Viral Mimicry to Usurp Ubiquitin and SUMO Host Pathways
Abstract
:1. Introduction
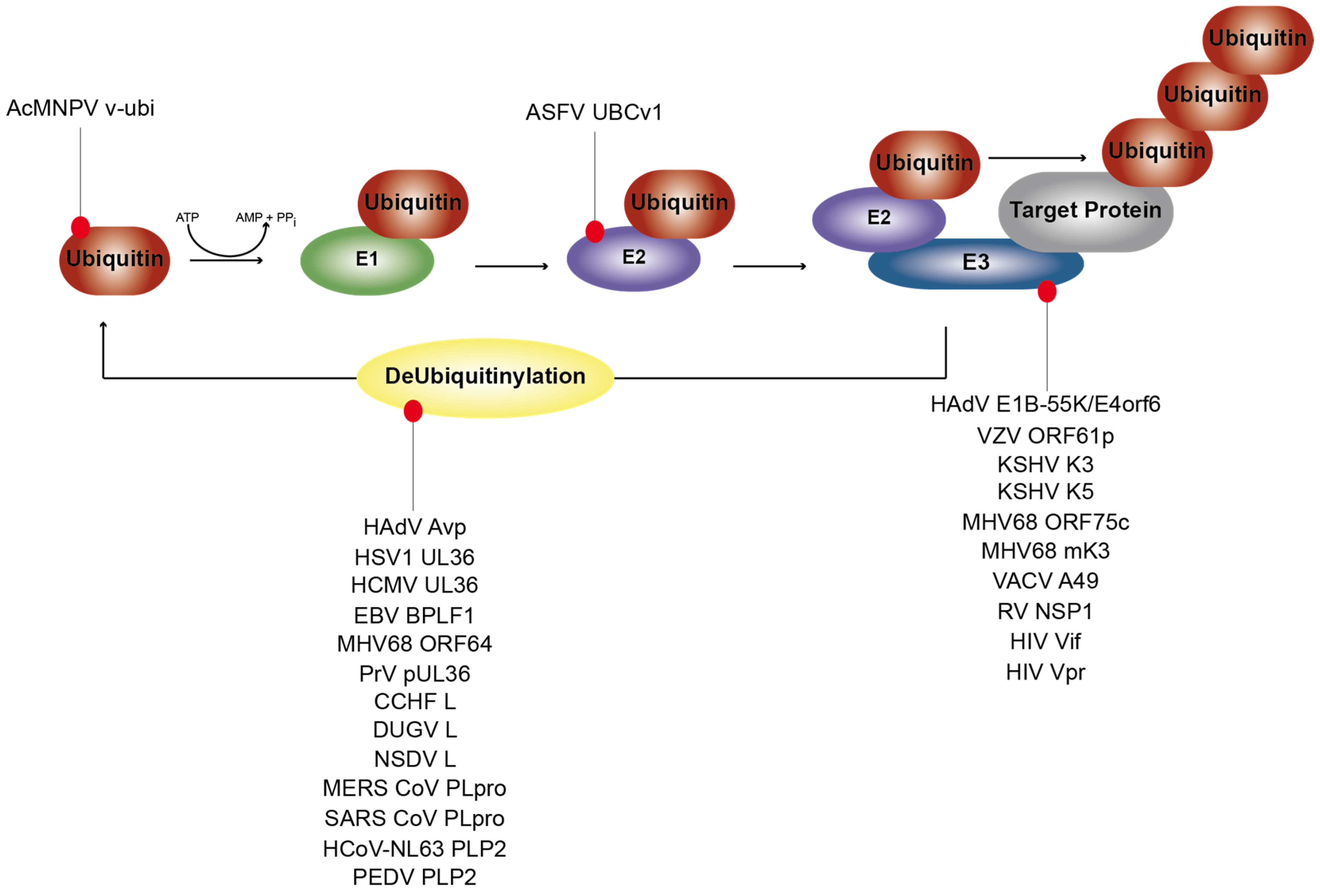
1.1. Ubiquitinylation and De-Ubiquitinylation Processes
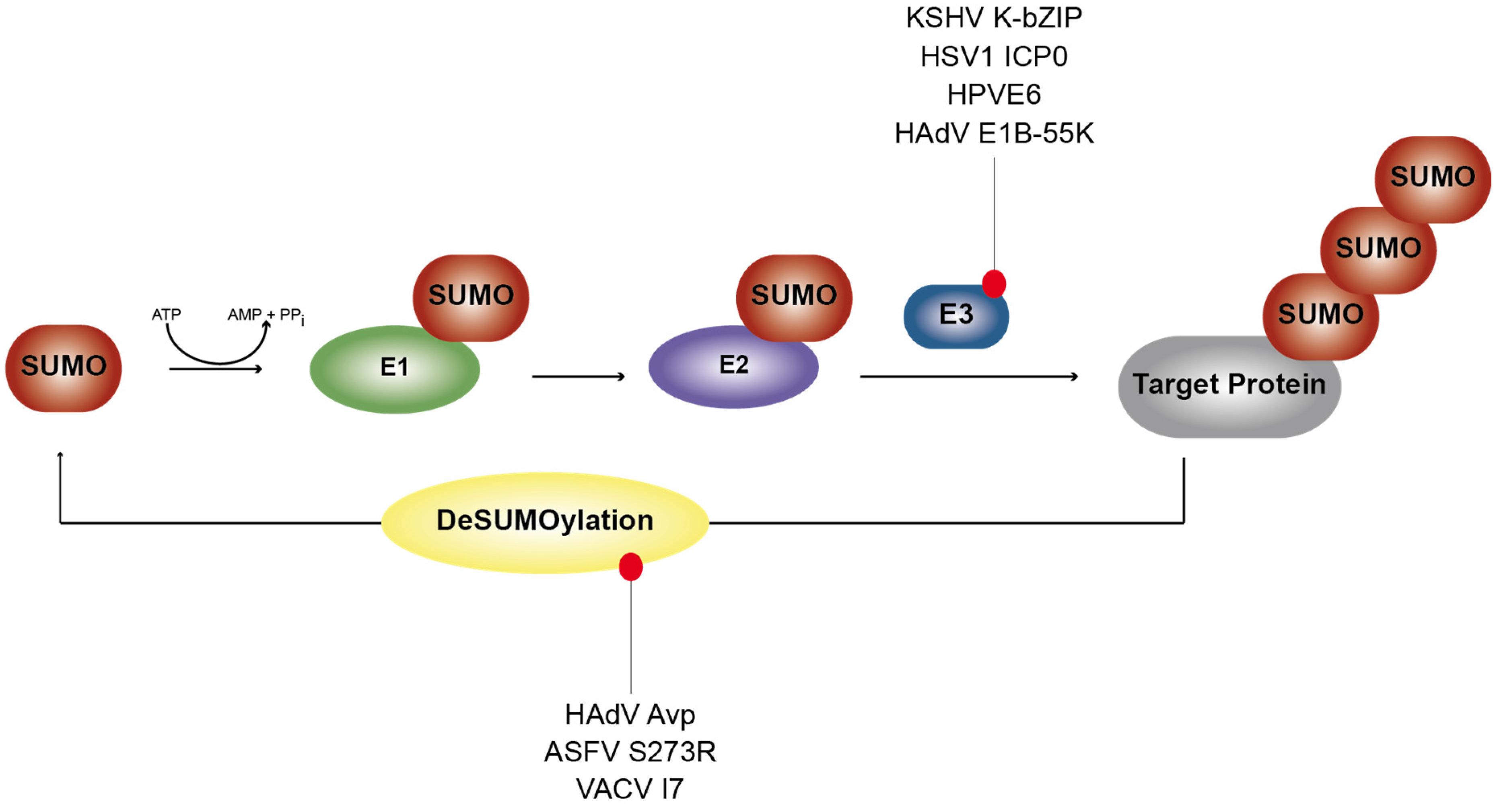
1.2. SUMOylation and De-SUMOylation Processes
2. Viral Mimicry of Ubiquitin or Ubiquitin E2/E3 Ligases
2.1. Viruses Expressing Viral Ubiquitin (v-ubi) Analogs
2.2. Viruses Encoding Viral Ubiquitin E2 Enzyme Counterparts
2.3. Viruses Synthesizing Ubiquitin E3 Enzyme-Like Proteins
2.4. Viruses Blocking Host Ubiquitin E3 Enzymes by Virus-Encoded Inhibitors
3. Virus-Mediated Cross-Talk between SUMO and Ubiquitin PTM
4. Viruses Hijacking De-Ubiquitinylation Processes for Efficient Replication
4.1. DNA Viruses Synthesize Viral DUBs to Counteract Ubiquitinylation Events
| Virus | Viral Homologue | Host Cell Function | Putative Consequence | References | |
|---|---|---|---|---|---|
| Human Adenoviruses (HAdV) | Avp | DUB | efficient viral replication | [101] | |
| Human Adenoviruses (HAdV) | E1B-55K E4orf6 | E3 ubiquitin ligase | efficient viral replication oncogenic potential | [25] | |
| Human Adenoviruses (HAdV) | Avp | SENP | efficient viral replication | [103,104] | |
| Human Adenoviruses (HAdV) | E1B-55K | E3 SUMO ligase | p53 SUMOylation oncogenic potential | [105,106,107] | |
| Human Papillomaviruses (HPV) | E6 | E3 ubiquitin ligase | efficient viral replication oncogenic potential | [48,49,50,51] | |
| Human herpes simplex virus type 1 (HSV1) | ICP0 | E3 ubiquitin ligase | efficient viral replication | [89,90] | |
| Human herpes simplex virus type 1 (HSV1) | UL36 | DUB | [108] | ||
| Human Cytomegalovirus (HCMV) | UL36 | DUB | [109] | ||
| Kaposi’s sarcoma associated herpesvirus (KSHV) | K3 K5 | E3 ubiquitin ligase | immune evasion | [38,39,40] | |
| Kaposi’s sarcoma associated herpesvirus (KSHV) | K-bZIP | E3 SUMO ligase | [110,111] | ||
| Epstein-Barr virus (EBV) | BPLF1 | DUB | [112] | ||
| Murine gammaherpesvirus 68 (MHV68) | ORF75c | E3 ubiquitin ligase | efficient viral replication | [96] | |
| Murine gammaherpesvirus 68 (MHV68) | ORF64 | DUB | [113] | ||
| Murine gammaherpesvirus 68 (MHV68) | mK3 | E3 ubiquitin ligase | immune evasion | [44,47] | |
| Varicella zoster virus (VZV) | ORF61p | E3 ubiquitin ligase | NF-κB inhibition | [92,93,95] | |
| Pseudorabies viruses (PrV) | pUL36 | DUB | [114] | ||
| African swine fever virus (ASFV) | S273R | SENP | efficient viral replication | [115] | |
| African swine fever virus (ASFV) | UBCv1 | ubiquitin conjugating activity | uncoating/assembly early/late transition virus DNA replication virus-mediated DNA repair | [23,24] | |
| Vaccinia virus (VACV) | I7 | SENP | [103,104] | ||
| Vaccinia virus (VACV) | A49 | E3 ubiquitin ligase inhibition | NF-κB inhibition viral virulence immune evasion | [77] | |
| Autographa californica nuclear polyhedrosis virus (AcMNPV) | v-ubi | ubiquitin homologue | substrate for ubiquitin processing enzymes | [21] | |
| Rotavirus (RV) | NSP1 | E3 ubiquitin ligase inhibition | NF-κB inhibition | [80,81,82] | |
| Human immunodeficiency virus (HIV) | Vif | E3 ubiquitin ligase | immune evasion | [63,64,65] | |
| Human immunodeficiency virus (HIV) | Vpr | E3 ubiquitin ligase | immune evasion | [69] | |
| Crimean–Congo hemorrhagic fever virus (CCHF) | L | DUB | viral replication | [116] | |
| Dugbe virus (DUGV) | L | DUB | viral replication | [116] | |
| Nairobi sheep disease virus (NSDV) | L | DUB | viral replication | [116] | |
| Severe acute respiratory syndrome coronavirus (SARS CoV) | PLpro | DUB | de-ISGylation immune evasion | [117,118] | |
| Middle East respiratory syndrome coronavirus (MERS-CoV) | PLpro | DUB | de-ISGylation immune evasion | [117,119,120,121] | |
| Human coronavirus NL63 (HCoV-NL63) | PLP2 | DUB | de-ISGylation immune evasion | [117,119,120,121] | |
| Porcine epidemic diarrhea virus (PEDV) | PLP2 | DUB | de-ISGylation immune evasion | [117,119,120,121] | |
| Mouse hepatitis virus (MHV) | PLP2 | DUB | de-ISGylation immune evasion | [117,119,120,121] | |
4.2. DUBs Encoded by RNA Viruses Limiting Host Ubiquitinylation
5. Viral Analogs of the Host SUMOylation Pathway
5.1. Viruses Mimicking Cellular SENPs
5.2. Viruses Expressing SUMO Ligase-Like Activity
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Hunter, T.; Sun, H. Crosstalk between the SUMO and ubiquitin pathways. Ernst Scher. Found. Symp. Proc. 2009, 2008/1, 1–16. [Google Scholar]
