimer Formation
4.2. Maturation of the p66’ Subunit Involves a Molecular Tug-of-War
4.3. Tyr427-Triggered RH Domain Unfolding
4.4. Kinetic Barriers Stabilize Transient Intermediates
5. Indirect Effects on Dimer Stability
5.1. The Structural Equilibrium of the Polymerase Domain
5.2. Effects of NNRTIs on RT Maturation
5.3. Mutational Effects on RT Maturation
5.4. Mutational Perturbation of the Polymerase-RH Tug-of-War
6. Maturation within the Virion
7. Maturation of HIV-2 Reverse Transcriptase
8. Reverse Transcriptase Maturation as an Attractive Drug Target
Acknowledgments
Conflicts of Interest
References
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 2003, 11, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Loeb, L.A.; Preston, B.D. Lethal mutagenesis of HIV. Virus Res. 2005, 107, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.D.; Bebenek, K.; Kunkel, T.A. The Accuracy of Reverse-Transcriptase from Hiv-1. Science 1988, 242, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Sierra, S.; Kupfer, B.; Kaiser, R. Basics of the virology of HIV-1 and its replication. J. Clin. Virol. 2005, 34, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Belshaw, R.; Pybus, O.G.; Rambaut, A. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 2007, 17, 1496–1504. [Google Scholar] [CrossRef] [PubMed]
- Biswas, P.; Jiang, X.; Pacchia, A.L.; Dougherty, J.P.; Peltz, S.W. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J. Virol. 2004, 78, 2082–2087. [Google Scholar] [CrossRef] [PubMed]
- Jacks, T.; Power, M.D.; Masiarz, F.R.; Luciw, P.A.; Barr, P.J.; Varmus, H.E. Characterization of Ribosomal Frameshifting in Hiv-1 Gag-Pol Expression. Nature 1988, 331, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Mouzakis, K.D.; Lang, A.L.; Vander Meulen, K.A.; Easterday, P.D.; Butcher, S.E. HIV-1 frameshift efficiency is primarily determined by the stability of base pairs positioned at the mRNA entrance channel of the ribosome. Nucleic Acids Res. 2013, 41, 1901–1913. [Google Scholar] [CrossRef] [PubMed]
- Geyer, M.; Fackler, O.T.; Peterlin, B.M. Structure-function relationships in HIV-1 Nef. EMBO Rep. 2001, 2, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Malim, M.H.; Emerman, M. HIV-1 accessory proteins—Ensuring viral survival in a hostile environment. Cell Host Microbe 2008, 3, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Schubert, U.; Bour, S.; Ferrer-Montiel, A.V.; Montal, M.; Maldarell, F.; Strebel, K. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 1996, 70, 809–819. [Google Scholar] [PubMed]
- Katz, R.A.; Skalka, A.M. The Retroviral Enzymes. Annu. Rev. Biochem. 1994, 63, 133–173. [Google Scholar] [CrossRef] [PubMed]
- Wlodawer, A.; Miller, M.; Jaskólski, M.; Sathyanarayana, B.K.; Baldwin, E.; Weber, I.T.; Selk, L.M.; Clawson, L.; Schneider, J.; Kent, S.B. Conserved Folding in Retroviral Proteases—Crystal-Structure of a Synthetic Hiv-1 Protease. Science 1989, 245, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.; Gupta, S.S.; Valkov, E.; Engelman, A.; Cherepanov, P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 2010, 464, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Pedersen, L.C.; Gabel, S.A.; Mueller, G.A.; Cuneo, M.J.; DeRose, E.F.; Krahn, J.M.; London, R.E. Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation. Nucleic Acids Res. 2014, 42, 5361–5377. [Google Scholar] [CrossRef] [PubMed]
- Jochmans, D. Novel HIV-1 reverse transcriptase inhibitors. Virus Res. 2008, 134, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Sarafianos, S.G.; Marchand, B.; Das, K.; Himmel, D.M.; Parniak, M.A.; Hughes, S.H.; Arnold, E. Structure and Function of HIV-1 Reverse Transcriptase: Molecular Mechanisms of Polymerization and Inhibition. J. Mol. Biol. 2009, 385, 693–713. [Google Scholar] [CrossRef] [PubMed]
- Menendez-Arias, L. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 2008, 134, 124–146. [Google Scholar] [CrossRef] [PubMed]
- Lewis, W.; Day, B.J.; Copeland, W.C. Mitochondrial toxicity of NRTI antiviral drugs: An integrated cellular perspective. Nat. Rev. Drug Discov. 2003, 2, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Jacobo-Molina, A.; Tantillo, C.; Lu, X.; Nanni, R.G.; Arnold, E. Buried surface analysis of HIV-1 reverse transcriptase p66/p51 heterodimer and its interaction with dsDNA template/primer. J. Mol. Recognit. 1994, 7, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Kohlstaedt, L.A.; Wang, J.; Friedman, J.M.; Rice, P.A.; Steitz, T.A. Crystal-Structure at 3.5 Angstrom Resolution of Hiv-1 Reverse-Transcriptase Complexed with an Inhibitor. Science 1992, 256, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.F.; Hostomska, Z.; Hostomsky, Z.; Jordan, S.R.; Matthews, D.A. Crystal-Structure of the Ribonuclease-H Domain of Hiv-1 Reverse-Transcriptase. Science 1991, 252, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Perera, L.; Mueller, G.A.; DeRose, E.F.; London, R.E. Asymmetric conformational maturation of HIV-1 reverse transcriptase. eLife 2015, 4, e06359. [Google Scholar]
