Wolbachia Do Not Induce Reactive Oxygen Species-Dependent Immune Pathway Activation in Aedes albopictus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mosquito and Cell Lines
2.2. Gene Transcription qRT-PCR Assays
| Gene | Forward Primer | Reverse Primer | Product | Transcript |
|---|---|---|---|---|
| CAC | TGTTCAGCTCGTCTTCGTCA | GGACTGGTGGTACTGGTGCT | 71bp | 140339 |
| Rel1 | GCTATCGGACACTGGACGTT | TGCGTTTCTTTCGTTTGGCT | 132bp | 145853 |
| CSP | AGAATGCGTAGCGGAGTGTC | GACCGGTGAGAACATAACGAA | 137bp | 082813 |
| Rel2 | CCGCATCTTCATTCAGCTTT | TTTCGATACCAATCGGAGATG | 81bp | 008087 |
| PIAS | TCAAACCGGCAGATTACACA | CGGGAAGGTCTTCTTGCTTT | 57bp | 135921 |
| STAT | ATCACCTGCCCTATCAACCG | ACGACGACGCAAACATATCG | 66bp | 146249 |
| TAK1 | GTTCGGAAAATCCCAACTCAGG | CTCCGCAACAAACCATGGAAG | 62bp | 099738 |
| Duox | GGGAATAGCAAGGCTTCCGT | ACCAAGCGTTTGTGATTGGC | 71bp | 144190 |
| DuoxA | TCAGCAATGGTGGGAACCTC | ACAAAGTCAAAGCGCAGCAG | 115bp | 102149 |
| OXR1 | CTACTCGTGGTCCCTGGTGT | ATTGGGCTTTCCAGTTTGTG | 90bp | 078559 |
| Gpx | CCAACGAGGAGATCAAGCAC | TCAAACTTGGCTCCCTTCTG | 52bp | 063197 |
| CuZnSOD | ATGTCAAGGGCACCATCTTC | GGCTTGAGTCCAGTCACCTC | 82bp | 143681 |
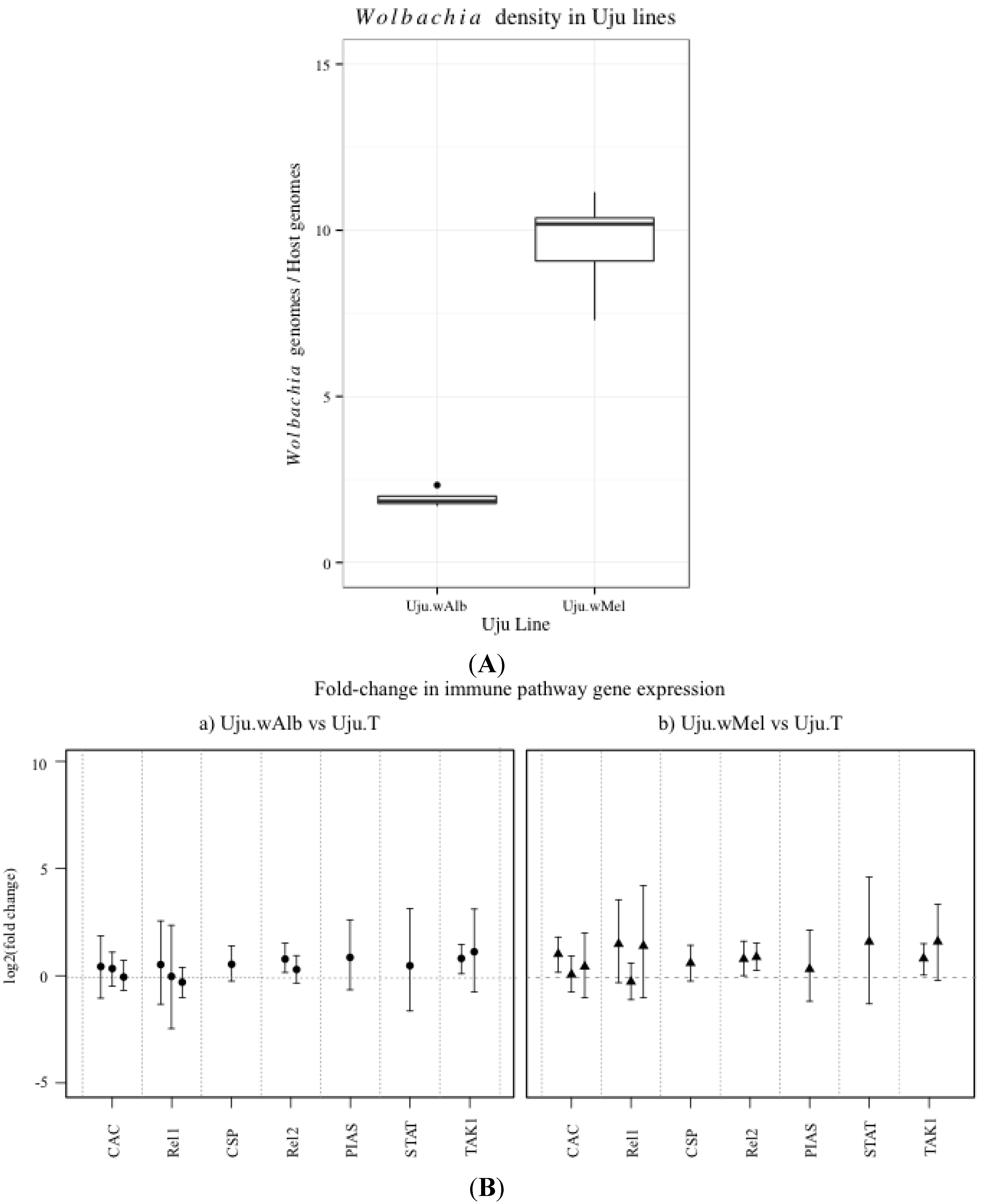
2.3. Wolbachia Density qPCR Assay
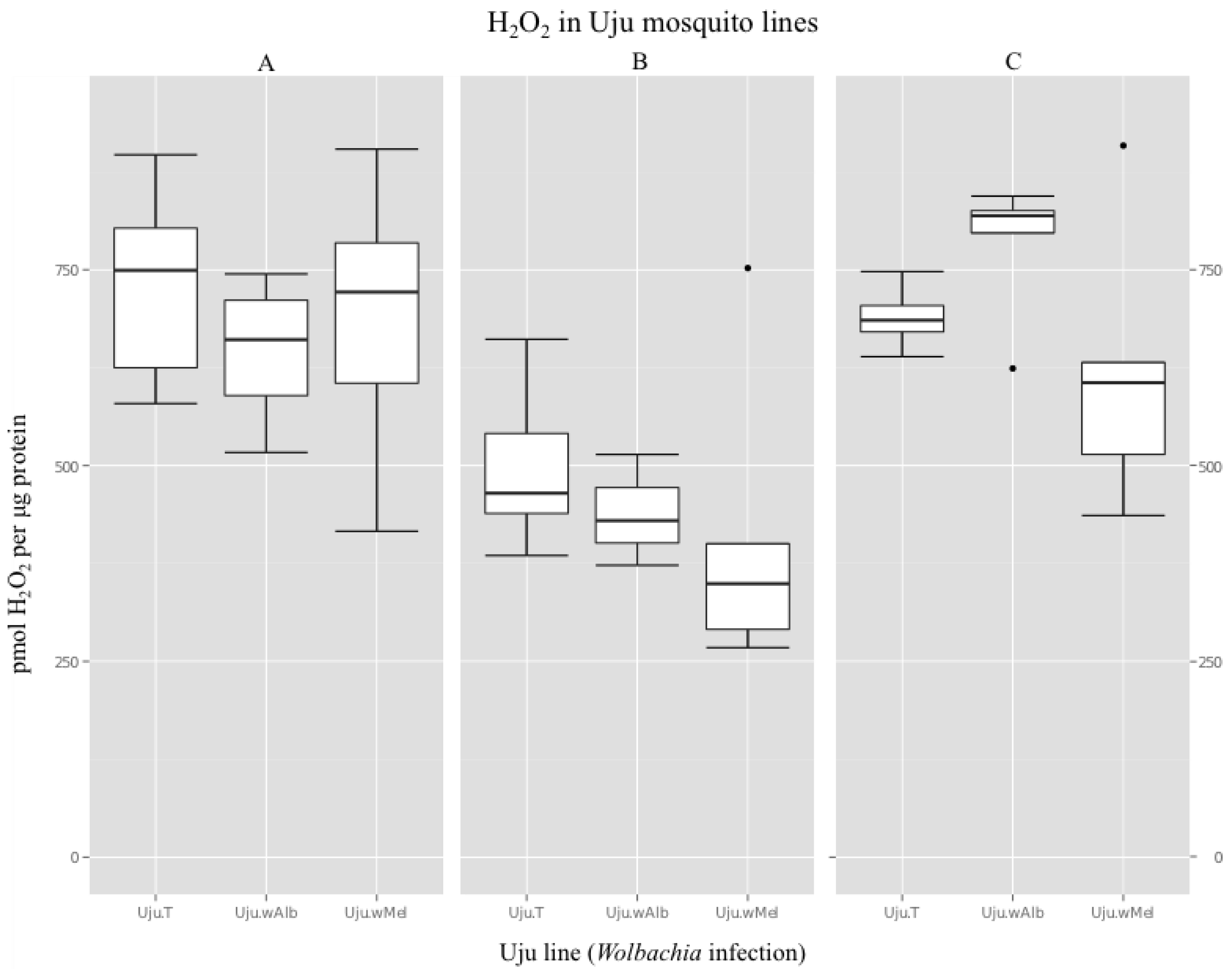
2.4. Hydrogen Peroxide Assays
3. Results
3.1. Innate Immune Pathway Gene Transcription

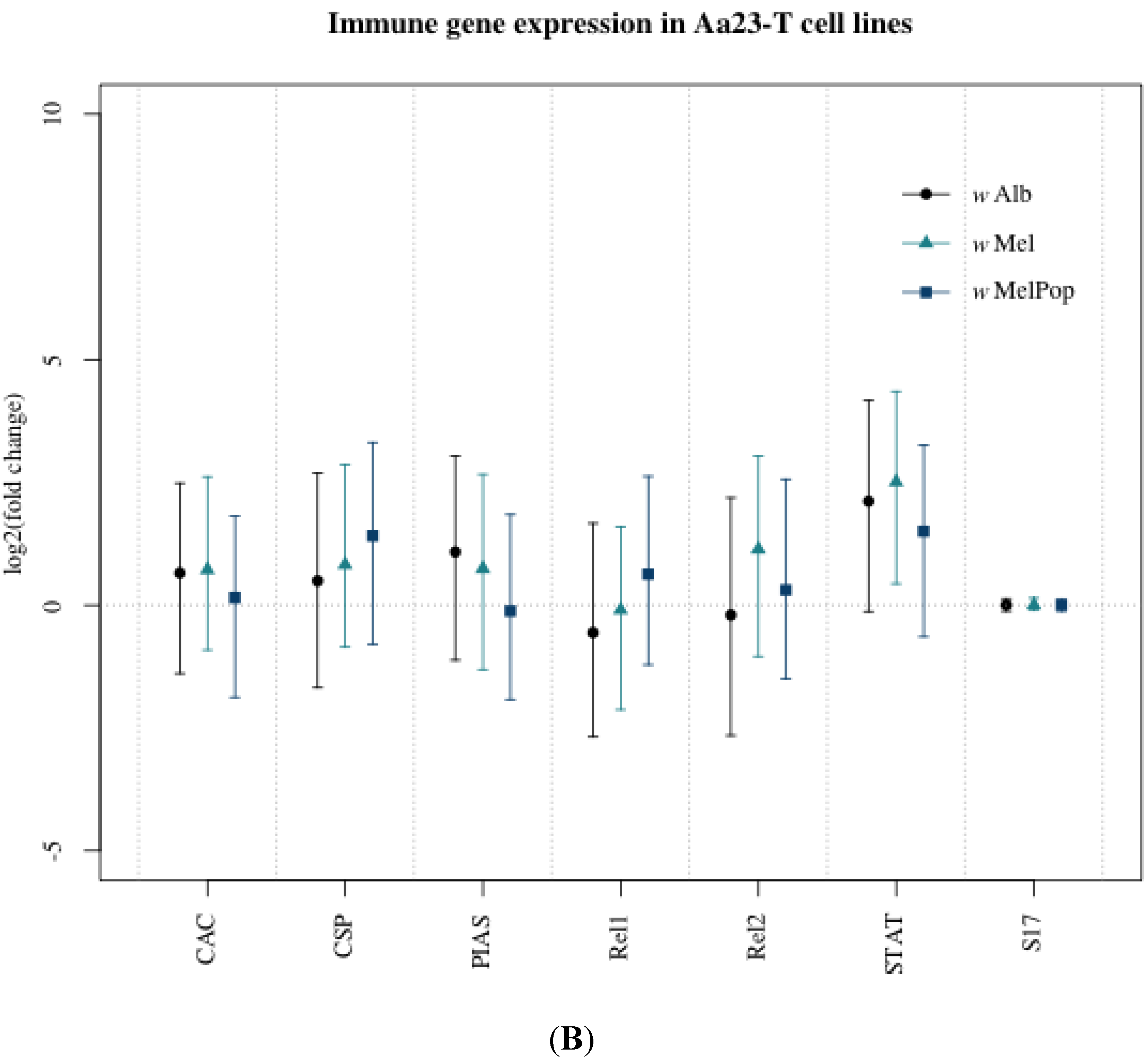
3.2. H2O2 Levels in Ae. albopictus Adults and Cell Lines

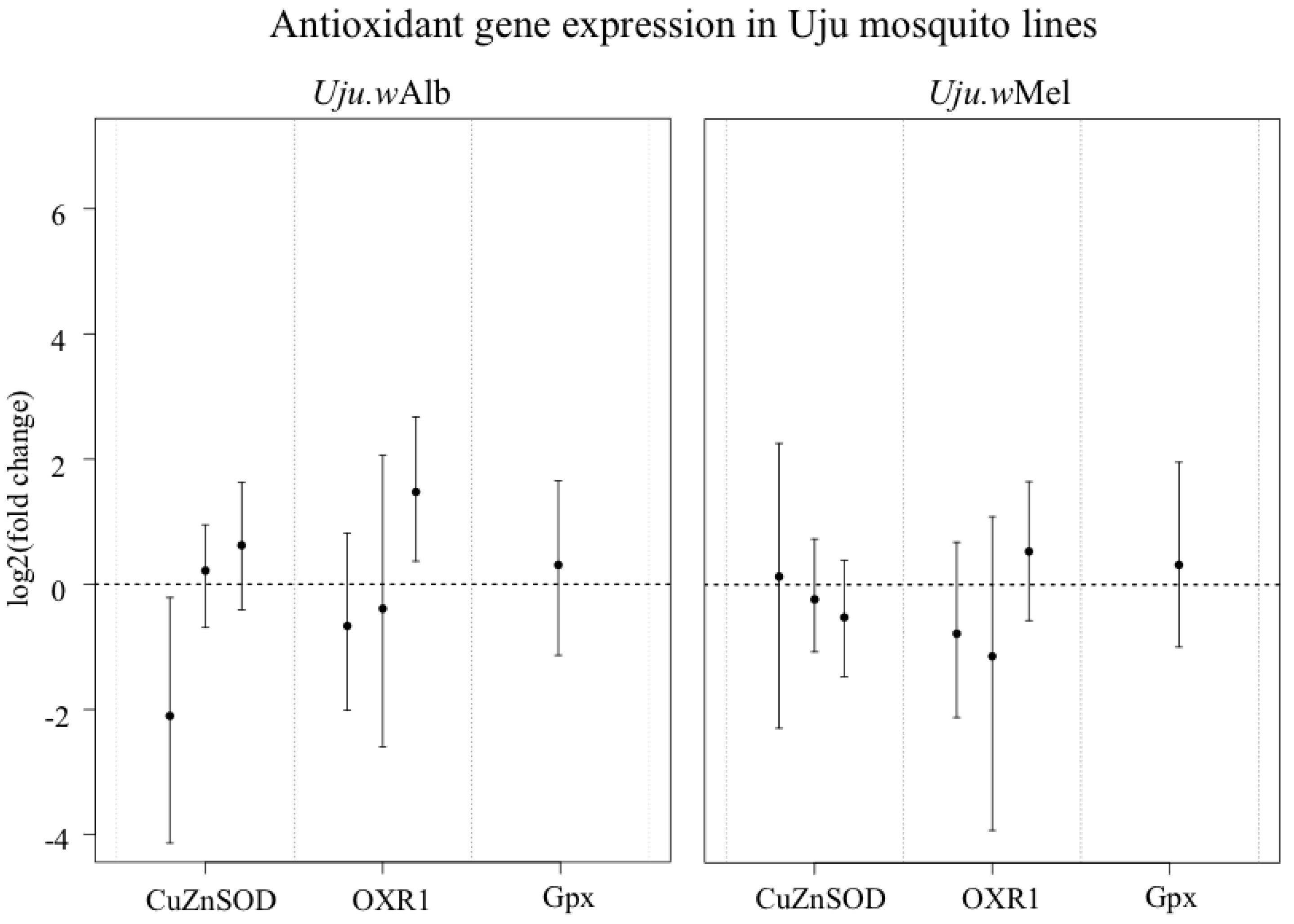
3.3. ROS-Generating Enzyme and Antioxidant Gene Transcription
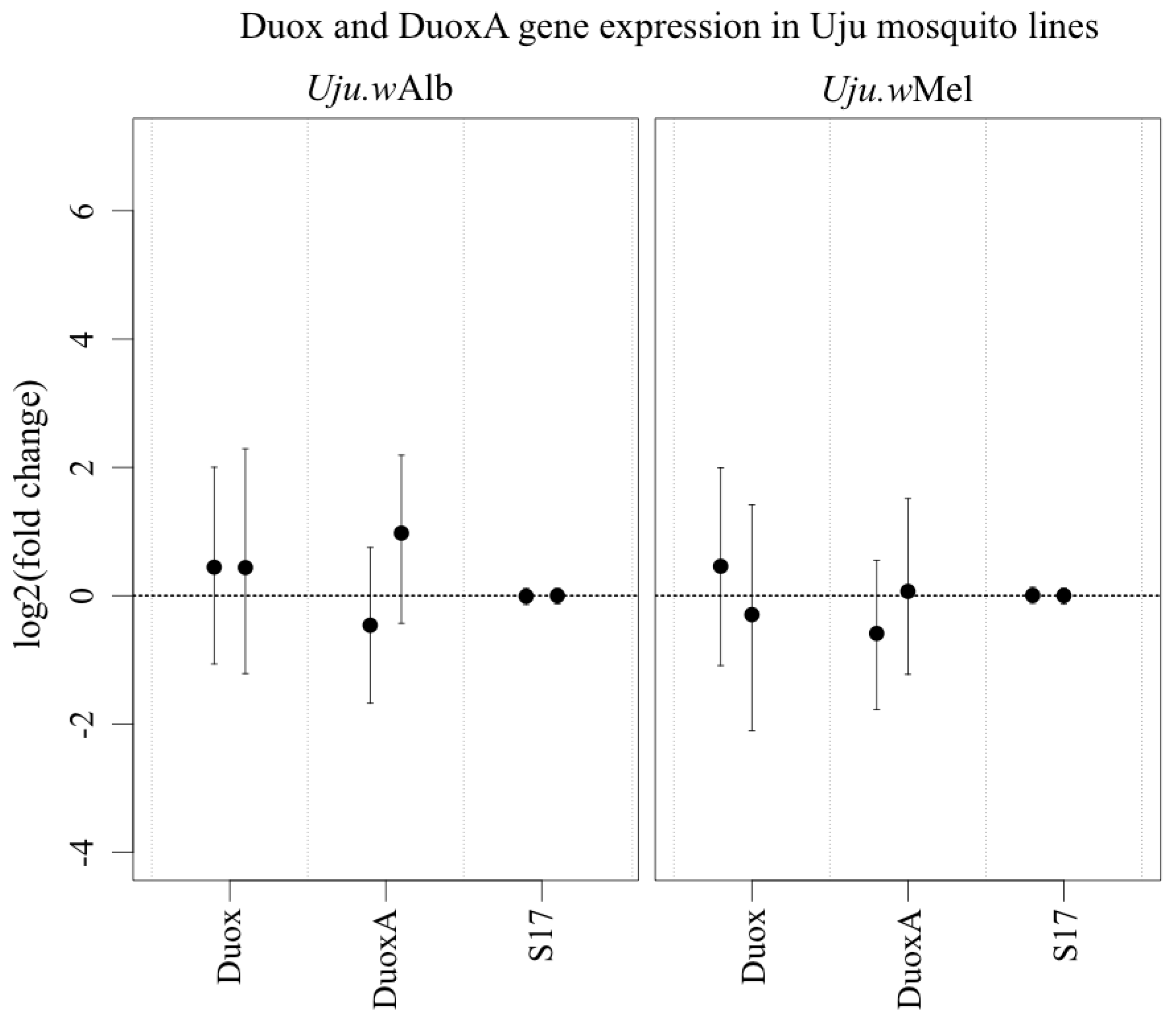