- Iniguez-Lluhi, J.A. For a healthy histone code, a little SUMO in the tail keeps the acetyl away. ACS Chem. Biol. 2006, 1, 204–206. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Chiang, C.M. SUMOylation in gene regulation, human disease, and therapeutic action. F1000Prime Rep. 2013, 5, e45. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, P.; Schreiner, S.; Dobner, T. Human pathogens and the host cell SUMOylation system. J. Virol. 2012, 86, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, D.H.; Goldstein, G.; Niall, H.D. The complete amino acid sequence of ubiquitin, an adenylate cyclase stimulating polypeptide probably universal in living cells. Biochemistry 1975, 14, 2214–2218. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, D.H.; Goldstein, G. Molecular conservation of 74 amino acid sequence of ubiquitin between cattle and man. Nature 1975, 255, 423–424. [Google Scholar] [CrossRef] [PubMed]
- McDowell, G.S.; Philpott, A. Non-canonical ubiquitylation: Mechanisms and consequences. Int. J. Biochem. Cell Biol. 2013, 45, 1833–1842. [Google Scholar] [CrossRef] [PubMed]
- Voutsadakis, I.A. Ubiquitination and the ubiquitin-proteasome system as regulators of transcription and transcription factors in epithelial mesenchymal transition of cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2012, 33, 897–910. [Google Scholar] [CrossRef] [PubMed]
- Trempe, J.F. Reading the ubiquitin postal code. Curr. Opin. Struct. Biol. 2011, 21, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and rRING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012, 125, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Ardley, H.C.; Robinson, P.A. E3 ubiquitin ligases. Essays Biochem. 2005, 41, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Spratt, D.E.; Walden, H.; Shaw, G.S. Rbr E3 ubiquitin ligases: New structures, new insights, new questions. Biochem. J. 2014, 458, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.L.; Jin, G.; Li, C.F.; Jeong, Y.S.; Moten, A.; Xu, D.; Feng, Z.; Chen, W.; Cai, Z.; Darnay, B.; et al. Cycles of ubiquitination and deubiquitination critically regulate growth factor-mediated activation of AKT signaling. Sci. Signal. 2013, 6. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Hannoun, Z.; Greenhough, S.; Jaffray, E.; Hay, R.T.; Hay, D.C. Post-Translational modification by SUMO. Toxicology 2010, 278, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Matunis, M.J.; Coutavas, E.; Blobel, G. A novel ubiquitin-like modification modulates the partitioning of the ran-gtpase-activating protein rangap1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996, 135, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Tatham, M.H.; Kim, S.; Jaffray, E.; Song, J.; Chen, Y.; Hay, R.T. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat. Struct. Mol. Biol. 2005, 12, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Bernier-Villamor, V.; Sampson, D.A.; Matunis, M.J.; Lima, C.D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 2002, 108, 345–356. [Google Scholar] [CrossRef]
- Vertegaal, A.C. Small ubiquitin-related modifiers in chains. Biochem. Soc. Trans. 2007, 35, 1422–1423. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, P.; Blanchette, P.; Schreiner, S.; Ching, W.; Groitl, P.; Berscheminski, J.; Branton, P.E.; Will, H.; Dobner, T. Cross-Talk between phosphorylation and SUMOylation regulates transforming activities of an adenoviral oncoprotein. Oncogene 2013, 32, 1626–1637. [Google Scholar] [CrossRef] [PubMed]
- Guarino, L.A. Identification of a viral gene encoding a ubiquitin-like protein. Proc. Natl. Acad. Sci. USA 1990, 87, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Chapman, D.A.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Hingamp, P.M.; Arnold, J.E.; Mayer, R.J.; Dixon, L.K. A ubiquitin conjugating enzyme encoded by african swine fever virus. EMBO J. 1992, 11, 361–366. [Google Scholar] [PubMed]
- Hingamp, P.M.; Leyland, M.L.; Webb, J.; Twigger, S.; Mayer, R.J.; Dixon, L.K. Characterization of a ubiquitinated protein which is externally located in african swine fever virions. J. Virol. 1995, 69, 1785–1793. [Google Scholar] [PubMed]
- Schreiner, S.; Wimmer, P.; Dobner, T. Adenovirus degradation of cellular proteins. Future Microbiol. 2012, 7, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.; Rohleder, K.J.; Hanakahi, L.A.; Ketner, G. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 2007, 81, 7034–7040. [Google Scholar] [CrossRef] [PubMed]
- Querido, E.; Blanchette, P.; Yan, Q.; Kamura, T.; Morrison, M.; Boivin, D.; Kaelin, W.G.; Conaway, R.C.; Conaway, J.W.; Branton, P.E. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a cullin-containing complex. Genes Dev. 2001, 15, 3104–3117. [Google Scholar] [CrossRef] [PubMed]