- Zheng, X.H.; Mueller, G.A.; Cuneo, M.J.; DeRose, E.F.; London, R.E. Homodimerization of the p51 Subunit of HIV-1 Reverse Transcriptase. Biochemistry 2010, 49, 2821–2833. [Google Scholar] [CrossRef] [PubMed]
- Kensch, O.; Restle, T.; WoÈhrl, B.M.; Goody, R.S.; Steinhoff, H.J. Temperature-dependent equilibrium between the open and closed conformation of the p66 subunit of HIV-1 reverse transcriptase revealed by site-directed spin labelling. J. Mol. Biol. 2000, 301, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Hsiou, Y.; Ding, J.; Das, K.; Clark, A.D.; Hughes, S.H.; Arnold, E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 angstrom resolution: Implications of conformational changes for polymerization and inhibition mechanisms. Structure 1996, 4, 853–860. [Google Scholar] [CrossRef]
- Das, K.; Clark, A.D.; Lewi, P.J.; Heeres, J.; de Jonge, M.R.; Koymans, L.M.H.; Vinkers, H.M.; Daeyaert, F.; Ludovici, D.W.; Kukla, M.J.; et al. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 2004, 47, 2550–2560. [Google Scholar] [PubMed]
- Unge, T.; Knight, S.; Bhikhabhai, R.; Lovgren, S.; Dauter, Z.; Wilson, K.; Strandberg, B. 2.2 A resolution structure of the amino-terminal half of HIV-1 reverse transcriptase (fingers and palm subdomains). Structure 1994, 2, 953–961. [Google Scholar] [CrossRef]
- Himmel, D.M.; Maegley, K.A.; Pauly, T.A.; Bauman, J.D.; Das, K.; Dharia, C.; Clark, A.D.; Ryan, K.; Hickey, M.J.; Love, R.A.; et al. Structure of HIV-1 Reverse Transcriptase with the Inhibitor beta-Thujaplicinol Bound at the RNase H Active Site. Structure 2009, 17, 1625–1635. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Pedersen, L.C.; Gabel, S.A.; Mueller, G.A.; DeRose, E.F.; London, R.E. Unfolding the HIV-1 reverse transcriptase RNase H domain - how to lose a molecular tug-of-war. Nucleic Acids Res. 2016, 44, 1776–1788. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Milton, J.; Weaver, K.L.; Short, S.A.; Stuart, D.I.; Stammers, D.K. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure 2000, 8, 1089–1094. [Google Scholar] [CrossRef]
- Esnouf, R.; Ren, J.S.; Ross, C.; Jones, Y.; Stammers, D.; Stuart, D. Mechanism of Inhibition of Hiv-1 Reverse-Transcriptase by Nonnucleoside Inhibitors. Nat. Struct. Biol. 1995, 2, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Bird, L.E.; Chamberlain, P.P.; Stewart-Jones, G.B.; Stuart, D.I.; Stammers, D.K. Structure of HIV-2 reverse transcriptase at 2.35-angstrom resolution and the mechanism of resistance to non-nucleoside inhibitors. Proc. Natl. Acad. Sci. USA 2002, 99, 14410–14415. [Google Scholar] [CrossRef] [PubMed]
- Schulze, T.; Nawrath, M.; Moelling, K. Cleavage of the Hiv-1 P66 Reverse-Transcriptase Rnase-H by the P9 Protease in vitro Generates Active P15 Rnase-H. Arch. Virol. 1991, 118, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Tomasselli, A.G.; Sarcich, J.L.; Barrett, L.J.; Reardon, I.M.; Howe, W.J.; Evans, D.B.; Sharma, S.K.; Heinrikson, R.L. Human-Immunodeficiency-Virus Type-1 Reverse-Transcriptase and Ribonuclease-H as Substrates of the Viral Protease. Protein Sci. 1993, 2, 2167–2176. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, D.; Evans, D.B.; Deibel, M.R., Jr.; Vosters, A.F.; Eckenrode, F.M.; Einspahr, H.M.; Hui, J.O.; Tomasselli, A.G.; Zurcher-Neely, H.A.; Heinrikson, R.L. Purification and Characterization of Heterodimeric Human-Immunodeficiency-Virus Type-1 (Hiv-1) Reverse-Transcriptase Produced by in vitro Processing of P66 with Recombinant Hiv-1 Protease. J. Biol. Chem. 1992, 267, 14227–14232. [Google Scholar] [PubMed]
- Slack, R.L.; Spiriti, J.; Ahn, J.; Parniak, M.A.; Zuckerman, D.M.; Ishima, R. Structural integrity of the ribonuclease H domain in HIV-1 reverse transcriptase. Proteins-Struct. Funct. Bioinform. 2015, 83, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Navarro, J.M.; Damier, L.; Boretto, J.; Priet, S.; Canard, B.; Quérat, G.; Sire, J. Glutamic residue 438 within the protease-sensitive subdomain of HIV-1 reverse transcriptase is critical for heterodimer processing in viral particles. Virology 2001, 290, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Wapling, J.; Moore, K.L.; Sonza, S.; Mak, J.; Tachedjian, G. Mutations that abrogate human immunodeficiency virus type 1 reverse transcriptase dimerization affect maturation of the reverse transcriptase heterodimer. J. Virol. 2005, 79, 10247–10257. [Google Scholar] [CrossRef] [PubMed]