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hoffmann, A.A.; Turelli, M. Cytoplasmic incompatibility in insects. In Influential Passengers: Microorganisms and Invertebrate Reproduction; O’Neill, S.L., Hoffmann, A.A., Werren, J., Eds.; Oxford University Press: Oxford, UK, 1997. [Google Scholar]
- Moreira, L.A.; Iturbe-Ormaetxe, I.; Jeffery, J.A.; Lu, G.; Pyke, A.T.; Hedges, L.M.; Rocha, B.C.; Hall-Mendelin, S.; Day, A.; Riegler, M.; et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and Plasmodium. Cell 2009, 139, 1268–1278. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.; Xu, Y.; Lu, P.; Xie, Y.; Xi, Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 2010, 6, e1000833. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.; Johnson, P.H.; Moreira, L.A.; Iturbe-Ormaetxe, I.; Frentiu, F.D.; McMeniman, C.J.; Leong, Y.S.; Dong, Y.; Axford, J.; Kriesner, P.; et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 2011, 476, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Blagrove, M.S.C.; Arias-Goeta, C.A.; Failloux, A.; Sinkins, S.P. The Wolbachia strain wMel induces cytoplasmic incompatibility in and blocks dengue transmission by Aedes albopictus. Proc. Natl. Acad. Sci. USA 2012, 109, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Blagrove, M.S.C.; Arias-Goeta, C.A.; Di Genua, C.; Failloux, A.-B.; Sinkins, S.P. A Wolbachia wMel transinfection in Aedes albopictus is not detrimental to host fitness and inhibits chikungunya virus. PLoS Negl. Trop. Dis. 2013, 7, e2152. [Google Scholar] [CrossRef] [PubMed]
