Abstract
Plant viruses recruit cellular translation factors not only to translate their viral RNAs but also to regulate their replication and potentiate their local and systemic movement. Because of the virus dependence on cellular translation factors, it is perhaps not surprising that many natural plant recessive resistance genes have been mapped to mutations of translation initiation factors eIF4E and eIF4G or their isoforms, eIFiso4E and eIFiso4G. The partial functional redundancy of these isoforms allows specific mutation or knock-down of one isoform to provide virus resistance without hindering the general health of the plant. New possible targets for antiviral strategies have also been identified following the characterization of other plant translation factors (eIF4A-like helicases, eIF3, eEF1A and eEF1B) that specifically interact with viral RNAs and proteins and regulate various aspects of the infection cycle. Emerging evidence that translation repression operates as an alternative antiviral RNA silencing mechanism is also discussed. Understanding the mechanisms that control the development of natural viral resistance and the emergence of virulent isolates in response to these plant defense responses will provide the basis for the selection of new sources of resistance and for the intelligent design of engineered resistance that is broad-spectrum and durable.
1. Introduction
Although plant viruses encode a number of essential proteins (e.g., coat proteins, movement proteins, replication enzymes) their coding capacity is limited and they must rely on host factors for every stage of the infection cycle [,,]. An early and critical step of this cycle is the translation of viral RNAs. Viruses do not normally encode canonical translation factors, but have developed a wide array of strategies to highjack translation factors from their hosts and favor the translation of viral RNAs to the detriment of endogenous mRNAs [,]. Usurped translation factors have also been shown to play critical roles in the replication of viral genomes and even in assisting viral movement in the plant. Perhaps not surprisingly, many naturally occurring plant recessive resistance genes have been mapped to mutations in isoforms of translation initiation factors eIF4E and eIF4G. These mutations generally hinder the interactions of host factors with viral RNAs or proteins [,,,]. Non-host resistance or resistance to non-adapted virus strains has also been linked with incompatibility between viruses and cellular translation factors [,]. New sources of resistance to a number of plant viruses have been developed using a more directed approach aimed at mutating or down-regulating translation factors [,]. Finally, translation repression has recently emerged as an endogenous antiviral RNA silencing mechanism [], reinforcing the idea that hindering the translation of viruses or their ability to interact with translation factors is a promising avenue for virus control. In this review, I will discuss recent advances in our understanding of the interactions between plant viruses and translation factors, the utility of various natural or engineered resistance mechanisms based on the manipulation of plant translation factors, the emergence of virulent virus isolates in response to these new selection pressures and possible future strategies to engineer durable virus resistance.
2. Biological Functions of Translation Factors in Plants
2.1. Translation of Cellular mRNAs
Eukaryotic mRNA translation has been reviewed by others [,] and will not be described in detail here. Rather, this section is focused on the biological function(s) of key translation factors that have also been implicated in virus resistance. Translation initiation is a crucial step of protein synthesis that requires a large number of eukaryotic initiation factors (eIFs, Figure 1A). Translation of eukaryotic mRNAs depends on the binding of translation eukaryotic initiation factor 4E (eIF4E) to their 5′ m7G cap structure and is also enhanced by interaction of the polyA-binding protein (PABP) with their 3′ polyA tail. The large eIF4G scaffold protein binds to both eIF4E and PABP promoting circularization of the mRNA. The tight association between eIF4E and eIF4G constitute the core of the eIF4F complex. eIF4G also interacts with eIF4A, a DEAD-box ATPase and ATP-dependent RNA helicase. DEAD-box helicases are so-named because they contain a conserved Asp-Glu-Ala-Asp (DEAD) motif. eIF4A unwinds the mRNA to assist ribosome scanning. RNAs with long structured 5′ untranslated regions (UTR) are particularly dependent on eIF4A or other DEAD-box helicases. The eIF4G-eIF4A interaction is looser than that between eIF4G and eIF4E and although eIF4A is considered a component of eIF4F, it is easily lost during purification. Other DEAD-box RNA helicases such as Dhx29 and Ded1/Ddx3 also participate in mRNA translation and may have distinct specificities for mRNAs with specific features in their 5′ UTR. eIF4E, eIF4G, eIF4A and other DEAD-box RNA helicases are key translation factors frequently usurped/manipulated by viruses and are attractive targets for antiviral strategies (Section 3, Figure 1A). The eIF3 protein complex (11–12 subunits, depending on the organism) interacts with both eIF4G and the 40S ribosome subunit, thereby bringing the 43S pre-initiation complex (which includes the 40S subunit and the eIF2-GTP-tRNAMet ternary complex) to the mRNA. As discussed below, a subunit of eIF3 has been implicated in the translation reinitiation mechanism of cauliflower mosaic virus (CaMV) polycistronic mRNA.