- Sluis-Cremer, N.; Arion, D.; Abram, M.E.; Parniak, M.A. Proteolytic processing of an HIV-1 pol polyprotein precursor: Insights into the mechanism of reverse transcriptase p66/p51 heterodimer formation. Int. J. Biochem. Cell Biol. 2004, 36, 1836–1847. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G. Biochemistry—Metamorphic proteins. Science 2008, 320, 1725–1726. [Google Scholar] [CrossRef] [PubMed]
- Bryan, P.N.; Orban, J. Proteins that switch folds. Curr. Opin. Struct. Biol. 2010, 20, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Doublie, S.; Sawaya, M.R.; Ellenberger, T. An open and closed case for all polymerases. Struct Fold. Des. 1999, 7, R31–R35. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Pelletier, H.; Kumar, A.; Wilson, S.H.; Kraut, J. Crystal-Structure of Rat DNA-Polymerase-Beta—Evidence for a Common Polymerase Mechanism. Science 1994, 264, 1930–1935. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Fiala, K.A.; Suo, Z.; Ling, H. Snapshots of a Y-family DNA polymerase in replication: Substrate-induced conformational transitions and implications for fidelity of Dpo4. J. Mol. Biol. 2008, 379, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Lansdon, E.B.; Samuel, D.; Lagpacan, L.; Brendza, K.M.; White, K.L.; Hung, M.; Ray, A.S. Visualizing the Molecular Interactions of a Nucleotide Analog, GS-9148, with HIV-1 Reverse Transcriptase-DNA Complex. J. Mol. Biol. 2010, 397, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Smerdon, S.J.; Jäger, J.; Kohlstaedt, L.A.; Rice, P.A.; Friedman, J.M.; Steitz, T.A. Structural Basis of Asymmetry in the Human-Immunodeficiency-Virus Type-1 Reverse-Transcriptase Heterodimer. Proc. Natl. Acad. Sci. USA 1994, 91, 7242–7246. [Google Scholar] [CrossRef] [PubMed]
- Cabodevilla, J.F.; Odriozola, L.; Santiago, E.; Martínez-Irujo, J.J. Factors affecting the dimerization of the p66 form of HIV-1 reverse transcriptase. Eur. J. Biochem. 2001, 268, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Venezia, C.F.; Howard, K.J.; Ignatov, M.E.; Holladay, L.A.; Barkley, M.D. Effects of efavirenz binding on the subunit equilibria of HIV-1 reverse transcriptase. Biochemistry 2006, 45, 2779–2789. [Google Scholar] [CrossRef] [PubMed]
- Jacobo-Molina, A.; Arnold, E. Hiv Reverse-Transcriptase Structure-Function-Relationships. Biochemistry 1991, 30, 6351–6361. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.P.; Das, K.; Hsiou, Y.; Sarafianos, S.G.; Clark, A.D.; Jacobo-Molina, A.; Arnold, E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 angstrom resolution. J. Mol. Biol. 1998, 284, 1095–1111. [Google Scholar] [CrossRef] [PubMed]
- Hostomska, Z.; Matthews, D.A.; Davies, J.F.; Nodes, B.R.; Hostomsky, Z. Proteolytic Release and Crystallization of the Rnase-H Domain of Human-Immunodeficiency-Virus Type-1 Reverse-Transcriptase. J. Biol. Chem. 1991, 266, 14697–14702. [Google Scholar] [PubMed]
- Pattyn, E.; Lavens, D.; Van der Heyden, J.; Verhee, A.; Lievens, S.; Lemmens, I.; Tavernier, J. MAPPIT (MAmmalian Protein-Protein Interaction Trap) as a tool to study HIV reverse transcriptase dimerization in intact human cells. J. Virol. Meth. 2008, 153, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Divita, G.; Restle, T.; Goody, R.S. Characterization of the Dimerization Process of Hiv-1 Reverse-Transcriptase Heterodimer Using Intrinsic Protein Fluorescence. FEBS Lett. 1993, 324, 153–158. [Google Scholar] [CrossRef]
- Venezia, C.F.; Meany, B.J.; Braz, V.A.; Barkley, M.D. Kinetics of Association and Dissociation of HIV-1 Reverse Transcriptase Subunits. Biochemistry 2009, 48, 9084–9093. [Google Scholar] [CrossRef] [PubMed]
- Paris, K.A.; Haq, O.; Felts, A.K.; Das, K.; Arnold, E.; Levy, R.M. Conformational Landscape of the Human Immunodeficiency Virus Type 1 Reverse Transcriptase Non-Nucleoside Inhibitor Binding Pocket: Lessons for Inhibitor Design from a Cluster Analysis of Many Crystal Structures. J. Med. Chem. 2009, 52, 6413–6420. [Google Scholar] [CrossRef] [PubMed]
- Tantillo, C.; Ding, J.; Jacobo-Molina, A.; Nanni, R.G.; Boyer, P.L.; Hughes, S.H.; Arnold, E. Locations of Anti-Aids Drug-Binding Sites and Resistance Mutations in the 3-Dimensional Structure of Hiv-1 Reverse-Transcriptase—Implications for Mechanisms of Drug-Inhibition and Resistance. J. Mol. Biol. 1994, 243, 369–387. [Google Scholar] [CrossRef] [PubMed]
- Chou, P.Y.; Fasman, G.D. Prediction of Beta-Turns. Biophys. J. 1979, 26, 367–383. [Google Scholar] [CrossRef]
- Goel, R.; Beard, W.A.; Kumar, A.; Casas-Finet, J.R.; Strub, M.P.; Stahl, S.J.; Becerra, S.P. Structure-Function Studies of Hiv-1(1) Reverse-Transcriptase—Dimerization-Defective Mutant L289k. Biochemistry 1993, 32, 13012–13018. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.K.; Kaushik, N.; Singh, K.; Sharma, B.; Upadhyay, A.K.; Kumar, S.; Pandey, V.N. Insertion of a small peptide of six amino acids into the beta 7-beta 8 loop of the p51 subunit of HIV-1 reverse transcriptase perturbs the heterodimer and affects its activities. BMC Biochemistry 2002, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Mulky, A.; Vu, B.C.; Conway, J.A.; Hughes, S.H.; Kappes, J.C. Analysis of amino acids in the beta 7-beta 8 loop of human immunodeficiency virus type 1 reverse transcriptase for their role in virus replication. J. Mol. Biol. 2007, 365, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Fan, N.S.; Evans, D.B. Human-Immunodeficiency-Virus Type-1 (Hiv-1) Recombinant Reverse-Transcriptase—Asymmetry in P66 Subunits of the P66/P66 Homodimer. FEBS Lett. 1994, 343, 125–130. [Google Scholar] [CrossRef]