- Van den Hurk, A.F.; Hall-Mendelin, S.; Pyke, A.T.; Frentiu, F.D.; McElroy, K.; Day, A.; Higgs, S.; O’Neill, S.L. Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PLoS Negl. Trop. Dis 2012, 6, e1892. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Montgomery, B.L.; Popovici, J.; Iturbe-Ormaetxe, I.; Johnson, P.H.; Muzzi, F.; Greenfield, M.; Durkan, M.; Leong, Y.S.; Dong, Y.; et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 2011, 476, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Iturbe-Ormaetxe, I.; Callahan, A.G.; Phillips, B.L.; Billington, K.; Axford, J.K.; Montgomery, B.; Turley, A.P.; O’Neill, S.L. Stability of the wMel Wolbachia infection following invasion into Aedes aegypti populations. PLoS Negl. Trop. Dis. 2014, 8, e3115. [Google Scholar] [CrossRef] [PubMed]
- Kambris, Z.; Cook, P.E.; Phuc, H.K.; Sinkins, S.P. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 2009, 326, 134–136. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhou, G.; Wu, J.; Bian, G.; Lu, P.; Raikhel, A.S.; Xi, Z. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2012, 109, E23–E31. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008, 4, e1000098. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.L.; Dimopoulos, G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev. Comp. Immunol. 2010, 34, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2004, 2, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Molina-Cruz, A.; DeJong, R.J.; Charles, B.; Gupta, L.; Kumar, S.; Jaramillo-Gutierrez, G.; Barillas-Mury, C. Reactive oxygen species modulate Anopheles gambiae immunity against bacteria and Plasmodium. J. Biol. Chem. 2008, 283, 3217–3223. [Google Scholar] [CrossRef] [PubMed]