- Stracker, T.H.; Carson, C.T.; Weitzman, M.D. Adenovirus oncoproteins inactivate the Mre11 Rad50 NBS1 DNA repair complex. Nature 2002, 418, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Orazio, N.I.; Naeger, C.M.; Karlseder, J.; Weitzman, M.D. The adenovirus e1b55k/e4orf6 complex induces degradation of the bloom helicase during infection. J. Virol. 2011, 85, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Dallaire, F.; Blanchette, P.; Groitl, P.; Dobner, T.; Branton, P.E. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton e3 ubiquitin ligase complex. J. Virol. 2009, 83, 5329–5338. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, S.; Kinkley, S.; Bürck, C.; Mund, A.; Wimmer, P.; Schubert, T.; Groitl, P.; Will, H.; Dobner, T. SPOC1-mediated antiviral host cell response is antagonized early in human adenovirus type 5 infection. PLoS Pathog. 2013, 9, e1003775. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, S.; Bürck, C.; Glass, M.; Groitl, P.; Wimmer, P.; Kinkley, S.; Mund, A.; Everett, R.D.; Dobner, T. Control of human adenovirus type 5 (Ad5) gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res. 2013, 41, 3532–3550. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Jha, S.; Engel, D.A.; Ornelles, D.A.; Dutta, A. Tip60 degradation by adenovirus relieves transcriptional repression of viral transcriptional activator E1A. Oncogene 2013, 32, 5017–5025. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Patel, R.N.; Forrester, N.A.; Theil, K.; Groitl, P.; Stewart, G.S.; Taylor, A.M.; Morgan, I.M.; Dobner, T.; Grand, R.J.; et al. Adenovirus 12 E4orf6 inhibits atr activation by promoting topbp1 degradation. Proc. Natl. Acad. Sci. USA 2010, 107, 12251–12256. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, S.; Wimmer, P.; Sirma, H.; Everett, R.D.; Blanchette, P.; Groitl, P.; Dobner, T. Proteasome-dependent degradation of daxx by the viral E1b-55K protein in human adenovirus-infected cells. J. Virol. 2010, 84, 7029–7038. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Gilson, T.; Dallaire, F.; Ketner, G.; Branton, P.E.; Blanchette, P. The E4orf6/E1B55k E3 ubiquitin ligase complexes of human adenoviruses exhibit heterogenity in composition and substrate specificity. J. Virol. 2011, 85, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Forrester, N.A.; Sedgwick, G.G.; Thomas, A.; Blackford, A.N.; Speiseder, T.; Dobner, T.; Byrd, P.J.; Stewart, G.S.; Turnell, A.S.; Grand, R.J. Serotype-specific inactivation of the cellular DNA damage response during adenovirus infection. J. Virol. 2010, 85, 2201–2211. [Google Scholar] [CrossRef] [PubMed]
- Coscoy, L.; Ganem, D. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 2000, 97, 8051–8056. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Ueda, K.; Nakano, K.; Hirata, Y.; Parravicini, C.; Corbellino, M.; Yamanishi, K. Major histocompatibility complex class i molecules are down-regulated at the cell surface by the K5 protein encoded by kaposi's sarcoma-associated herpesvirus/human herpesvirus-8. J. Gen. Virol. 2001, 82, 1175–1180. [Google Scholar] [PubMed]
- Boname, J.M.; Lehner, P.J. What has the study of the K3 and K5 viral ubiquitin E3 ligases taught us about ubiquitin-mediated receptor regulation? Viruses 2011, 3, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Duncan, L.M.; Piper, S.; Dodd, R.B.; Saville, M.K.; Sanderson, C.M.; Luzio, J.P.; Lehner, P.J. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006, 25, 1635–1645. [Google Scholar] [CrossRef] [PubMed]
- Bruce, A.G.; Ryan, J.T.; Thomas, M.J.; Peng, X.; Grundhoff, A.; Tsai, C.C.; Rose, T.M. Next-generation sequence analysis of the genome of rfhvmn, the macaque homolog of Kaposi's Sarcoma (KS)-associated herpesvirus, from a KS-like tumor of a pig-tailed macaque. J. Virol. 2013, 87, 13676–13693. [Google Scholar] [CrossRef] [PubMed]
- Virgin, H.W.T.; Latreille, P.; Wamsley, P.; Hallsworth, K.; Weck, K.E.; Dal Canto, A.J.; Speck, S.H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 1997, 71, 5894–5904. [Google Scholar] [PubMed]
- Brulois, K.; Jung, J.U. Interplay between kaposi's sarcoma-associated herpesvirus and the innate immune system. Cytokine Growth Factor Rev. 2014, 25, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Searles, R.P.; Bergquam, E.P.; Axthelm, M.K.; Wong, S.W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 1999, 73, 3040–3053. [Google Scholar] [PubMed]