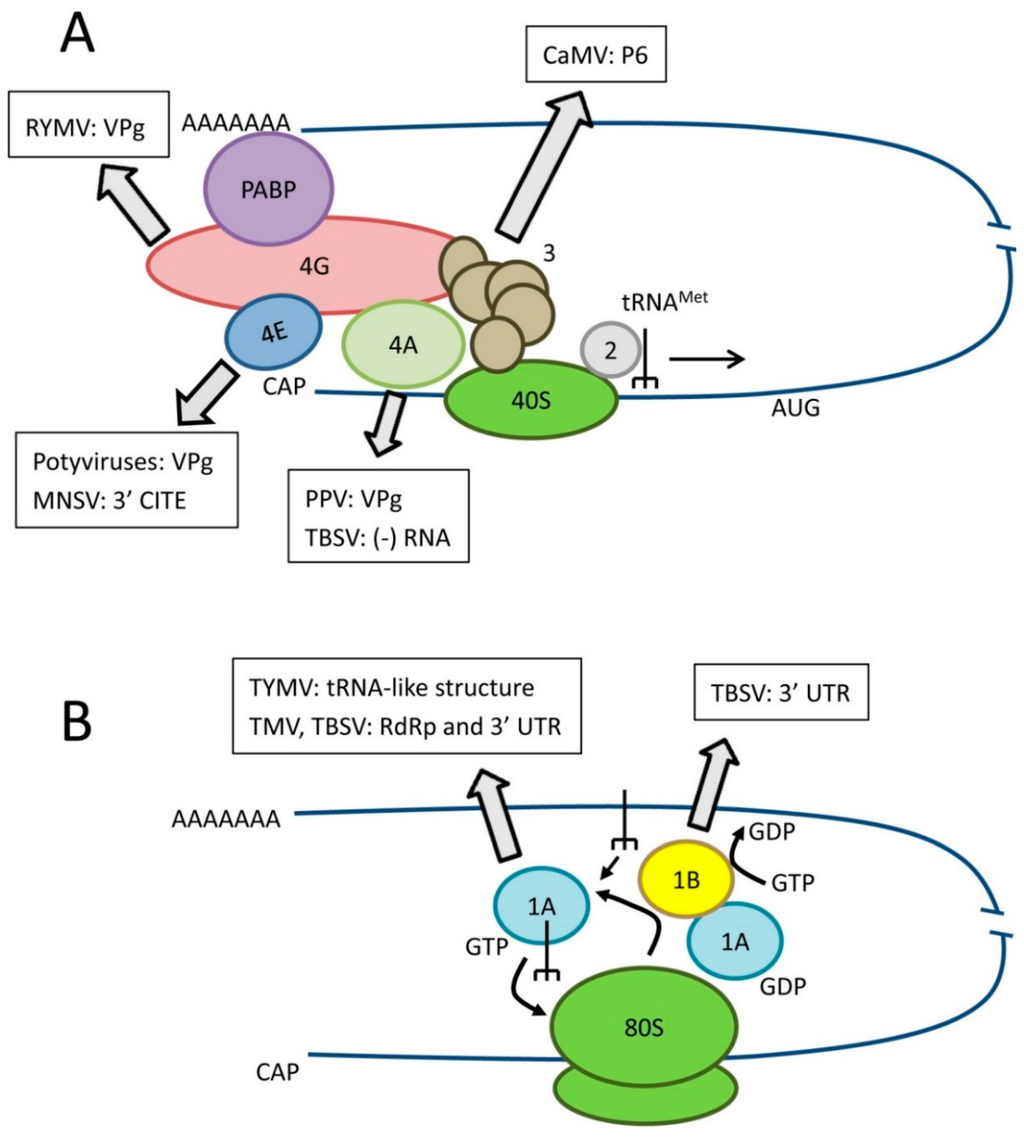
Figure 1.