- Cirimotich, C.M.; Dong, Y.; Clayton, A.M.; Sandiford, S.L.; Souza-Neto, J.A.; Mulenga, M.; Dimopoulos, G. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 2011, 332, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Molina-Cruz, A.; Gupta, L.; Rodrigues, J.; Barillas-Mury, C. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science 2010, 327, 1644–1648. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Christophides, G.K.; Cantera, R.; Charles, B.; Han, Y.S.; Meister, S.; Dimopoulos, G.; Kafatos, F.C.; Barillas-Mury, C. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2003, 100, 14139–14144. [Google Scholar] [CrossRef] [PubMed]
- De Almeida Oliveira, G.; Lieberman, J.; Barillas-Mury, C. Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science 2012, 335, 856–859. [Google Scholar] [CrossRef] [PubMed]
- Surachetpong, W.; Pakpour, N.; Cheung, K.W.; Luckhart, S. Reactive oxygen species-dependent cell signaling regulates the mosquito immune response to Plasmodium falciparum. Antioxid. Redox Signal. 2011, 14, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.J.; Liu, Z. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2010, 21, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Nakajima, A.; Sakon-Komazawa, S.; Piao, J.; Xue, X.; Okumura, K. Reactive oxygen species mediate crosstalk between NF-κB and JNK. Cell Death Differ. 2005, 13, 730–737. [Google Scholar] [CrossRef] [PubMed]
- DeJong, R.J.; Miller, L.M.; Molina-Cruz, A.; Gupta, L.; Kumar, S.; Barillas-Mury, C. Reactive oxygen species detoxification by catalase is a major determinant of fecundity in the mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2007, 104, 2121–2126. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo-Gutierrez, G.; Molina-Cruz, A.; Kumar, S.; Barillas-Mury, C. The Anopheles gambiae oxidation resistance 1 (OXR1) gene regulates expression of enzymes that detoxify reactive oxygen species. PLoS ONE 2010, 5, e11168. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.H.M.; Gonçalves, R.L.; Lara, F.A.; Dias, F.A.; Gandara, A.C.P.; Menna-Barreto, R.F.; Edwards, M.C.; Laurindo, F.R.; Silva-Neto, M.A.; Sorgine, M.H.; et al. Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog. 2011, 7, e1001320. [Google Scholar] [CrossRef] [PubMed]
- Frentiu, F.D.; Robinson, J.; Young, P.R.; McGraw, E.A.; O’Neill, S.L. Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PLoS ONE 2010, 5, e13398. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Bian, G.; Pan, X.; Xi, Z. Wolbachia induces density-dependent inhibition to dengue virus in mosquito cells. PLoS Negl. Trop. Dis. 2012, 6, e1754. [Google Scholar] [CrossRef] [PubMed]
- Brennan, L.J.; Keddie, B.A.; Braig, H.R.; Harris, H.L. The endosymbiont Wolbachia pipientis induces the expression of host antioxidant proteins in an Aedes albopictus cell line. PLoS ONE 2008, 3, e2083. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, S.; Pettigrew, M.; Sinkins, S.P.; Braig, H.R.; Andreadis, T.; Tesh, R. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Mol. Biol. 1997, 6, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Pinto, S.B.; Mariconti, M.; Bazzocchi, C.; Bandi, C.; Sinkins, S.P. Wolbachia surface protein induces innate immune responses in mosquito cells. BMC Microbiol. 2012, 12, eS11. [Google Scholar] [CrossRef] [PubMed]