- Ishido, S.; Choi, J.K.; Lee, B.S.; Wang, C.; DeMaria, M.; Johnson, R.P.; Cohen, G.B.; Jung, J.U. Inhibition of natural killer cell-mediated cytotoxicity by kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 2000, 13, 365–374. [Google Scholar] [CrossRef]
- Stevenson, P.G.; May, J.S.; Smith, X.G.; Marques, S.; Adler, H.; Koszinowski, U.H.; Simas, J.P.; Efstathiou, S. K3-mediated evasion of CD8(+) T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 2002, 3, 733–740. [Google Scholar] [PubMed]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991, 10, 4129–4135. [Google Scholar] [PubMed]
- Scheffner, M.; Huibregtse, J.M.; Howley, P.M. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc. Natl. Acad. Sci. USA 1994, 91, 8797–8801. [Google Scholar] [CrossRef] [PubMed]
- Nuber, U.; Schwarz, S.; Kaiser, P.; Schneider, R.; Scheffner, M. Cloning of human ubiquitin-conjugating enzymes Ubch6 and Ubch7 (e2-f1) and characterization of their interaction with E6-AP and Rsp5. J. Biol. Chem. 1996, 271, 2795–2800. [Google Scholar] [CrossRef] [PubMed]
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 1995, 373, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Scheffner, M.; Takahashi, T.; Huibregtse, J.M.; Minna, J.D.; Howley, P.M. Interaction of the human papillomavirus type 16 E6 oncoprotein with wild-type and mutant human p53 proteins. J. Virol. 1992, 66, 5100–5105. [Google Scholar] [PubMed]
- Band, V.; De Caprio, J.A.; Delmolino, L.; Kulesa, V.; Sager, R. Loss of p53 protein in human papillomavirus type 16 E6-immortalized human mammary epithelial cells. J. Virol. 1991, 65, 6671–6676. [Google Scholar] [PubMed]
- Gross-Mesilaty, S.; Reinstein, E.; Bercovich, B.; Tobias, K.E.; Schwartz, A.L.; Kahana, C.; Ciechanover, A. Basal and human papillomavirus e6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 8058–8063. [Google Scholar] [CrossRef] [PubMed]
- Kuhne, C.; Banks, L. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the l2g box. J. Biol. Chem. 1998, 273, 34302–34309. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Banks, L. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene 1998, 17, 2943–2954. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Srinivasan, S.; Boyer, S.N.; Wazer, D.E.; Band, V. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative gap protein, E6tp1, and target it for degradation. Mol. Cell. Biol. 1999, 19, 733–744. [Google Scholar] [PubMed]
- Gewin, L.; Myers, H.; Kiyono, T.; Galloway, D.A. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004, 18, 2269–2282. [Google Scholar] [CrossRef] [PubMed]
- Berezutskaya, E.; Yu, B.; Morozov, A.; Raychaudhuri, P.; Bagchi, S. Differential regulation of the pocket domains of the retinoblastoma family proteins by the HPV 16 E7 oncoprotein. Cell. Growth Differ. 1997, 8, 1277–1286. [Google Scholar] [PubMed]
- Boyer, S.N.; Wazer, D.E.; Band, V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996, 56, 4620–4624. [Google Scholar] [PubMed]
- Huh, K.; Zhou, X.; Hayakawa, H.; Cho, J.Y.; Libermann, T.A.; Jin, J.; Harper, J.W.; Munger, K. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J. Virol. 2007, 81, 9737–9747. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Yu, Y.; Liu, B.; Luo, K.; Kong, W.; Mao, P.; Yu, X.F. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 2003, 302, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Mehle, A.; Strack, B.; Ancuta, P.; Zhang, C.; McPike, M.; Gabuzda, D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2004, 279, 7792–7798. [Google Scholar] [CrossRef] [PubMed]
- Mehle, A.; Thomas, E.R.; Rajendran, K.S.; Gabuzda, D. A zinc-binding region in vif binds Cul5 and determines cullin selection. J. Biol. Chem. 2006, 281, 17259–17265. [Google Scholar] [CrossRef] [PubMed]
- Balliet, J.W.; Kolson, D.L.; Eiger, G.; Kim, F.M.; McGann, K.A.; Srinivasan, A.; Collman, R. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: Mutational analysis of a primary HIV-1 isolate. Virology 1994, 200, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Jowett, J.B.; Planelles, V.; Poon, B.; Shah, N.P.; Chen, M.L.; Chen, I.S. The human immunodeficiency virus type 1 Vpr gene arrests infected t cells in the G2 + M phase of the cell cycle. J. Virol. 1995, 69, 6304–6313. [Google Scholar] [PubMed]
- Rogel, M.E.; Wu, L.I.; Emerman, M. The human immunodeficiency virus type 1 Vpr gene prevents cell proliferation during chronic infection. J. Virol. 1995, 69, 882–888. [Google Scholar] [PubMed]
- Casey, L.; Wen, X.; de Noronha, C.M. The functions of the HIV1 protein Vpr and its action through the DCAF1.DDB1.Cullin4 ubiquitin ligase. Cytokine 2010, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shiyanov, P.; Nag, A.; Raychaudhuri, P. Cullin 4a associates with the uv-damaged DNA-binding protein DDB. J. Biol. Chem. 1999, 274, 35309–35312. [Google Scholar] [CrossRef] [PubMed]