Role of translation factors in canonical eukaryotic translation and interactions with viral RNAs or proteins. (A) Simplified diagram of translation initiation. Key initiation factors are shown as well as the best characterized interactions with viral components. eIF3 is shown as a complex of subunits. Please refer to the text for details of the specific isoform of eIF4E or eIF4G or specific eIF4A-like helicase recruited by various plant viruses; (B) Simplified diagram of translation elongation. Key elongation factors are shown as well as the best characterized interactions with viral components.
In plants, two isoforms of eIF4F are present: eIF4F, which consists of eIF4E and eIF4G, and eIFiso4F, which includes eIFiso4E and eIFiso4G [,,,]. To add to the complexity, these isoforms are often encoded by multi-gene families. For example, there are 1–4 genes coding for eIFiso4G depending on the plant species (two for Arabidopsis thaliana) []. eIF4E and eIFiso4E, although of similar size, only share 50% amino acid sequence identity. eIF4G (~180 kDa) and eIFiso4G (~86 kDa), share common binding motifs for eIF4E, eIF4A and eIF3 in their C-terminal domains but differ in size due to a large truncation of the N-terminal region in eIFiso4G. In wild-type A. thaliana cells, only cognate complexes (i.e. eIF4E-eIF4G and eIFiso4E-eIFiso4G) accumulate in detectable amounts [] and in wheat germ extracts, eIFiso4F is 3–5 time more abundant than eIF4F []. Knock-out of eIFiso4G in A. thaliana causes severe defects in plant health. In contrast, knock-out or down-regulation of eIF4E or eIFiso4E is tolerated and plants show little signs of impairment [,]. Interestingly, knock-out of eIFiso4E is compensated by increased expression of eIF4E, suggesting functional redundancy between the two isoforms []. Indeed, mixed complexes can be formed in vitro, although with lower binding activity than that shown for the cognate complexes, and these hetero-complexes are active in translation []. In spite of some functional redundancy, eIF4F and eIFiso4F exhibit distinct specificity for mRNAs in vitro []. As will be described below, viruses have adapted to use either eIF4F or eIFiso4F and mutation of one isoform of eIF4E or eIF4G can lead to resistance to plant viruses without affecting the plant health, making them attractive sources of natural or engineered resistance. Similarly to mammalian cells, plant translation initiation complexes utilize not only eIF4A (eIF4A-1 or eIF4A-2, which share 96% amino acid sequence identity) but also a variety of other DEAD-box RNA helicases. In proliferating A. thaliana cells, eIF4A1/2 is the predominant helicase found in association with both eIF4F and eIFiso4F complexes []. In quiescent cells, other DEAD-box helicases also co-purify with these complexes. The partial functional redundancy of eIF4A-like helicases in plants also makes them amenable to manipulation towards virus resistance.
Once translation is initiated, eukaryotic elongation factors (eEFs) allow continued synthesis of the nascent polypeptide chain [] (Figure 1B). Eukaryotic elongation factor 1A (eEF1A also referred to as eEF1α in the literature) is an abundant cytoplasmic protein that forms a ternary complex with GTP and aminoacylated tRNAs (all tRNAs but the initiator tRNAMet) and delivers these tRNAs to the elongating ribosome. As discussed below, eEF1A is also frequently found in association with the replication complexes of positive-strand RNA viruses (Section 3.2). eEF1A functions with the eEF1B complex, a guanine nucleotide exchange factor, which helps recycling eEF1A into its active GTP-bound form.
2.2. Additional Biological Functions of Translation Factors
In addition to their roles in the canonical translation of cellular mRNAs, translation factors have other biological functions that are of interest for viruses. In mammalian cells, a sub-population of eIF4E accumulates in nuclear bodies and has been implicated in the regulated nuclear export of mRNAs that contain a specific structure (the 4E-sensitivity element) [,,]. In A. thaliana cultured cells, eIF4E is predominantly cytoplasmic although its partition to the nucleus is regulated by the cell growth cycle []. eIFiso4E is equally distributed in nucleus and cytoplasm. Thus, a role for the plant eIF4E and/or eIFiso4E in nuclear export is likely. The plant eIFiso4F complex also associates with microtubules and facilitates their end-to-end annealing, suggesting a role in the regulation of microtubule dynamics [,]. The microtubule localization of eIFiso4F depends on a direct interaction between eIFiso4G and microtubules []. eEF1A, another multitask protein, may regulate nuclear export, cytoskeleton and microtubule stabilization/organization and protein stability, likely via interactions with the proteasome []. Viruses exploit these activities to control the stability of their proteins, regulate their replication and facilitate their movement in the cell and from cell-to-cell.