- McMeniman, C.J.; Lane, A.M.; Fong, A.W.; Voronin, D.A.; Iturbe-Ormaetxe, I.; Yamada, R.; McGraw, E.A.; O’Neill, S.L. Host adaptation of a Wolbachia strain after long-term serial passage in mosquito cell lines. Appl. Environ. Microbiol. 2008, 74, 6963–6969. [Google Scholar] [CrossRef] [PubMed]
- Poelchau, M.F.; Reynolds, J.A.; Denlinger, D.L.; Elsik, C.G.; Armbruster, P.A. A de novo transcriptome of the Asian tiger mosquito, Aedes albopictus, to identify candidate transcripts for diapause preparation. BMC Genomics 2011, 12, e619. [Google Scholar] [CrossRef] [PubMed]
- Glazov, E.A.; Pheasant, M.; McGraw, E.A.; Bejerano, G.; Mattick, J.S. Ultraconserved elements in insect genomes: A highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res. 2005, 15, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Kalyanaraman, B.; Darley-Usmar, V.; Davies, K.J.; Dennery, P.A.; Forman, H.J.; Grisham, M.B.; Mann, G.E.; Moore, K.; Roberts, L.J.; Ischiropoulos, H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Rad. Biol. Med. 2012, 52, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.; Quinn, M.T.; Lambeth, J.D. Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evolutionary Biology 2007, 7, e109. [Google Scholar] [CrossRef] [PubMed]
- Grasberger, H.; Refetoff, S. Identification of the maturation factor for dual oxidase evolution of an eukaryotic operon equivalent. J. Biol. Chem. 2006, 281, 18269–18272. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Hu, J.; Liu, X.; Qin, H.; Percival-Smith, A.; Rao, Y.; Li, S.S. NIP/DuoxA is essential for Drosophila embryonic development and regulates oxidative stress response. Int. J. Biol. Sci. 2010, 6, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Rancès, E.; Yixin, H.Y.; Woolfit, M.; McGraw, E.A.; O’Neill, S.L. The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathog. 2012, 8, e1002548. [Google Scholar] [CrossRef] [PubMed]