- Schrofelbauer, B.; Yu, Q.; Zeitlin, S.G.; Landau, N.R. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 2005, 79, 10978–10987. [Google Scholar] [CrossRef] [PubMed]
- Okumura, A.; Alce, T.; Lubyova, B.; Ezelle, H.; Strebel, K.; Pitha, P.M. HIV-1 accessory proteins Vpr and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology 2008, 373, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Fenner, F. A successful eradication campaign. Global eradication of smallpox. Rev. Infect. Dis 1982, 4, 916–930. [Google Scholar] [CrossRef] [PubMed]
- Seet, B.T.; Johnston, J.B.; Brunetti, C.R.; Barrett, J.W.; Everett, H.; Cameron, C.; Sypula, J.; Nazarian, S.H.; Lucas, A.; McFadden, G. Poxviruses and immune evasion. Annu. Rev. Immunol 2003, 21, 377–423. [Google Scholar] [CrossRef] [PubMed]
- Aguado, B.; Selmes, I.P.; Smith, G.L. Nucleotide sequence of 21.8 kbp of variola major virus strain harvey and comparison with vaccinia virus. J. Gen. Virol. 1992, 73, 2887–2902. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, E.; Greenbaum, J.A.; Sundstrom, M.; Schaffer, L.; Hammond, J.A.; Pasquetto, V.; Oseroff, C.; Hendrickson, R.C.; Lefkowitz, E.J.; Tscharke, D.C.; et al. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc. Natl. Acad. Sci. USA 2008, 105, 2140–2145. [Google Scholar] [CrossRef] [PubMed]
- Mansur, D.S.; Maluquer de Motes, C.; Unterholzner, L.; Sumner, R.P.; Ferguson, B.J.; Ren, H.; Strnadova, P.; Bowie, A.G.; Smith, G.L. Poxvirus targeting of E3 ligase beta-trcp by molecular mimicry: A mechanism to inhibit NFkB activation and promote immune evasion and virulence. PLoS Pathog. 2013, 9, e1003183. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Steele, A.D.; Duque, J.; Parashar, U.D.; WHO-coordinated Global Rotavirus Surveillance Network. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 136–141. [Google Scholar] [CrossRef]
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef] [PubMed]
- Broquet, A.H.; Hirata, Y.; McAllister, C.S.; Kagnoff, M.F. RIG-I/MDA5/MAVS are required to signal a protective ifn response in rotavirus-infected intestinal epithelium. J. Immunol. 2011, 186, 1618–1626. [Google Scholar] [CrossRef] [PubMed]
- Uzri, D.; Greenberg, H.B. Characterization of rotavirus RNAs that activate innate immune signaling through the RIG-I-like receptors. PLoS ONE 2013, 8, e69825. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Pruijssers, A.J.; Dermody, T.S.; Garcia-Sastre, A.; Greenberg, H.B. The early interferon response to rotavirus is regulated by PKR and depends on MAVS/IPS-1, RIG-I, MDA5, and IRF3. J. Virol. 2011, 85, 3717–3732. [Google Scholar] [CrossRef] [PubMed]
- Graff, J.W.; Ettayebi, K.; Hardy, M.E. Rotavirus NSP1 inhibits nfkappab activation by inducing proteasome-dependent degradation of beta-TRCP: A novel mechanism of ifn antagonism. PLoS Pathog. 2009, 5, e1000280. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, P.; Bhowmick, R.; Nandi, S.; Kant Nayak, M.; Chawla-Sarkar, M. Rotavirus nsp1 inhibits interferon induced non-canonical NFkB activation by interacting with TNF receptor associated factor 2. Virology 2013, 444, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Sriramachandran, A.M.; Dohmen, R.J. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 2014, 1843, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Tatham, M.H.; Geoffroy, M.C.; Shen, L.; Plechanovova, A.; Hattersley, N.; Jaffray, E.G.; Palvimo, J.J.; Hay, R.T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008, 10, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Leverson, J.D.; Hunter, T. Conserved function of RNF4 family proteins in eukaryotes: Targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007, 26, 4102–4112. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Hunter, T. Poly-small ubiquitin-like modifier (polysumo)-binding proteins identified through a string search. J. Biol. Chem. 2012, 287, 42071–42083. [Google Scholar] [CrossRef] [PubMed]
- Boutell, C.; Orr, A.; Everett, R.D. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J. Virol. 2003, 77, 8686–8694. [Google Scholar] [CrossRef] [PubMed]
- Boutell, C.; Cuchet-Lourenco, D.; Vanni, E.; Orr, A.; Glass, M.; McFarlane, S.; Everett, R.D. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 2011, 7, e1002245. [Google Scholar] [CrossRef] [PubMed]