- Sharaf, N.G.; Poliner, E.; Slack, R.L.; Christen, M.T.; Byeon, I.J. L.; Parniak, M.A.; Ishima, R. The p66 immature precursor of HIV-1 reverse transcriptase. Proteins 2014, 82, 2343–2352. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.F.; Lengruber, R.B.; Soares, E.A.; Jere, A.; Sprinz, E.; Martinez, A.M.; Soares, M.A. Conservation patterns of HIV-1 RT connection and RNase H domains: Identification of new mutations in NRTI-treated patients. PLoS ONE 2008, 3, e1781. [Google Scholar] [CrossRef] [PubMed]
- Kern, G.; Handel, T.; Marqusee, S. Characterization of a folding intermediate from HIV-1 ribonuclease H. Protein Sci. 1998, 7, 2164–2174. [Google Scholar] [CrossRef] [PubMed]
- Byeon, I.J.; Louis, J.M.; Gronenborn, A.M. A captured folding intermediate involved in dimerization and domain-swapping of GB1. J. Mol. Biol. 2004, 340, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Newcomer, M.E. Protein folding and three-dimensional domain swapping: A strained relationship? Curr. Opin. Struct. Biol. 2002, 12, 48–53. [Google Scholar] [CrossRef]
- Rousseau, F.; Schymkowitz, J.; Itzhaki, L.S. Implications of 3D domain swapping for protein folding, misfolding and function. Adv. Exp. Med. Biol. 2012, 747, 137–152. [Google Scholar] [PubMed]
- Rousseau, F.; Schymkowitz, J.W.; Itzhaki, L.S. The unfolding story of three-dimensional domain swapping. Structure 2003, 11, 243–251. [Google Scholar] [CrossRef]
- Pari, K.; Mueller, G.A.; DeRose, E.F.; Kirby, T.W.; London, R.E. Solution structure of the RNase H domain of the HIV-1 reverse transcriptase in the presence of magnesium. Biochemistry 2003, 42, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Christen, M.T.; Menon, L.; Myshakina, N.S.; Ahn, J.; Parniak, M.A.; Ishima, R. Structural basis of the allosteric inhibitor interaction on the HIV-1 reverse transcriptase RNase H domain. Chem. Biol. Drug. Des. 2012, 80, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Divita, G.; Rittinger, K.; Geourjon, C.; Deléage, G.; Goody, R.S. Dimerization Kinetics of Hiv-1 and Hiv-2 Reverse-Transcriptase—A 2-Step Process. J. Mol. Biol. 1995, 245, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Rowley, G.L.; Ma, Q.F.; Bathurst, I.C.; Barr, P.J.; Kenyon, G.L. Stabilization and Activation of Recombinant Human Immunodeficiency Virus-1 Reverse Transcriptase-P66. Biochem. Biophys. Res. Commun. 1990, 167, 673–679. [Google Scholar] [CrossRef]
- Tachedjian, G.; Orlova, M.; Sarafianos, S.G.; Arnold, E.; Goff, S.P. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 2001, 98, 7188–7193. [Google Scholar] [CrossRef] [PubMed]
- Tachedjian, G.; Moore, K.L.; Goff, S.P.; Sluis-Cremer, N. Efavirenz enhances the proteolytic processing of an HIV-1 pol polyprotein precursor and reverse transcriptase homodimer formation. FEBS Lett. 2005, 579, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Sluis-Cremer, N.; Arion, D.; Parniak, M.A. Destabilization of the HIV-1 reverse transcriptase dimer upon interaction with N-acyl hydrazone inhibitors. Mol. Pharmacol. 2002, 62, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Camarasa, M.J.; Velazquez, S.; San-Felix, A.; Perez-Perez, M.J.; Bonache, M.C.; Castro, S.D. TSAO derivatives, inhibitors of HIV-1 reverse transcriptase dimerization: Recent progress. Curr. Pharm. Des. 2006, 12, 1895–1907. [Google Scholar] [CrossRef] [PubMed]
- Sluis-Cremer, N.; Hamamouch, N.; San Félix, A.; Velazquez, S.; Balzarini, J.; Camarasa, M.J. Structure-activity relationships of [2′,5′-bis-O-(tert-butyldimethylsilyl)-beta-D-ribofuranosyl]-3′-spiro-5″-(4″-amino-1″,2″-oxathiole-2″,2″-dioxide) thymine derivatives as inhibitors of HIV-1 reverse transcriptase dimerization. J. Med. Chem. 2006, 49, 4834–4841. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Martinez, S.E.; Bauman, J.D.; Arnold, E. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat. Struct. Mol. Biol. 2012, 19, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.K.; Kaushik, N.; Talele, T.T.; Yadav, P.N.S.; Pandey, V.N. Insertion of a peptide from MuLV RT into the connection subdomain of HIV-1 RT results in a functionally active chimeric enzyme in monomeric conformation. Mol. Cell. Biochem. 2001, 225, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Tachedjian, G.; Radzio, J.; Sluis-Cremer, N. Relationship between enzyme activity and dimeric structure of recombinant HIV-1 reverse transcriptase. Proteins-Struct. Funct. Bioinform. 2005, 60, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Bauman, J.D.; Rim, A.S.; Dharia, C.; Clark, A.D., Jr.; Camarasa, M.J.; Arnold, E. Crystal Structure of tert-Butyldimethylsilyl-spiroaminooxathioledioxide-thymine (TSAO-T) in Complex with HIV-1 Reverse Transcriptase (RT) Redefines the Elastic Limits of the Non-nucleoside Inhibitor-Binding Pocket. J. Med. Chem. 2011, 54, 2727–2737. [Google Scholar] [CrossRef] [PubMed]