- Wong, Z.S.; Hedges, L.M.; Brownlie, J.C.; Johnson, K.N. Wolbachia-mediated antibacterial protection and immune gene regulation in Drosophila. PLoS ONE 2011, 6, e25430. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.; Ferreira, Á.; Ashburner, M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008, 6, e1000002. [Google Scholar] [CrossRef] [PubMed]
- Hedges, L.M.; Brownlie, J.C.; O’Neill, S.L.; Johnson, K.N. Wolbachia and virus protection in insects. Science 2008, 322, 702–702. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Whiteman, M. Measuring reactive species and oxidative damage in vivo and in cell culture: How should you do it and what do the results mean? Br. J. Pharmacol. 2004, 142, 231–255. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Nichol, H.; Law, J.H.; Winzerling, J.J. Iron metabolism in insects. Ann. Rev. Entomol. 2002, 47, 535–559. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, G.D.; Kurtti, T.J.; Munderloh, U.G. Susceptibility of Rickettsia monacensis and Rickettsia peacockii to Cecropin A, Ceratotoxin A, and Lysozyme. Curr. Microbiol. 2005, 51, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Dai, J.; Zhao, Y.O.; Narasimhan, S.; Yang, Y.; Zhang, L.; Fikrig, E. Ixodes scapularis JAK-STAT pathway regulates tick antimicrobial peptides, thereby controlling the agent of human granulocytic anaplasmosis. J. Infect. Dis. 2012, 206, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Login, F.H.; Balmand, S.; Vallier, A.; Vincent-Monégat, C.; Vigneron, A.; Weiss-Gayet, M.; Rochat, D.; Heddi, A. Antimicrobial peptides keep insect endosymbionts under control. Science 2011, 334, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Hedges, L.M.; Yamada, R.; O’Neill, S.L.; Johnson, K.N. The small interfering RNA pathway is not essential for Wolbachia-mediated antiviral protection in Drosophila melanogaster. Appl. Environ. Microbiol. 2012, 78, 6773–6776. [Google Scholar] [CrossRef] [PubMed]
- Caragata, E.P.; Rances, E.; Hedges, L.M.; Gofton, A.W.; Johnson, K.N.; O’Neill, S.L.; McGraw, E.A. Dietary cholesterol modulates pathogen blocking by Wolbachia. PLoS Pathog. 2013, 9, e1003459. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molloy, J.C.; Sinkins, S.P. Wolbachia Do Not Induce Reactive Oxygen Species-Dependent Immune Pathway Activation in Aedes albopictus. Viruses 2015, 7, 4624-4639. https://doi.org/10.3390/v7082836
Molloy JC, Sinkins SP. Wolbachia Do Not Induce Reactive Oxygen Species-Dependent Immune Pathway Activation in Aedes albopictus. Viruses. 2015; 7(8):4624-4639. https://doi.org/10.3390/v7082836
Chicago/Turabian StyleMolloy, Jennifer C., and Steven P. Sinkins. 2015. "Wolbachia Do Not Induce Reactive Oxygen Species-Dependent Immune Pathway Activation in Aedes albopictus" Viruses 7, no. 8: 4624-4639. https://doi.org/10.3390/v7082836
APA StyleMolloy, J. C., & Sinkins, S. P. (2015). Wolbachia Do Not Induce Reactive Oxygen Species-Dependent Immune Pathway Activation in Aedes albopictus. Viruses, 7(8), 4624-4639. https://doi.org/10.3390/v7082836