- Chaurushiya, M.S.; Lilley, C.E.; Aslanian, A.; Meisenhelder, J.; Scott, D.C.; Landry, S.; Ticau, S.; Boutell, C.; Yates, J.R., 3rd; Schulman, B.A.; et al. Viral E3 ubiquitin ligase-mediated degradation of a cellular E3: Viral mimicry of a cellular phosphorylation mark targets the RNF8 fha domain. Mol. Cell 2012, 46, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Kyratsous, C.A.; Silverstein, S.J. Components of nuclear domain 10 bodies regulate varicella-zoster virus replication. J. Virol. 2009, 83, 4262–4274. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Oliver, S.L.; Sommer, M.; Rajamani, J.; Reichelt, M.; Arvin, A.M. Disruption of PML nuclear bodies is mediated by orf61 SUMO-interacting motifs and required for varicella-zoster virus pathogenesis in skin. PLoS Pathog. 2011, 7, e1002157. [Google Scholar] [CrossRef] [PubMed]
- Walters, M.S.; Kyratsous, C.A.; Silverstein, S.J. The ring finger domain of varicella-zoster virus orf61p has E3 ubiquitin ligase activity that is essential for efficient autoubiquitination and dispersion of sp100-containing nuclear bodies. J. Virol. 2010, 84, 6861–6865. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zheng, C.; Xing, J.; Wang, S.; Li, S.; Lin, R.; Mossman, K.L. Varicella-zoster virus immediate-early protein orf61 abrogates the IRF3-mediated innate immune response through degradation of activated IRF3. J. Virol. 2011, 85, 11079–11089. [Google Scholar] [CrossRef] [PubMed]
- Ling, P.D.; Tan, J.; Sewatanon, J.; Peng, R. Murine gammaherpesvirus 68 open reading frame 75c tegument protein induces the degradation of PML and is essential for production of infectious virus. J. Virol. 2008, 82, 8000–8012. [Google Scholar] [CrossRef] [PubMed]
- Mangel, W.F.; Baniecki, M.L.; McGrath, W.J. Specific interactions of the adenovirus proteinase with the viral DNA, an 11-amino-acid viral peptide, and the cellular protein actin. Cell. Mol. Life Sci. 2003, 60, 2347–2355. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.W. The proteinase polypeptide of adenovirus serotype 2 virions. Virology 1990, 177, 259–272. [Google Scholar] [CrossRef]
- Weber, J.M. Adenovirus endopeptidase and its role in virus infection. Curr. Top. Microbiol. Immunol. 1995, 199, 227–235. [Google Scholar] [PubMed]
- Cotten, M.; Weber, J.M. The adenovirus protease is required for virus entry into host cells. Virology 1995, 213, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Balakirev, M.Y.; Jaquinod, M.; Haas, A.L.; Chroboczek, J. Deubiquitinating function of adenovirus proteinase. J. Virol. 2002, 76, 6323–6331. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.; Ornelles, D.A.; Shenk, T. The adenovirus l3 23-kilodalton proteinase cleaves the amino-terminal head domain from cytokeratin 18 and disrupts the cytokeratin network of hela cells. J. Virol. 1993, 67, 3507–3514. [Google Scholar] [PubMed]
- Li, S.J.; Hochstrasser, M. A new protease required for cell-cycle progression in yeast. Nature 1999, 398, 246–251. [Google Scholar] [PubMed]
- Mukhopadhyay, D.; Dasso, M. Modification in reverse: The SUMO proteases. Trends Biochem. Sci. 2007, 32, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, P.; Berscheminski, J.; Blanchette, P.; Groitl, P.; Branton, P.E.; Hay, R.T.; Dobner, D.; Schreiner, S. Pml isoforms IV and V contribute to adenovirus-mediated oncogenic transformation by functional inhibition of the tumor suppressor p53. Oncogene 2015. manuscript accepted. [Google Scholar] [CrossRef] [PubMed]
- Pennella, M.A.; Liu, Y.; Woo, J.L.; Kim, C.A.; Berk, A.J. Adenovirus e1b 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies. J. Virol. 2010, 84, 12210–12225. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Dobner, T. The adenovirus E1B-55K oncoprotein induces sumo modification of p53. Cell Cycle 2008, 7, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Kattenhorn, L.M.; Korbel, G.A.; Kessler, B.M.; Spooner, E.; Ploegh, H.L. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family herpesviridae. Mol. Cell 2005, 19, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Loveland, A.N.; Kattenhorn, L.M.; Ploegh, H.L.; Gibson, W. High-molecular-weight protein (pul48) of human cytomegalovirus is a competent deubiquitinating protease: Mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 2006, 80, 6003–6012. [Google Scholar] [CrossRef] [PubMed]
- Izumiya, Y.; Ellison, T.J.; Yeh, E.T.; Jung, J.U.; Luciw, P.A.; Kung, H.J. Kaposi's sarcoma-associated herpesvirus K-bZip represses gene transcription via sumo modification. J. Virol. 2005, 79, 9912–9925. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.C.; Izumiya, Y.; Wu, C.Y.; Fitzgerald, L.D.; Campbell, M.; Ellison, T.J.; Lam, K.S.; Luciw, P.A.; Kung, H.J. Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J. Biol. Chem. 2010, 285, 5266–5273. [Google Scholar] [CrossRef] [PubMed]