- Sluis-Cremer, N.; Dmitrienko, G.I.; Balzarini, J.; Camarasa, M.J.; Parniak, M.A. Human immunodeficiency virus type 1 reverse transcriptase dimer destabilization by 1-{spiro[4″-amino-2″,2″-dioxo-1″,2″-oxathiole-5″,3′-[2′,5′-bis-O-(tert-butyldimethylsilyl)-beta-D-ribofuranosyl]]}-3-ethylthymine. Biochemistry 2000, 39, 1427–1433. [Google Scholar] [CrossRef] [PubMed]
- Divita, G.; Restle, T.; Goody, R.S.; Chermann, J.C.; Baillon, J.G. Inhibition of Human-Immunodeficiency-Virus Type-1 Reverse-Transcriptase Dimerization Using Synthetic Peptides Derived from the Connection Domain. J. Biol. Chem. 1994, 269, 13080–13083. [Google Scholar] [PubMed]
- Nissley, D.V.; Radzio, J.; Ambrose, Z.; Sheen, C.W.; Hamamouch, N.; Moore, K.L.; Sluis-Cremer, N. Characterization of novel non-nucleoside reverse transcriptase (RT) inhibitor resistance mutations at residues 132 and 135 in the 51 kDa subunit of HIV-1 RT. Biochem. J. 2007, 404, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.; Moore, K.L.; Mak, J.; Sluis-Cremer, N.; de Bethune, M.P.; Tachedjian, G. Potent nonnucleoside reverse transcriptase inhibitors target HIV-1 Gag-Pol. PLoS Pathog. 2006, 2, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Sarafianos, S.G.; Clark, A.D.; Boyer, P.L.; Hughes, S.H.; Arnold, E. Crystal structures of clinically relevant Lys103Asn/Tyr181Cys double mutant HIV-1 reverse transcriptase in complexes with ATP and non-nucleoside inhibitor HBY 097. J. Mol. Biol. 2007, 365, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chopra, R.; Verdine, G.L.; Harrison, S.C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 1998, 282, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.H.; Mueller, G.A.; DeRose, E.F.; London, R.E. Protein-Mediated Antagonism between HIV Reverse Transcriptase Ligands Nevirapine and MgATP. Biophys. J. 2013, 104, 2695–2705. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.; Zelina, S.; Sluis-Cremer, N.; Tachedjian, G. Impact of residues in the nonnucleoside reverse transcriptase inhibitor binding pocket on HIV-1 reverse transcriptase heterodimer stability. Curr. HIV Res. 2008, 6, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Tachedjian, G.; Aronson, H.E.G.; Goff, S.P. Analysis of mutations and suppressors affecting interactions between the subunits of the HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 2000, 97, 6334–6339. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Jacques, P.S.; Rodgers, D.W.; Ottman, M.; Darlix, J.L.; Le Grice, S.F. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry 1996, 35, 8553–8562. [Google Scholar] [CrossRef] [PubMed]
- Wohrl, B.M.; Krebs, R.; Thrall, S.H.; Le Grice, S.F.; Scheidig, A.J.; Goody, R.S. Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66—Implications for DNA synthesis and dimerization. J. Biol. Chem. 1997, 272, 17581–17587. [Google Scholar] [CrossRef] [PubMed]
- Mulky, A.; Sarafianos, S.G.; Jia, Y.; Arnold, E.; Kappes, J.C. Identification of amino acid residues in the human immunodeficiency virus type-1 reverse transcriptase tryptophan-repeat motif that are required for subunit interaction using infectious virions. J. Mol. Biol. 2005, 349, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Tachedjian, G.; Aronson, H.E.G.; de los Santos, M.; Seehra, J.; McCoy, J.M.; Goff, S.P. Role of residues in the tryptophan repeat motif for HIV-1 reverse transcriptase dimerization. J. Mol. Biol. 2003, 326, 381–396. [Google Scholar] [CrossRef]
- Auwerx, J.; Van Nieuwenhove, J.; Rodríguez-Barrios, F.; de Castro, S.; Velázquez, S.; Ceccherini-Silberstein, F.; Balzarini, J. The N137 and P140 amino acids in the p51 and the P95 amino acid in the p66 subunit of human immunodeficiency virus type I (HIV-1) reverse transcriptase are instrumental to maintain catalytic activity and to design new classes of anti-HIV-1 drugs. FEBS Lett. 2005, 579, 2294–2300. [Google Scholar] [CrossRef] [PubMed]