- Sompallae, R.; Gastaldello, S.; Hildebrand, S.; Zinin, N.; Hassink, G.; Lindsten, K.; Haas, J.; Persson, B.; Masucci, M.G. Epstein-barr virus encodes three bona fide ubiquitin-specific proteases. J. Virol. 2008, 82, 10477–10486. [Google Scholar] [CrossRef] [PubMed]
- Gredmark, S.; Schlieker, C.; Quesada, V.; Spooner, E.; Ploegh, H.L. A functional ubiquitin-specific protease embedded in the large tegument protein (orf64) of murine gammaherpesvirus 68 is active during the course of infection. J. Virol. 2007, 81, 10300–10309. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, W.; Klupp, B.G.; Granzow, H.; Mettenleiter, T.C. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 2004, 78, 11879–11889. [Google Scholar] [CrossRef] [PubMed]
- Andres, G.; Alejo, A.; Simon-Mateo, C.; Salas, M.L. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 2001, 276, 780–787. [Google Scholar] [CrossRef] [PubMed]
- Honig, J.E.; Osborne, J.C.; Nichol, S.T. Crimean-congo hemorrhagic fever virus genome l RNA segment and encoded protein. Virology 2004, 321, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Barretto, N.; Jukneliene, D.; Ratia, K.; Chen, Z.; Mesecar, A.D.; Baker, S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005, 79, 15189–15198. [Google Scholar] [CrossRef] [PubMed]
- Lindner, H.A.; Fotouhi-Ardakani, N.; Lytvyn, V.; Lachance, P.; Sulea, T.; Menard, R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005, 79, 15199–15208. [Google Scholar] [CrossRef] [PubMed]
- Kass, R.S.; Arena, J.P.; Chin, S. Block of l-type calcium channels by charged dihydropyridines. Sensitivity to side of application and calcium. J. Gen. Physiol. 1991, 98, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Clementz, M.A.; Chen, Z.; Banach, B.S.; Wang, Y.; Sun, L.; Ratia, K.; Baez-Santos, Y.M.; Wang, J.; Takayama, J.; Ghosh, A.K.; et al. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010, 84, 4619–4629. [Google Scholar] [CrossRef] [PubMed]
- Mielech, A.M.; Kilianski, A.; Baez-Santos, Y.M.; Mesecar, A.D.; Baker, S.C. Mers-CoV papain-like protease has deisgylating and deubiquitinating activities. Virology 2014, 450, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Amerik, A.Y.; Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 2004, 1695, 189–207. [Google Scholar] [CrossRef] [PubMed]
- Schlieker, C.; Korbel, G.A.; Kattenhorn, L.M.; Ploegh, H.L. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 2005, 79, 15582–15585. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ortiz, D.A.; Gurczynski, S.J.; Khan, F.; Pellett, P.E. Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex. J. Virol. 2014, 88, 9086–9099. [Google Scholar] [CrossRef] [PubMed]
- Granzow, H.; Klupp, B.G.; Mettenleiter, T.C. Entry of pseudorabies virus: An immunogold-labeling study. J. Virol. 2005, 79, 3200–3205. [Google Scholar] [CrossRef] [PubMed]
- Antinone, S.E.; Smith, G.A. Two modes of herpesvirus trafficking in neurons: Membrane acquisition directs motion. J. Virol. 2006, 80, 11235–11240. [Google Scholar] [CrossRef] [PubMed]
- Luxton, G.W.; Haverlock, S.; Coller, K.E.; Antinone, S.E.; Pincetic, A.; Smith, G.A. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 5832–5837. [Google Scholar] [CrossRef] [PubMed]
- Gill, M.B.; Kutok, J.L.; Fingeroth, J.D. Epstein-barr virus thymidine kinase is a centrosomal resident precisely localized to the periphery of centrioles. J. Virol. 2007, 81, 6523–6535. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, S.; Maresch, C.; Granzow, H.; Klupp, B.G.; Teifke, J.P.; Mettenleiter, T.C. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J. Virol. 2008, 82, 6009–6016. [Google Scholar] [CrossRef] [PubMed]
- Burt, F.J.; Leman, P.A.; Abbott, J.C.; Swanepoel, R. Serodiagnosis of crimean-congo haemorrhagic fever. Epidemiol. Infect. 1994, 113, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Deyde, V.M.; Khristova, M.L.; Rollin, P.E.; Ksiazek, T.G.; Nichol, S.T. Crimean-congo hemorrhagic fever virus genomics and global diversity. J. Virol. 2006, 80, 8834–8842. [Google Scholar] [CrossRef] [PubMed]
- Marriott, A.C.; Nuttall, P.A. Large rna segment of dugbe nairovirus encodes the putative RNA polymerase. J. Gen. Virol. 1996, 77, 1775–1780. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, D.M.; Rozanov, M.N.; Hillman, B.I. Autocatalytic processing of the 223-kDa protein of blueberry scorch carlavirus by a papain-like proteinase. Virology 1995, 207, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Yang, J.; Sun, H.R.; Xin, X.; Wang, H.D.; Chen, J.P.; Adams, M.J. Genomic analysis of rice stripe virus zhejiang isolate shows the presence of an OTU-like domain in the RNA1 protein and a novel sequence motif conserved within the intergenic regions of ambisense segments of tenuiviruses. Arch. Virol. 2007, 152, 1917–1923. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Bredenbeek, P.J.; Dobbe, J.C.; Thiel, V.; Ziebuhr, J.; Poon, L.L.; Guan, Y.; Rozanov, M.; Spaan, W.J.; Gorbalenya, A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003, 331, 991–1004. [Google Scholar] [CrossRef]
- Kiel, C.; Serrano, L. The ubiquitin domain superfold: Structure-based sequence alignments and characterization of binding epitopes. J. Mol. Biol. 2006, 355, 821–844. [Google Scholar] [CrossRef] [PubMed]
- Ratia, K.; Saikatendu, K.S.; Santarsiero, B.D.; Barretto, N.; Baker, S.C.; Stevens, R.C.; Mesecar, A.D. Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 2006, 103, 5717–5722. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.J.; Lenschow, D.J. The antiviral activities of isg15. J. Mol. Biol. 2013, 425, 4995–5008. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, S.G.; Sawicki, D.L. The effect of overproduction of nonstructural proteins on alphavirus plus-strand and minus-strand RNA synthesis. Virology 1986, 152, 507–512. [Google Scholar] [CrossRef]
- Yamamoto, D.; Ishida, T.; Inoue, M. A comparison between the binding modes of a substrate and inhibitor to papain as observed in complex crystal structures. Biochem. Biophys. Res. Commun. 1990, 171, 711–716. [Google Scholar] [CrossRef]
- Kane, E.M.; Shuman, S. Vaccinia virus morphogenesis is blocked by a temperature-sensitive mutation in the I7 gene that encodes a virion component. J. Virol. 1993, 67, 2689–2698. [Google Scholar] [PubMed]
- Simon-Mateo, C.; Andres, G.; Vinuela, E. Polyprotein processing in african swine fever virus: A novel gene expression strategy for a DNA virus. EMBO J. 1993, 12, 2977–2987. [Google Scholar] [PubMed]
- Lopez-Otin, C.; Simon-Mateo, C.; Martinez, L.; Vinuela, E. Gly-Gly-x, a novel consensus sequence for the proteolytic processing of viral and cellular proteins. J. Biol. Chem. 1989, 264, 9107–9110. [Google Scholar] [PubMed]
- Endter, C.; Kzhyshkowska, J.; Stauber, R.; Dobner, T. Sumo-1 modification required for transformation by adenovirus type 5 early region 1b 55-kDa oncoprotein. Proc. Natl. Acad. Sci. USA 2001, 98, 11312–11317. [Google Scholar] [CrossRef] [PubMed]
- Kindsmuller, K.; Groitl, P.; Hartl, B.; Blanchette, P.; Hauber, J.; Dobner, T. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. USA 2007, 104, 6684–6689. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Shibata, H.; Kurihara, I.; Yokota, K.; Suda, N.; Saito, I.; Saruta, T. Ubc9 interacts with chicken ovalbumin upstream promoter-transcription factor i and represses receptor-dependent transcription. J. Mol. Endocrinol. 2004, 32, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Meyers, G.; Tautz, N.; Dubovi, E.J.; Thiel, H.J. Viral cytopathogenicity correlated with integration of ubiquitin-coding sequences. Virology 1991, 180, 602–616. [Google Scholar] [CrossRef]
- Rytkonen, A.; Holden, D.W. Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe 2007, 1, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Rytkonen, A.; Poh, J.; Garmendia, J.; Boyle, C.; Thompson, A.; Liu, M.; Freemont, P.; Hinton, J.C.; Holden, D.W. Ssel, a salmonella deubiquitinase required for macrophage killing and virulence. Proc. Natl. Acad. Sci. USA 2007, 104, 3502–3507. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Monack, D.M.; Kayagaki, N.; Wertz, I.; Yin, J.; Wolf, B.; Dixit, V.M. Yersinia virulence factor yopj acts as a deubiquitinase to inhibit NF-kappa b activation. J. Exp. Med. 2005, 202, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wimmer, P.; Schreiner, S. Viral Mimicry to Usurp Ubiquitin and SUMO Host Pathways. Viruses 2015, 7, 4854-4872. https://doi.org/10.3390/v7092849
Wimmer P, Schreiner S. Viral Mimicry to Usurp Ubiquitin and SUMO Host Pathways. Viruses. 2015; 7(9):4854-4872. https://doi.org/10.3390/v7092849
Chicago/Turabian StyleWimmer, Peter, and Sabrina Schreiner. 2015. "Viral Mimicry to Usurp Ubiquitin and SUMO Host Pathways" Viruses 7, no. 9: 4854-4872. https://doi.org/10.3390/v7092849
APA StyleWimmer, P., & Schreiner, S. (2015). Viral Mimicry to Usurp Ubiquitin and SUMO Host Pathways. Viruses, 7(9), 4854-4872. https://doi.org/10.3390/v7092849