- Malkov, S.N.; Živković, M.V.; Beljanski, M.V.; Hall, M.B.; Zarić, S.D. A reexamination of the propensities of amino acids towards a particular secondary structure: Classification of amino acids based on their chemical structure. J. Mol. Model. 2008, 14, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Ceccherini-Silberstein, F.; Gago, F.; Santoro, M.; Gori, C.; Svicher, V.; Rodríguez-Barrios, F.; Balzarini, J. High sequence conservation of human immunodeficiency virus-type 1 reverse transcriptase under drug pressure despite the continuous appearance of mutations. J. Virol. 2005, 79, 10718–10729. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Sato, A.; El-Farrash, M.; Miki, S.; Abe, K.; Isaka, Y.; Sugimoto, H. S-1153 inhibits replication of known drug-resistant strains of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 1998, 42, 1340–1345. [Google Scholar] [PubMed]
- Feng, M.Z.; Wang, D.; Grobler, J.A.; Hazuda, D.J.; Miller, M.D.; Lai, M.T. In Vitro Resistance Selection with Doravirine (MK-1439), a Novel Nonnucleoside Reverse Transcriptase Inhibitor with Distinct Mutation Development Pathways. Antimicrob. Agents Chemother. 2015, 59, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Hammond, J.; Alexander, T.N.; Graham, J.P.; Binford, S.; Sugita, K.I.; Patick, A.K. In vitro selection of mutations in human immunodeficiency virus type 1 reverse transcriptase that confer resistance to capravirine, a novel nonnucleoside reverse transcriptase inhibitor. Antivir. Res. 2006, 70, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.Q.; Felock, P.J.; Munshi, V.; Hrin, R.C.; Wang, Y.J.; Yan, Y.; Williams, T.M. Antiviral Activity and In Vitro Mutation Development Pathways of MK-6186, a Novel Nonnucleoside Reverse Transcriptase Inhibitor. Antimicrob. Agents Chemother. 2012, 56, 3324–3335. [Google Scholar] [CrossRef] [PubMed]
- Willard, L.; Ranjan, A.; Zhang, H.; Monzavi, H.; Boyko, R.F.; Sykes, B.D.; Wishart, D.S. VADAR: A web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003, 31, 3316–3319. [Google Scholar] [CrossRef] [PubMed]
- Mulky, A.; Sarafianos, S.G.; Arnold, E.; Wu, X.; Kappes, J.C. Subunit-specific analysis of the human immunodeficiency virus type 1 reverse transcriptase in vivo. J. Virol. 2004, 78, 7089–7096. [Google Scholar] [CrossRef] [PubMed]
- Bacheler, L.; Jeffrey, S.; Hanna, G.; D’Aquila, R.; Wallace, L.; Logue, K.; Baker, D. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 2001, 75, 4999–5008. [Google Scholar] [CrossRef] [PubMed]
- Kleim, J.P.; Bender, R.; Kirsch, R.; Meichsner, C.; Paessens, A.; Rösner, M.; Schneweis, K.E. Preclinical Evaluation of Hby-097, a New Nonnucleoside Reverse-Transcriptase Inhibitor of Human-Immunodeficiency-Virus Type-1 Replication. Antimicrob. Agents Chemother. 1995, 39, 2253–2257. [Google Scholar] [CrossRef] [PubMed]
- Kolomeets, A.N.; Varghese, V.; Lemey, P.; Bobkova, M.R.; Shafer, R.W. A uniquely prevalent nonnucleoside reverse transcriptase inhibitor resistance mutation in Russian subtype A HIV-1 viruses. AIDS 2014, 28, F1–F8. [Google Scholar] [CrossRef] [PubMed]
- Neogi, U.; Anita, S.H.E.T.; Shamsundar, R.; Ekstrand, M.L. Selection of nonnucleoside reverse transcriptase inhibitor-associated mutations in HIV-1 subtype C: Evidence of etravirine cross-resistance. AIDS 2011, 25, 1123–1126. [Google Scholar] [CrossRef] [PubMed]
- Paolucci, S.; Baldanti, F.; Campanini, G.; Cancio, R.; Belfiore, A.; Maga, G.; Gerna, G. NNRTI-selected mutations at codon 190 of human immunodeficiency virus type 1 reverse transcriptase decrease susceptibility to stavudine and zidovudine. Antivir. Res. 2007, 76, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dykes, C.; Domaoal, R.A.; Koval, C.E.; Bambara, R.A.; Demeter, L.M. The HIV-1 reverse transcriptase mutants G190S and G190A, which confer resistance to non-nucleoside reverse transcriptase inhibitors, demonstrate reductions in RNase H activity and DNA synthesis from tRNA(Lys,3) that correlate with reductions in replication efficiency. Virology 2006, 348, 462–474. [Google Scholar] [PubMed]
- Abram, M.E.; Parniak, M.A. Virion instability of human immunodeficiency virus type 1 reverse transcriptase (RT) mutated in the protease cleavage site between RT p51 and the RT RNase H domain. J. Virol. 2005, 79, 11952–11961. [Google Scholar] [CrossRef] [PubMed]
- Abram, M.E.; Sarafianos, S.G.; Parniak, M.A. The mutation T477A in HIV-1 reverse transcriptase (RT) restores normal proteolytic processing of RT in virus with Gag-Pol mutated in the p51-RNH cleavage site. Retrovirology 2010, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Chou, P.Y.; Fasman, G.D. Empirical Predictions of Protein Conformation. Annu. Rev. Biochem. 1978, 47, 251–276. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, L.; Ghirlando, R.; Clore, G.M. Conformation and dynamics of the Gag polyprotein of the human immunodeficiency virus 1 studied by NMR spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 3374–3379. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Dou, J.; Ding, L.; Spearman, P. Myristoylation is required for human immunodeficiency virus type 1 gag-gag multimerization in mammalian cells. J. Virol. 2007, 81, 12899–12910. [Google Scholar] [CrossRef] [PubMed]
- Sudo, S.; Haraguchi, H.; Hirai, Y.; Gatanaga, H.; Sakuragi, J.I.; Momose, F.; Morikawa, Y. Efavirenz Enhances HIV-1 Gag Processing at the Plasma Membrane through Gag-Pol Dimerization. J. Virol. 2013, 87, 3348–3360. [Google Scholar] [CrossRef] [PubMed]
- Konnyu, B.; Sadiq, S.K.; Turányi, T.; Hírmondó, R.; Müller, B.; Kräusslich, H.G.; Müller, V. Gag-Pol Processing during HIV-1 Virion Maturation: A Systems Biology Approach. PLoS Comput. Biol. 2013, 9, e1003103. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Mueller, G.A.; DeRose, E.F.; London, R.E. Metal and ligand binding to the HIV-RNase H active site are remotely monitored by Ile556. Nucleic Acids Res. 2012, 40, 10543–10553. [Google Scholar] [CrossRef] [PubMed]
- Powers, R.; Clore, G.M.; Stahl, S.J.; Wingfield, P.T.; Gronenborn, A. Analysis of the Backbone Dynamics of the Ribonuclease-H Domain of the Human-Immunodeficiency-Virus Reverse-Transcriptase Using N-15-Relaxation Measurements. Biochemistry 1992, 31, 9150–9157. [Google Scholar] [CrossRef] [PubMed]
- Mueller, G.A.; Pari, K.; DeRose, E.F.; Kirby, T.W.; London, R.E. Backbone dynamics of the RNase H domain of HIV-1 reverse transcriptase. Biochemistry 2004, 43, 9332–9342. [Google Scholar] [CrossRef] [PubMed]
- Pettit, S.C.; Lindquist, J.N.; Kaplan, A.H.; Swanstrom, R. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology 2005, 2, 66. [Google Scholar] [CrossRef] [PubMed]
- Louis, J.M.; Clore, G.M.; Gronenborn, A.M. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 1999, 6, 868–875. [Google Scholar] [PubMed]
- Agosto, L.M.; Zhong, P.; Munro, J.; Mothes, W. Highly Active Antiretroviral Therapies Are Effective against HIV-1 Cell-to-Cell Transmission. PLoS Pathog. 2014, 10, e1003982. [Google Scholar] [CrossRef] [PubMed]
- Warrilow, D.; Stenzel, D.; Harrich, D. Isolated HIV-1 core is active for reverse transcription. Retrovirology 2007, 4, 77. [Google Scholar] [CrossRef] [PubMed]
- Kleiman, L.; Jones, C.P.; Musier-Forsyth, K. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010, 584, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Lapadat-Tapolsky, M.; Pernelle, C.; Borie, C.; Darlix, J.L. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995, 23, 2434–2441. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.A.; Gorelick, R.J. Nucleocapsid protein function in early infection processes. Virus Res. 2008, 134, 39–63. [Google Scholar] [CrossRef] [PubMed]
- Druillennec, S.; Caneparo, A.; de Rocquigny, H.; Roques, B.P. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J. Biol. Chem. 1999, 274, 11283–11288. [Google Scholar] [CrossRef] [PubMed]
- Delviks-Frankenberry, K.A.; Nikolenko, G.N.; Barr, R.; Pathak, V.K. Mutations in human immunodeficiency virus type 1 RNase H primer grip enhance 3-Azido-3’-deoxythymidine resistance. J. Virol. 2007, 81, 6837–6845. [Google Scholar] [CrossRef] [PubMed]
- Sarafianos, S.G.; Das, K.; Tantillo, C.; Clark, A.D.; Ding, J.; Whitcomb, J.M.; Arnold, E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA : DNA. EMBO J. 2001, 20, 1449–1461. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Parra, S.; Marin, M.; Gahlaut, N.; Suter, R.; Kondo, N.; Melikyan, G.B. Fusion of Mature HIV-1 Particles Leads to Complete Release of a Gag-GFP-Based Content Marker and Raises the Intraviral pH. PLoS ONE 2013, 8, e71002. [Google Scholar]
- Ido, E.; Han, H.P.; Kezdy, F.J.; Tang, J. Kinetic-Studies of Human-Immunodeficiency-Virus Type-1 Protease and Its Active-Site Hydrogen-Bond Mutant A28s. J. Biol. Chem. 1991, 266, 24359–24366. [Google Scholar] [PubMed]
- Li, Y.F.; Breaker, R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2’-hydroxyl group. J. Am. Chem. Soc. 1999, 121, 5364–5372. [Google Scholar] [CrossRef]
- Hizi, A.; Tal, R.; Shaharabany, M.; Loya, S. Catalytic Properties of the Reverse Transcriptases of Human Immunodeficiency Viruses Type-1 and Type-2. J. Biol. Chem. 1991, 266, 6230–6239. [Google Scholar] [PubMed]
- Waheed, A.A.; Tachedjian, G. Why Do We Need New Drug Classes for HIV Treatment and Prevention? Curr. Top. Med. Chem. 2016, 16, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Sluis-Cremer, N.; Tachedjian, G. Modulation of the oligomeric structures of HIV-1 retroviral enzymes by synthetic peptides and small molecules. Eur. J. Biochem. 2002, 269, 5103–5111. [Google Scholar] [CrossRef] [PubMed]
- Andreola, M.L. Therapeutic Potential of Peptide Motifs Against HIV-1 Reverse Transcriptase and Integrase. Curr. Pharm. Des. 2009, 15, 2508–2519. [Google Scholar] [CrossRef] [PubMed]
- Restle, T.; Muller, B.; Goody, R.S. Dimerization of Human-Immunodeficiency-Virus Type-1 Reverse-Transcriptase—A Target for Chemotherapeutic Intervention. J. Biol. Chem. 1990, 265, 8986–8988. [Google Scholar] [PubMed]
- Agopian, A.; Gros, E.; Aldrian-Herrada, G.; Bosquet, N.; Clayette, P.; Divita, G. A New Generation of Peptide-based Inhibitors Targeting HIV-1 Reverse Transcriptase Conformational Flexibility. J. Biol. Chem. 2009, 284, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Wendeler, M.; Lee, H.F.; Bermingham, A.; Miller, J.T.; Chertov, O.; Bona, M.K.; Gotte, M. Vinylogous Ureas as a Novel Class of Inhibitors of Reverse Transcriptase-Associated Ribonuclease H Activity. ACS Chem. Biol. 2008, 3, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.M.; Wendeler, M.; Rausch, J.W.; Beilhartz, G.; Gotte, M.; O’Keefe, B.R.; Le Grice, S.F. Structure-Activity Analysis of Vinylogous Urea Inhibitors of Human Immunodeficiency Virus-Encoded Ribonuclease H. Antimicrob. Agents Chemother. 2010, 54, 3913–3921. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.M.; Miller, J.T.; Johnson, B.C.; Hughes, S.H.; Le Grice, S.F.J. Mutagenesis of Human Immunodeficiency Virus Reverse Transcriptase p51 Subunit Defines Residues Contributing to Vinylogous Urea Inhibition of Ribonuclease H Activity. J. Biol. Chem. 2012, 287, 4066–4075. [Google Scholar] [CrossRef] [PubMed]
- Masaoka, T.; Chung, S.; Caboni, P.; Rausch, J.W.; Wilson, J.A.; Taskent-Sezgin, H.; Le Grice, S.F. Exploiting Drug-Resistant Enzymes as Tools To Identify Thienopyrimidinone Inhibitors of Human Immunodeficiency Virus Reverse Transcriptase-Associated Ribonuclease H. J. Med. Chem. 2013, 56, 5436–5445. [Google Scholar] [CrossRef] [PubMed]
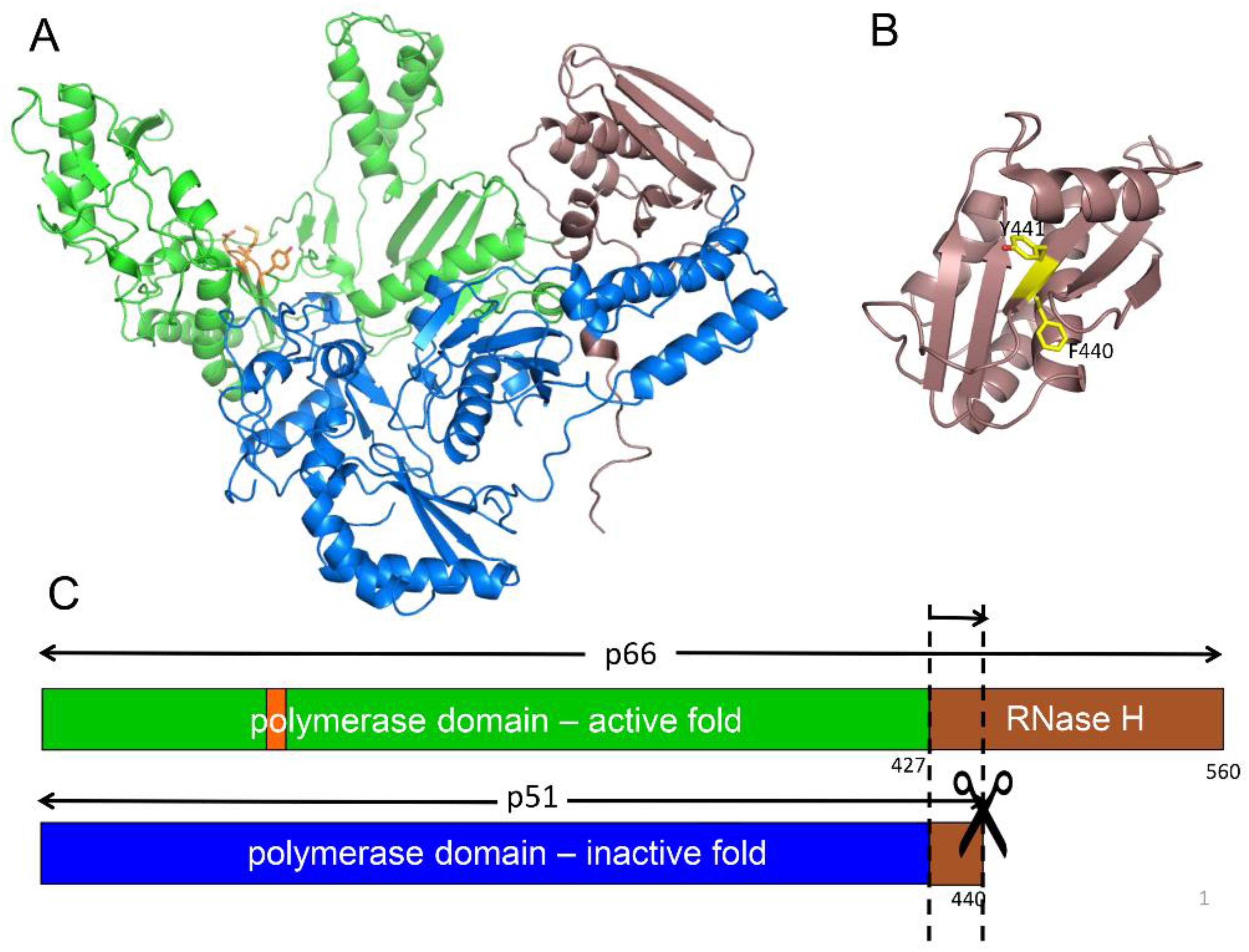
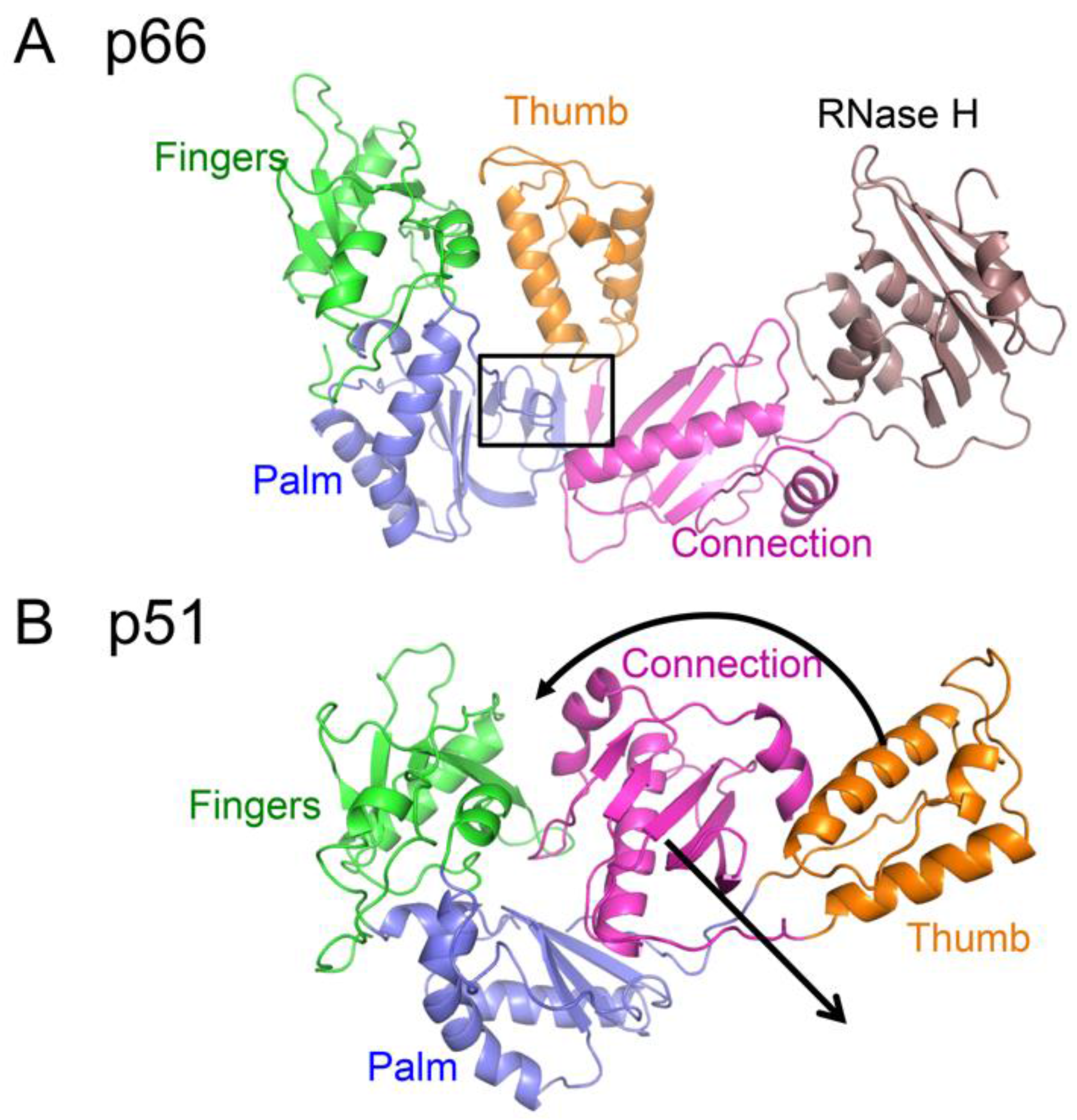
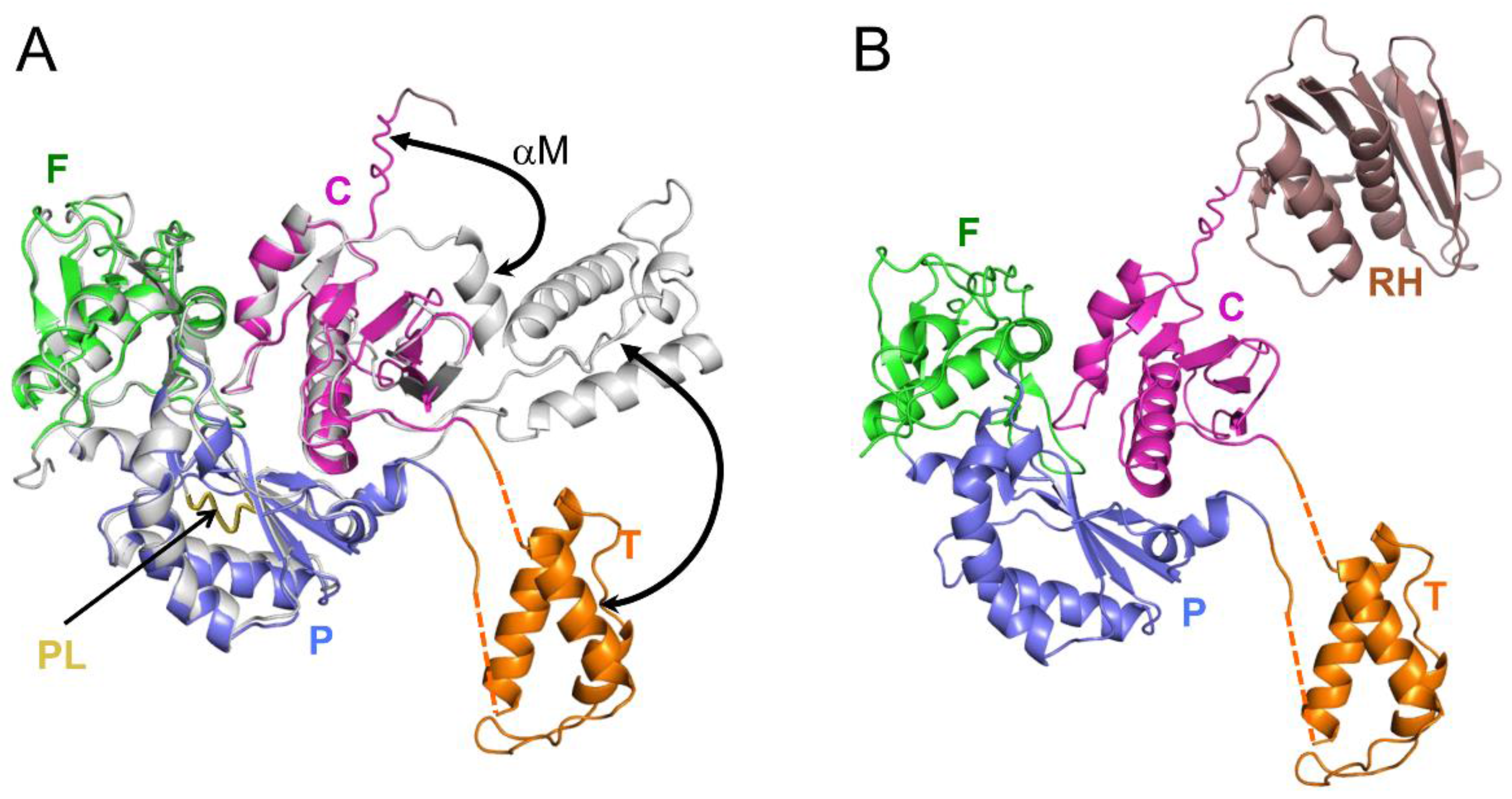
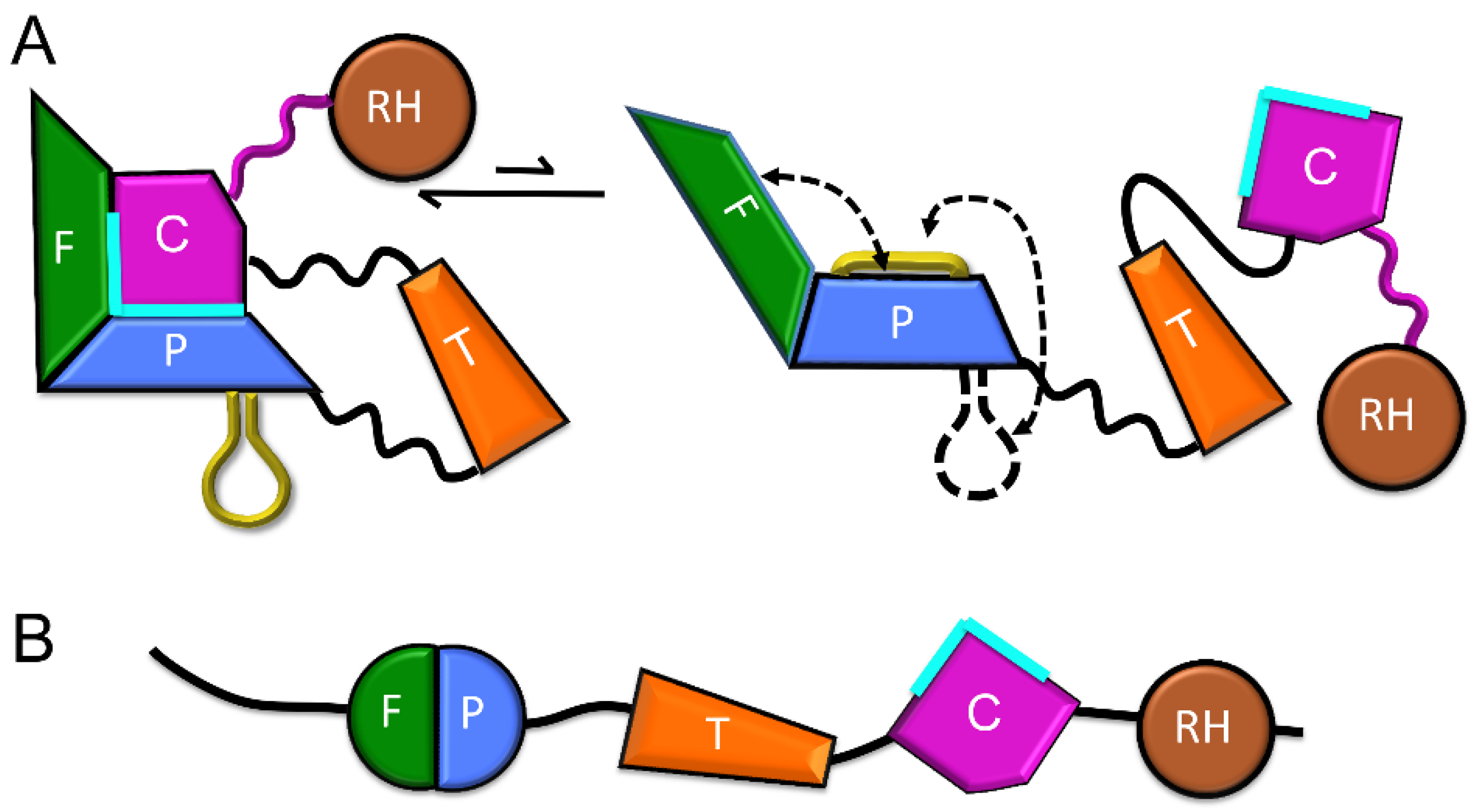
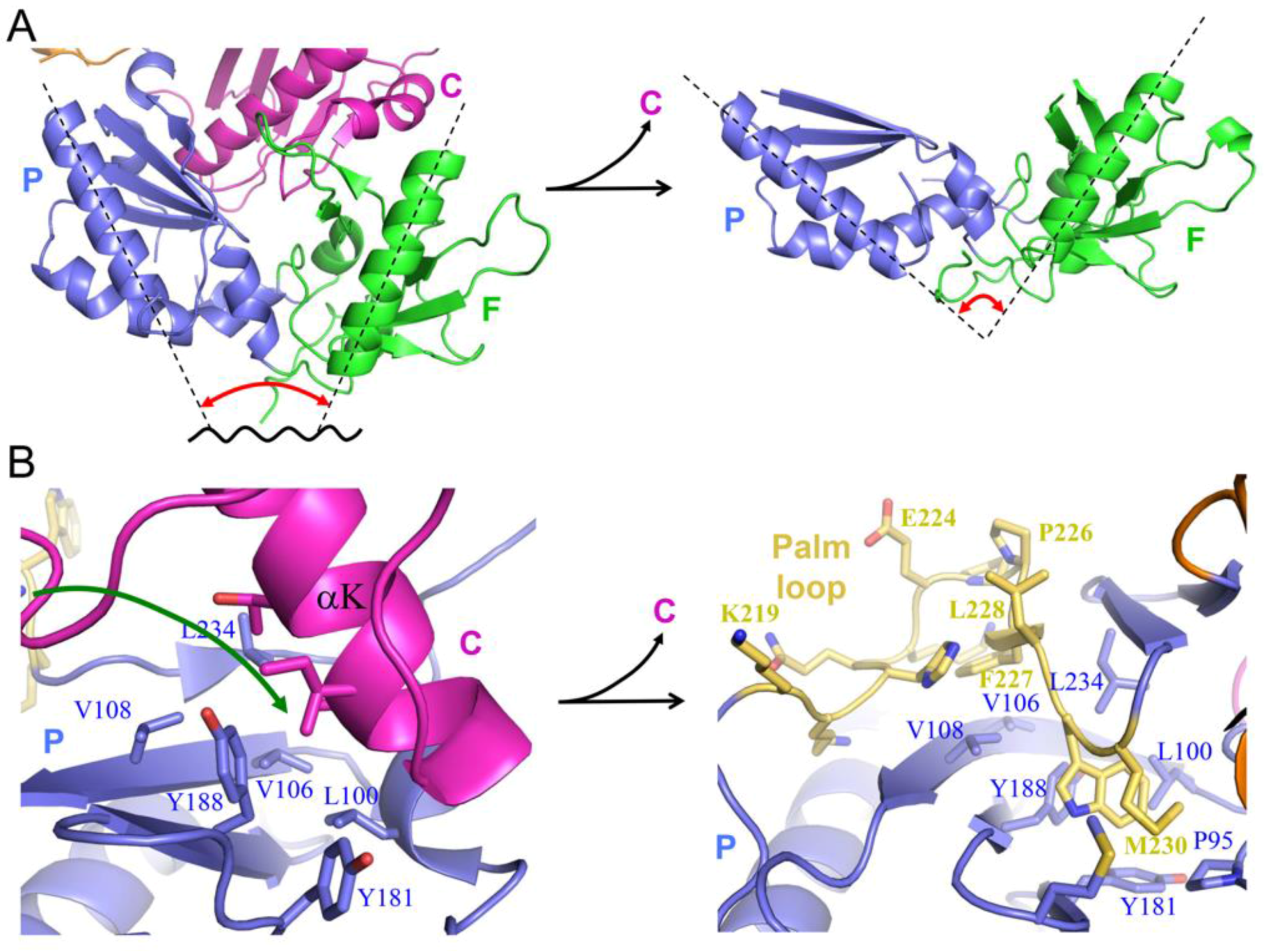
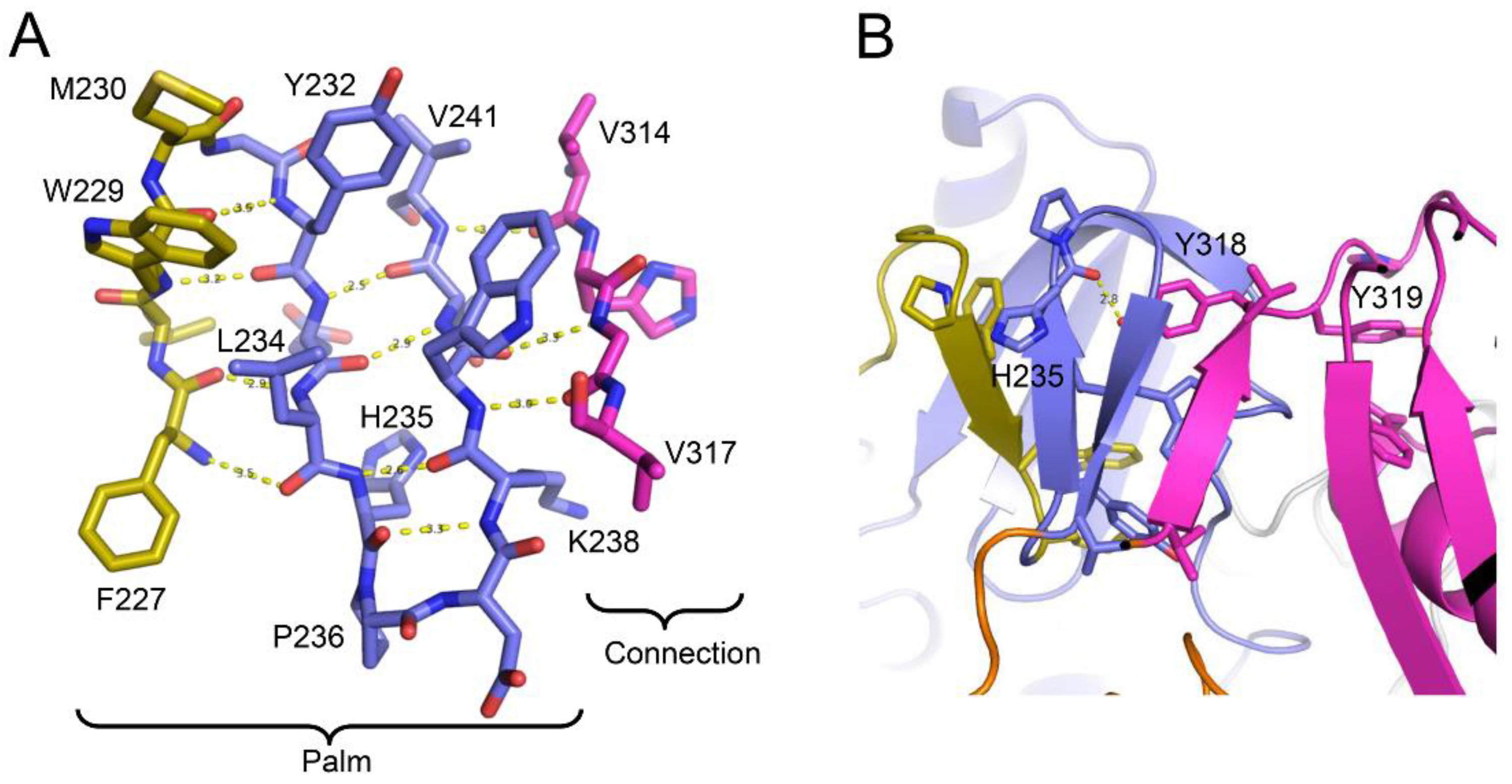
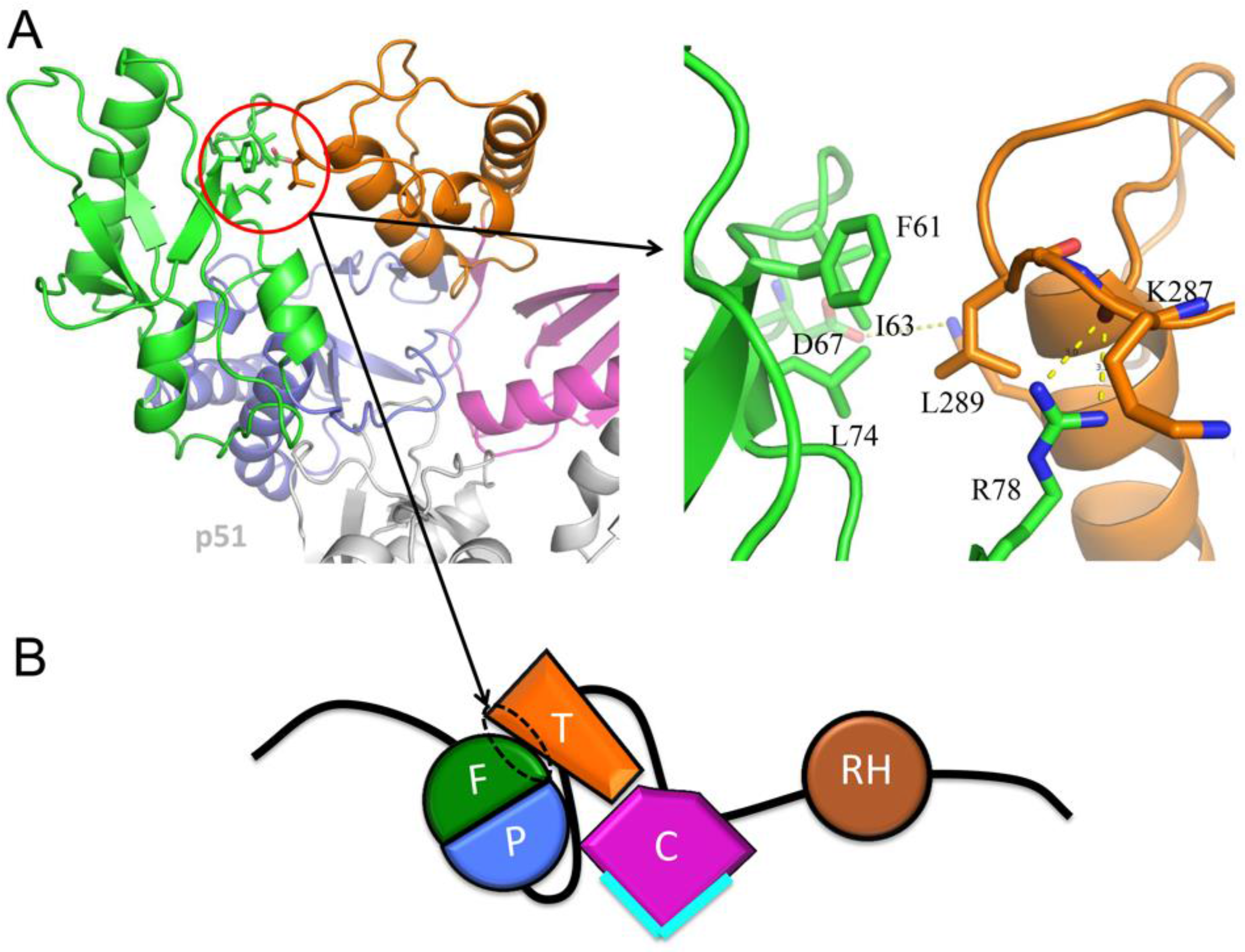
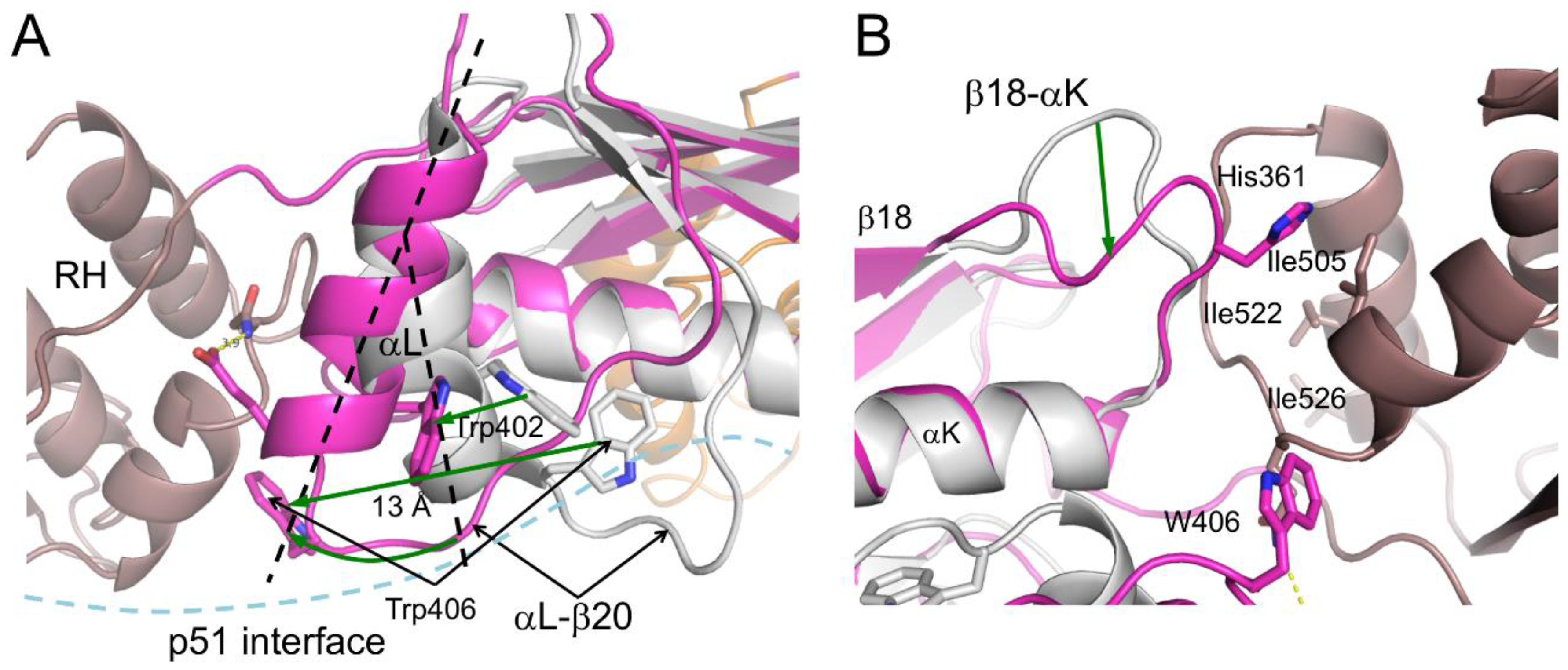
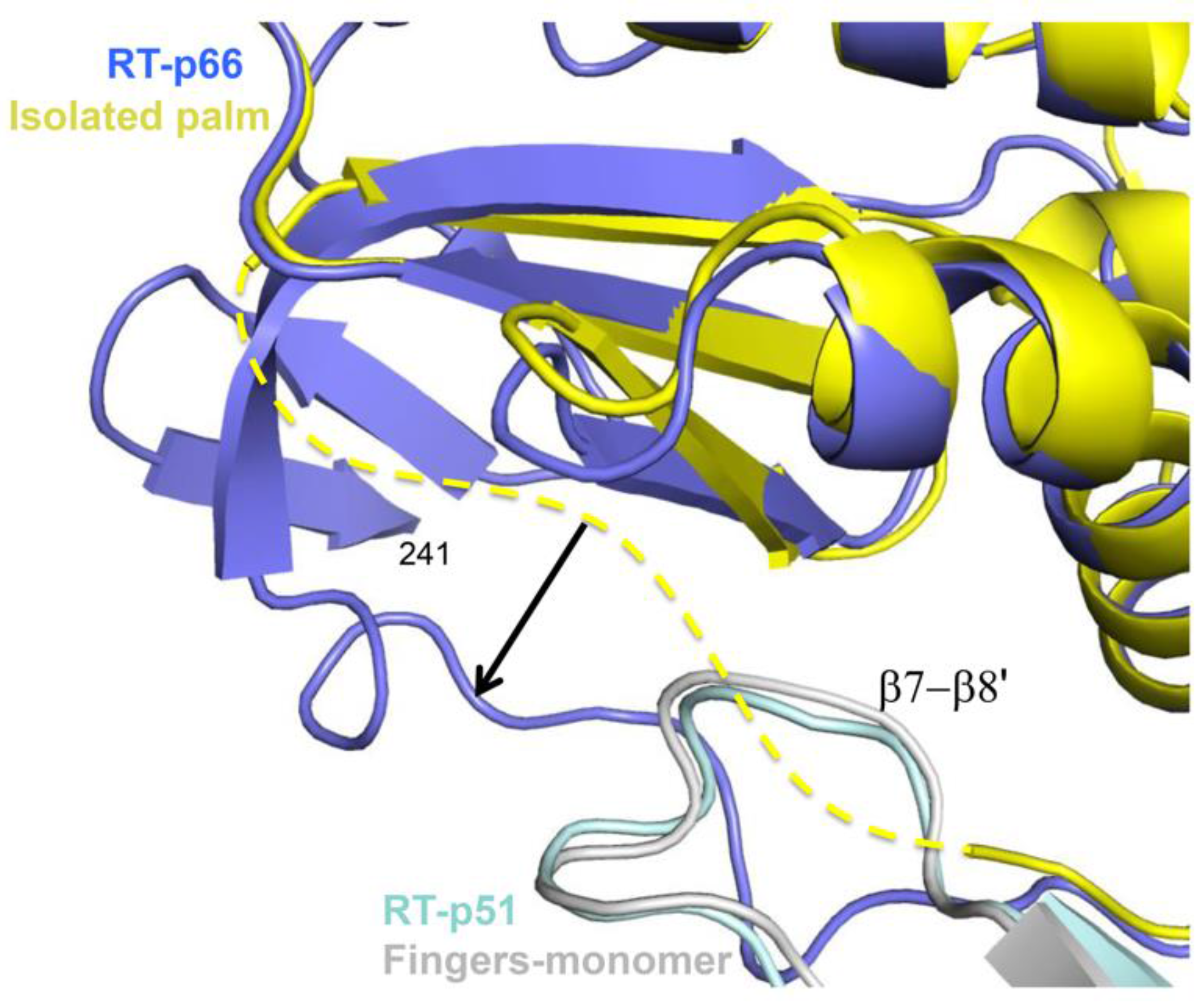
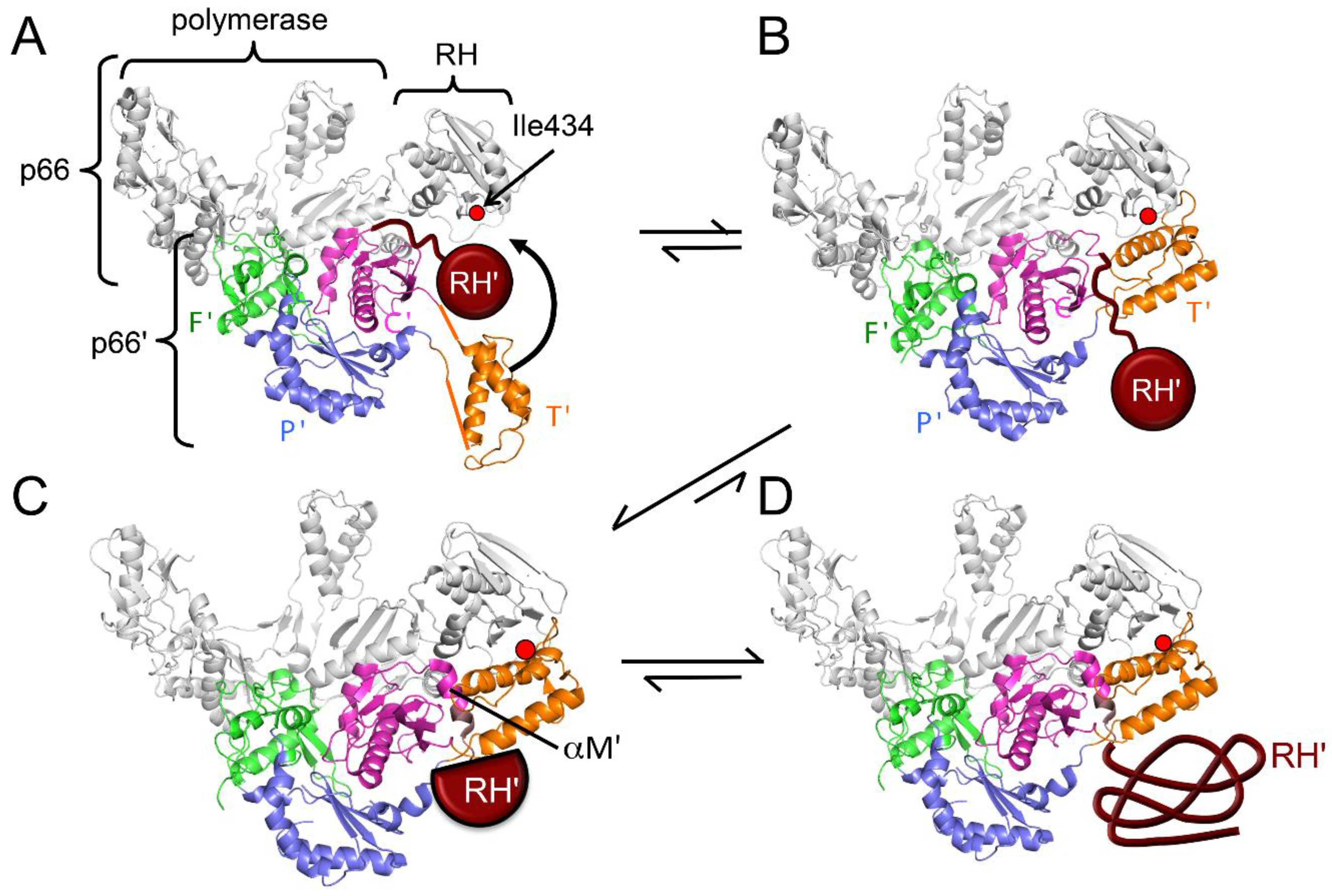
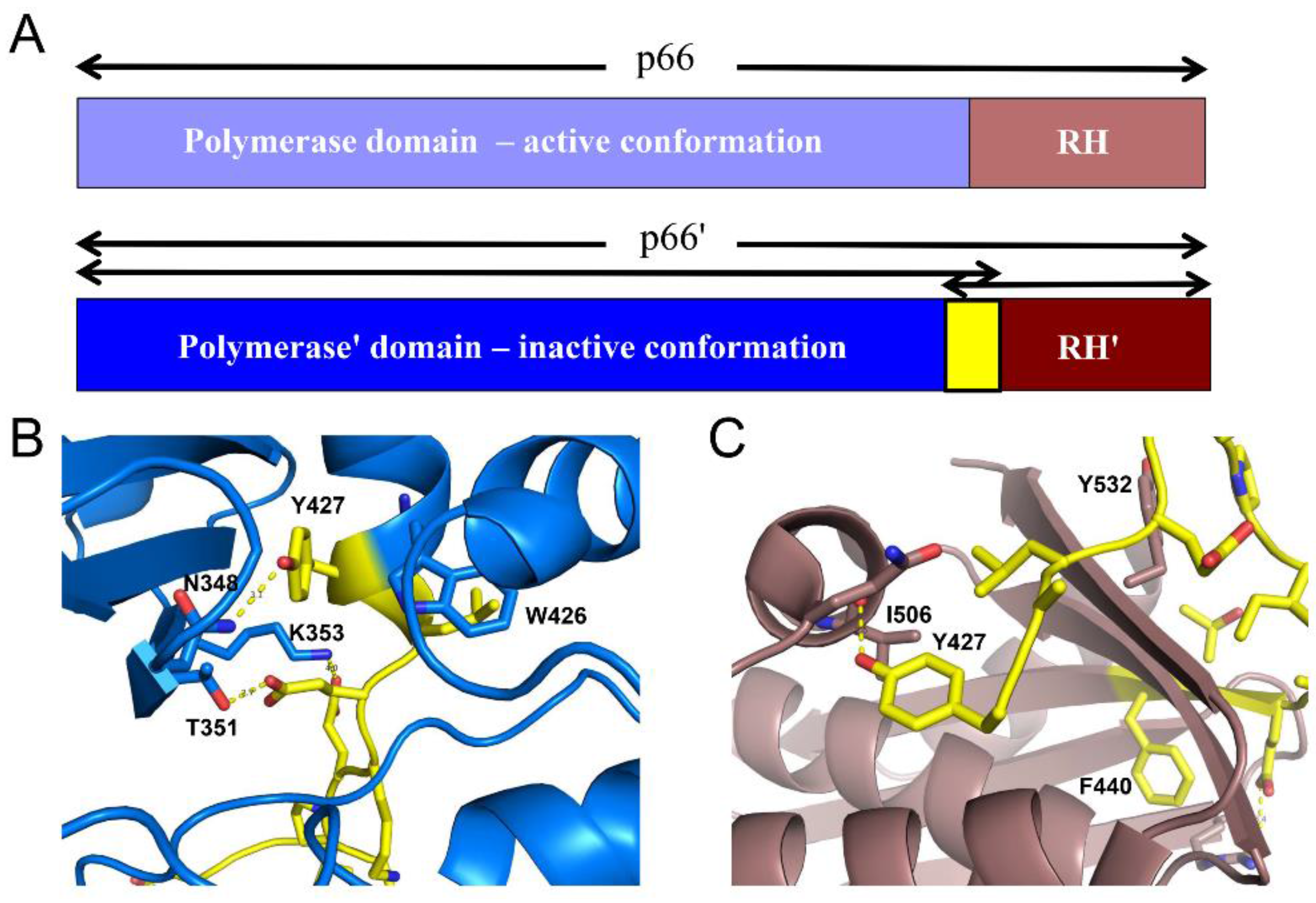
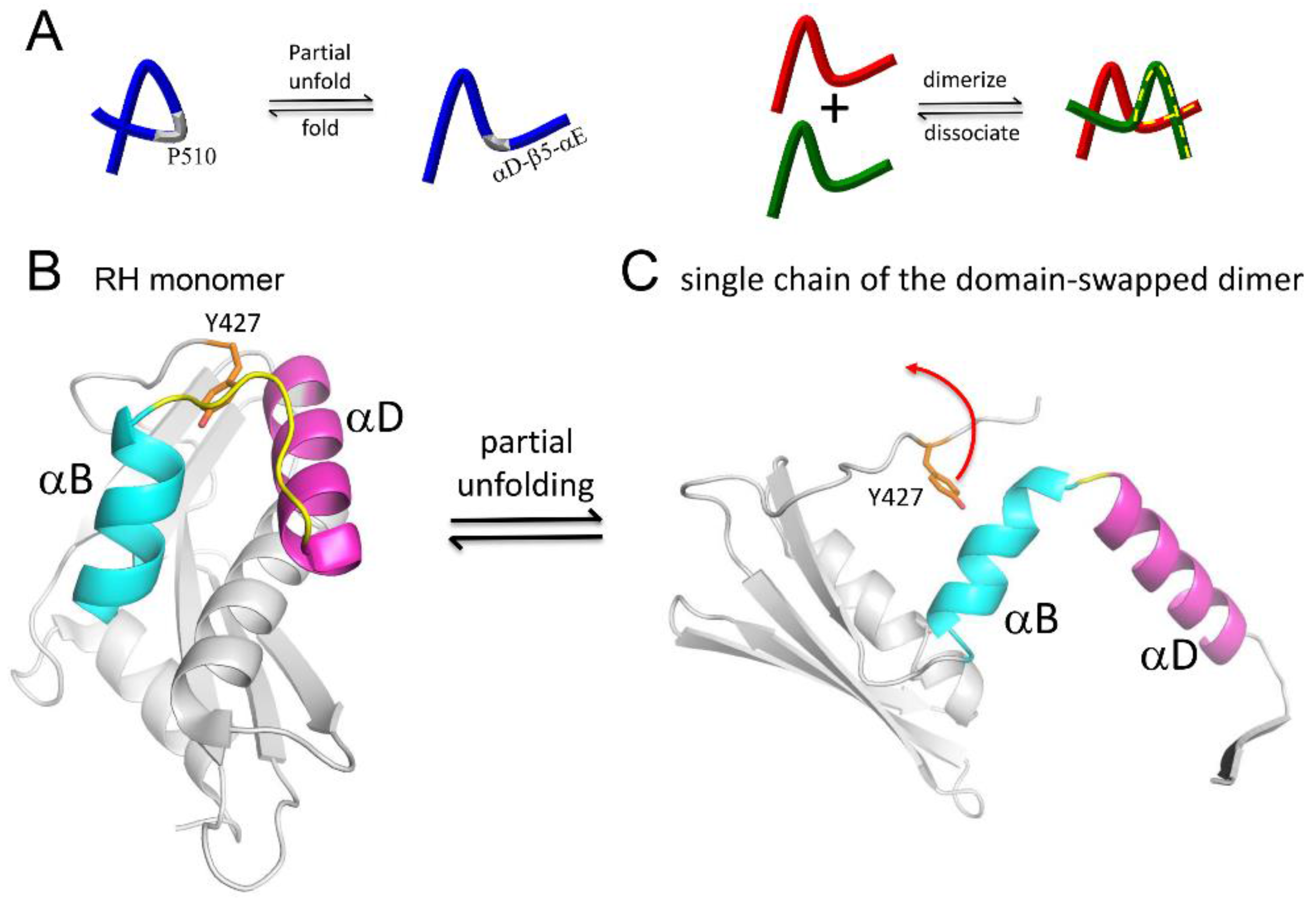
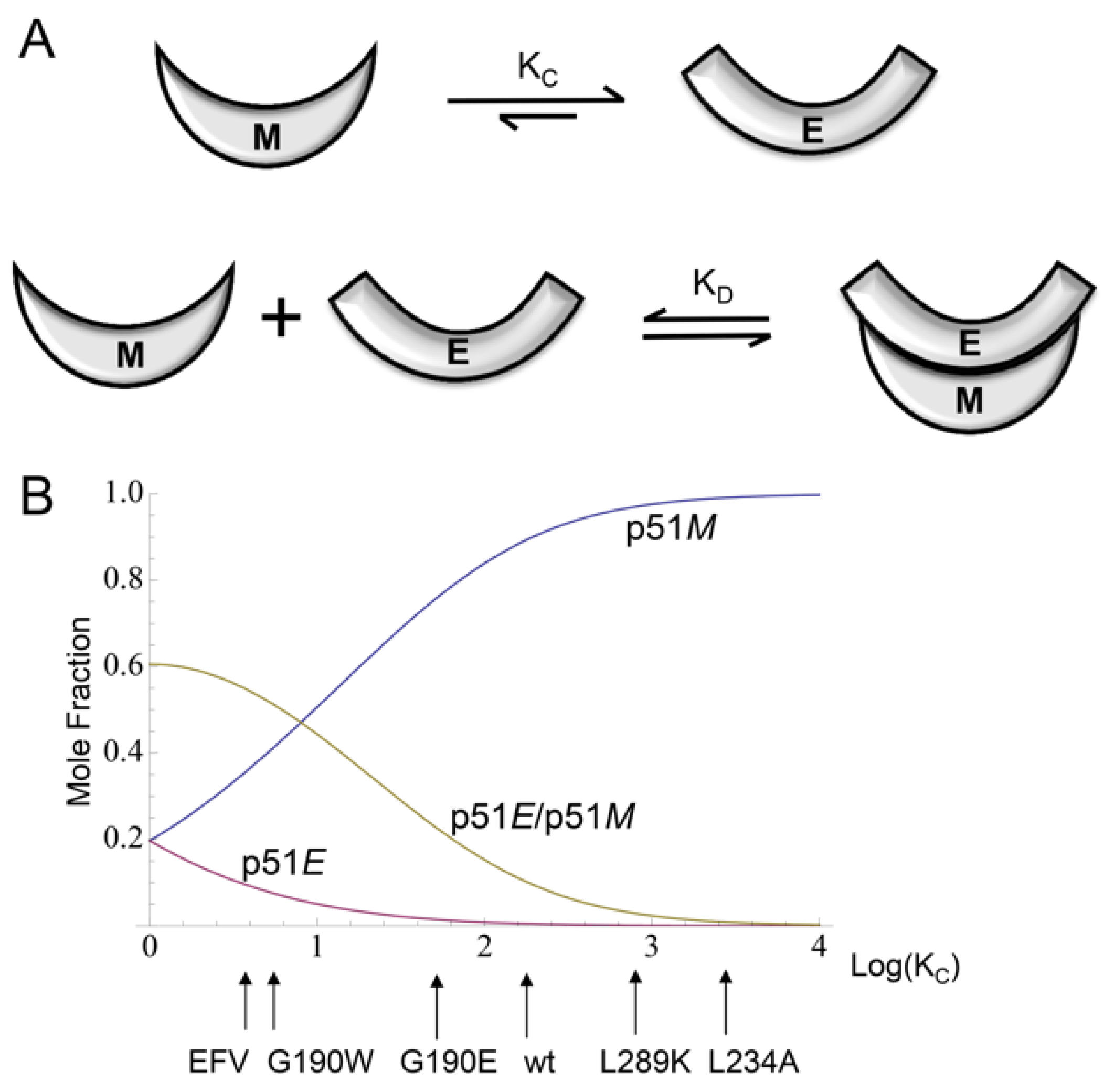
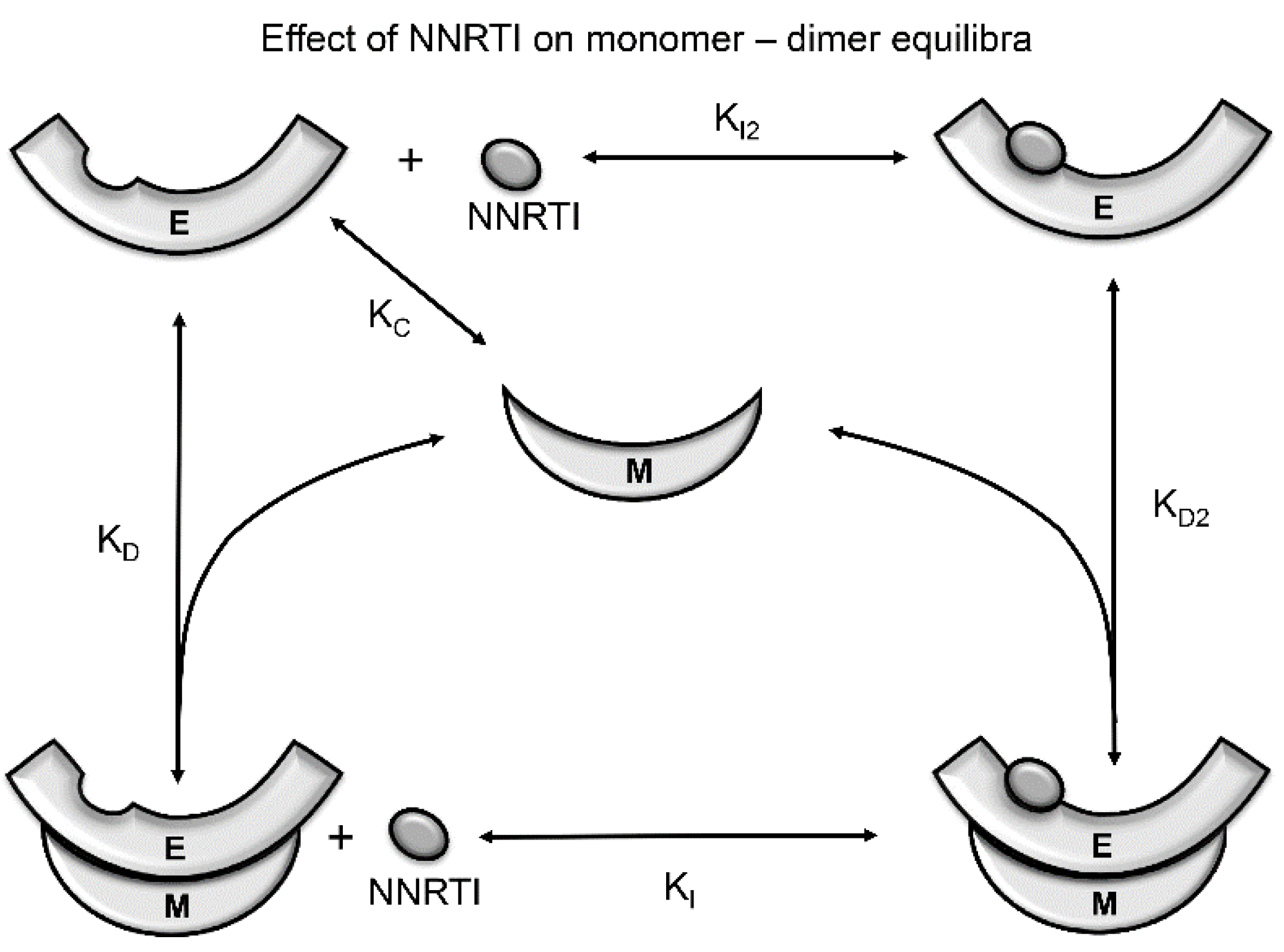
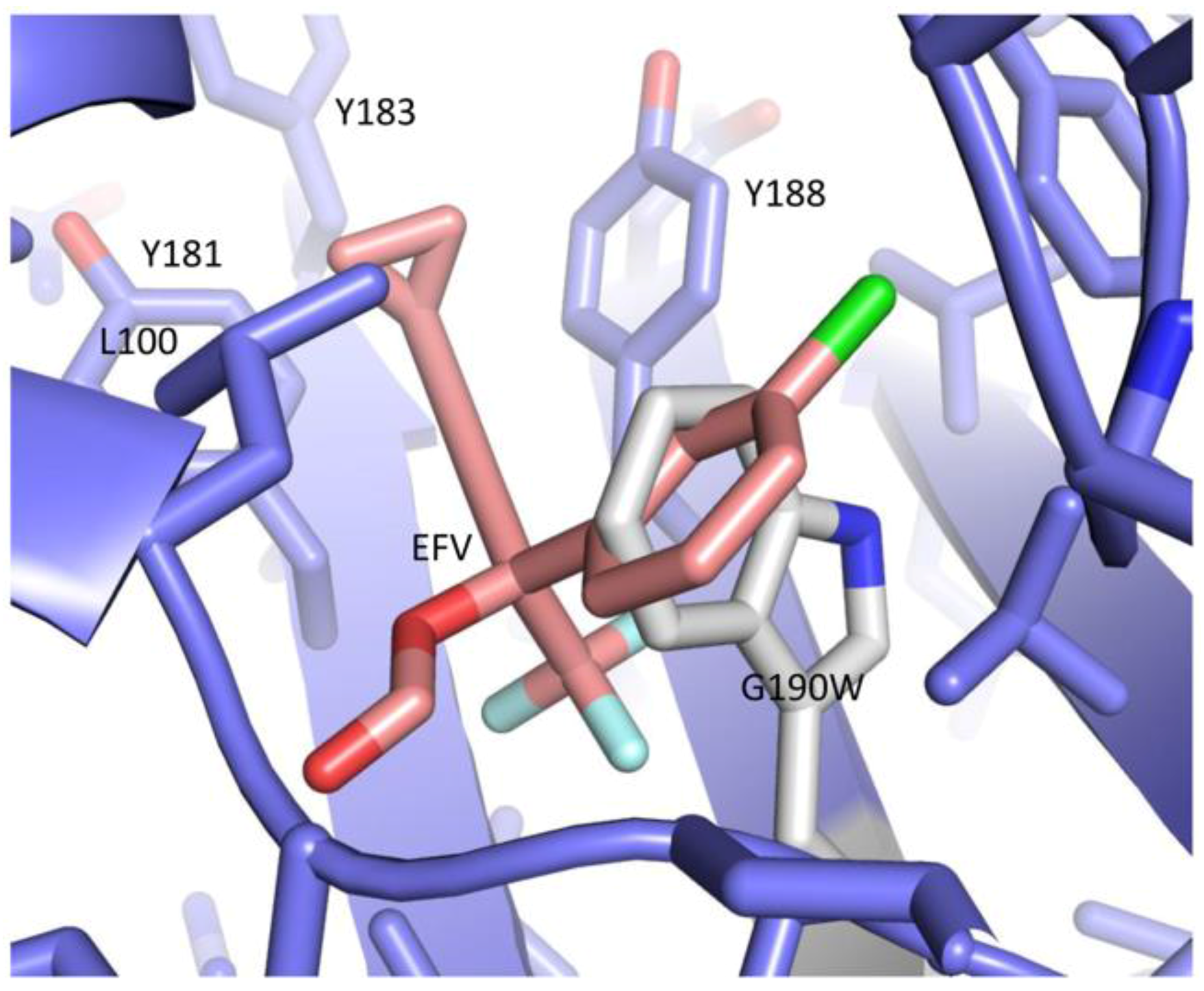
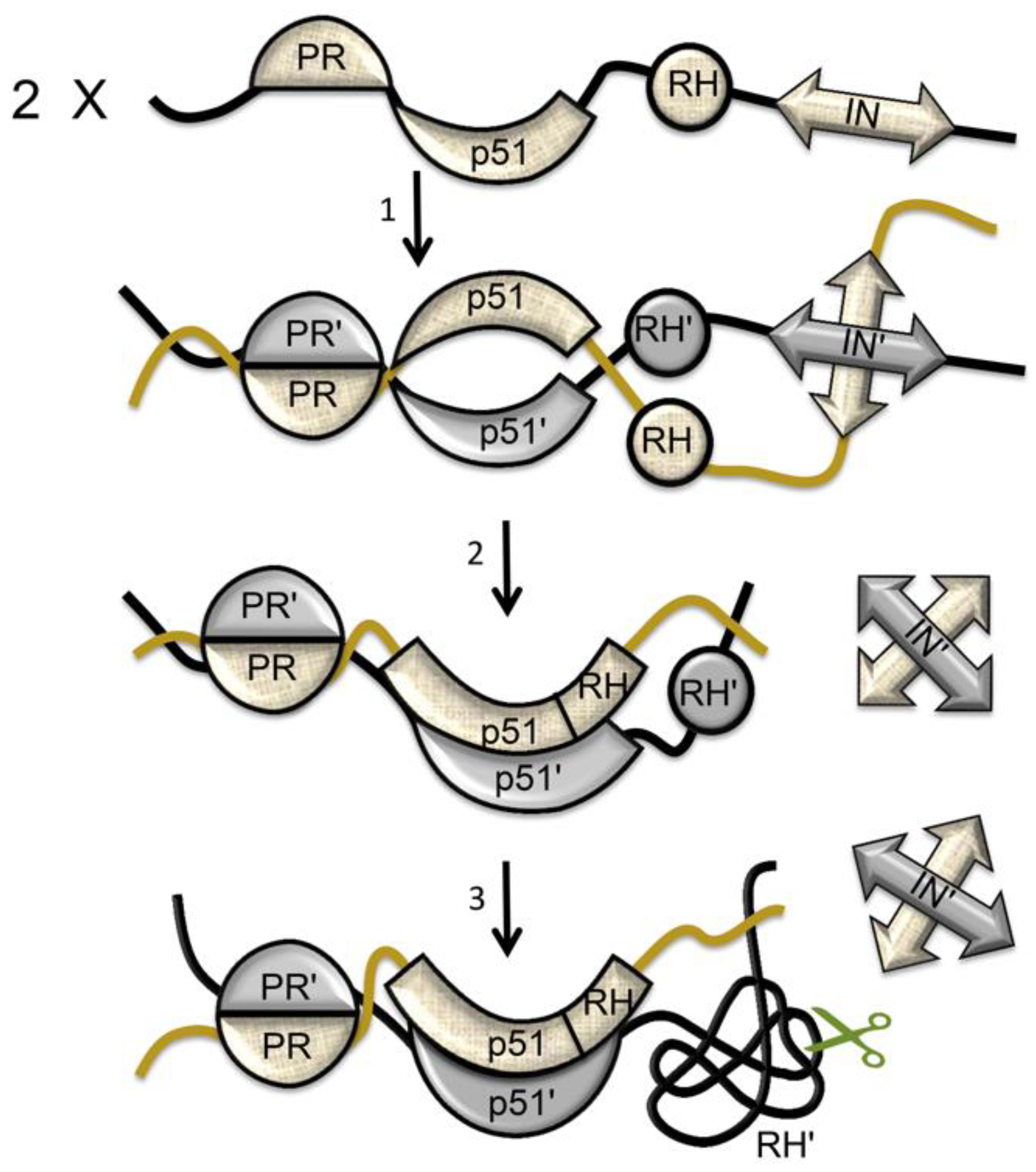
| Abbreviations | Definitions |
|---|---|
| Polymerase domain | Residues |
| Fingers (F) | 1–84; 119–154 |
| Palm (P)1 | 85–118; 155–241 |
| Thumb (T) | 242–313 |
| Connection (C) | 314–426 |
| RH—RNase H domain | 427–560 |
| Description/example | |
| p51E2 | Extended (E) structure of the active polymerase domain; corresponds to residues 1–426 of the p66 subunit of the reverse transcriptase (RT) heterodimer (Figure 1A) |
| p51C | Compact (C) structure of the polymerase domain; observed in the p51 subunit of RT (Figure 1A and Figure 2B) |
| p51M | Monomeric (M) p51 structure, observed in crystallized p51∆PL (Figure 3A) |
| p66E2 | p66 structure in the RT heterodimer (Figure 2A) |
| p66C3 | p66 structure containing an inactively-folded polymerase domain linked to a ribonuclease H (RH) domain |
| p66M | M structure of p66 (Figure 3B) |
| p66/p66’ | Homodimer in which the p66 subunit conformation approximates p66E, and p66’ matures from an initial p66M’-like conformation to a final conformation containing a p51C’ subunit linked to an unfolded RH’ domain |
| PDB ID | Description | Reference |
|---|---|---|
| 1DLO | Unliganded human immunodeficiency virus 1 (HIV-1) RT heterodimer | [27] |
| 1S9E | RT-NNRTI complex in which the often disordered residues of the p51 palm loop are observed | [28] |
| 4KSE | p51∆PL monomer, lacking the disordered palm loop—unable to adopt an E structure | [16] |
| 1HAR | RT216, a construct that includes the fingers and most of the palm domain | [29] |
| 1HRH | Isolated RH domain | [23] |
| 3K2P | Isolated RH-active site inhibitor complex | [30] |
| 5DZM | Domain-swapped RH dimer—each RH chain corresponds to a partially unfolded monomer | [31] |
| 1FK9 | RT heterodimer–efavirenz (EFV) complex | [32] |
| 1RTJ | RT heterodimer in which the p51 subunit extends to the Phe440 cleavage site residue | [33] |
| 1MU2 | Unliganded HIV-2 RT heterodimer | [34] |
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
London, R.E. Structural Maturation of HIV-1 Reverse Transcriptase—A Metamorphic Solution to Genomic Instability. Viruses 2016, 8, 260. https://doi.org/10.3390/v8100260
London RE. Structural Maturation of HIV-1 Reverse Transcriptase—A Metamorphic Solution to Genomic Instability. Viruses. 2016; 8(10):260. https://doi.org/10.3390/v8100260
Chicago/Turabian StyleLondon, Robert E. 2016. "Structural Maturation of HIV-1 Reverse Transcriptase—A Metamorphic Solution to Genomic Instability" Viruses 8, no. 10: 260. https://doi.org/10.3390/v8100260
APA StyleLondon, R. E. (2016). Structural Maturation of HIV-1 Reverse Transcriptase—A Metamorphic Solution to Genomic Instability. Viruses, 8(10), 260. https://doi.org/10.3390/v8100260