3. Characterization of the Interactions between Cellular Translation Factors and Plant Viruses and Identification of Potential Sources of Viral Resistance
3.1. Translation Factors Facilitating Non-Canonical Viral RNA Translation
As mentioned above, viruses depend on cellular translation factors to synthesize their proteins. The RNAs of positive-strand RNA viruses differ from plant mRNAs in that they lack the 5′ cap structure and/or the polyA tail. Viral RNAs often have highly structured 5′ UTRs with multiple stem-loops or pseudoknots that are necessary for other steps of the infection cycle (replication, encapsidation) and that can interfere with normal scanning of ribosomes. Finally, some viruses have polycistronic RNAs that require translation reinitiation mechanisms or internal ribosome entry sites to facilitate translation at downstream open reading frames (ORFs). Thus, viruses have developed a variety of non-canonical mechanisms to translate their RNAs. The reader is referred to a collection of excellent recent reviews that describe these mechanisms in detail [,,,,,,,].
3.1.1. Core eIF4F/iso4F Components Recruited by Viral RNAs or Viral VPg Proteins
One common strategy used for the translation of non-canonical viral RNAs is the recruitment of initiation factors (usually eIF4E/iso4E or eIF4G/iso4G) or ribosomal RNAs by cis-acting translation enhancing sequences (CITE) present on the viral RNAs. These cis-acting sequences include internal ribosome entry sites (IRES) in the 5′ untranslated region (UTR) or in intergenic regions, and other CITE sequences in the 3′ UTR or even within an ORF. Long-distance kissing loop interactions between cis-acting sequences in the 5′ and 3′ UTRs are often required for translation enhancement by promoting circularization of the RNA. A natural melon recessive resistance gene to melon necrotic spot virus (MNSV, a carmovirus) corresponds to a mutation of eIF4E that disrupts its interaction with 3′ CITE sequences [] and will be described in detail below (Section 4.2). In addition, non-host resistance to MNSV in Nicotiana benthamiana was recently reported to be caused by an incompatibility between the viral RNA and eIF4E [].
The uncapped RNAs of some viruses, notably members of the families Potyviridae and Secoviridae and of the genus Sobemovirus are covalently linked with a small protein (termed VPg) at their 5′ end []. Although variable in size and sequence, the VPg of these divergent viruses share the common property of being intrinsically disordered, possibly allowing flexible adaptation to various binding partners [,,,,]. An interaction between the turnip mosaic virus (TuMV, a potyvirus) VPg protein and eIFiso4E, first identified in a yeast two-hybrid screen [], was later shown to be essential for virus infection []. As will be discussed in detail below, recessive resistance to potyviruses has been mapped to mutations of eIF4E and/or eIFiso4E (Section 4.1, Table 1). In addition, resistance to one strain of plum pox virus (PPV, another potyvirus) in wild-type A. thaliana and Chenopodium foetidum was attributed to the inability of this isolate to recruit translation initiation factors []. Potyvirus RNA translation depends on an IRES located in the 5′ UTR, which recruits one of the eIF4F isoforms, eIF4F in the case of tobacco etch virus (TEV) []. The VPg protein promotes host translational shut-down by competing with eIF4E or eIFiso4E cap binding activity and enhances the affinity of eIF4E/iso4E for viral RNAs in vitro, thereby facilitating their translation [,,]. Ectopic expression of VPg was shown to inhibit the translation of cellular mRNAs and stimulate the translation of potato virus A RNA in planta []. In addition to VPg and eIFiso4E, the ribosomal stalk protein P0 was also implicated in viral RNA translation, and silencing of P0 reduces the translation of a reporter viral RNA []. In addition, analysis of A. thaliana T-DNA insertion lines revealed the dependence of potyviruses on specific eIF4G isoforms that work concomitantly with the corresponding eIF4E isoform [], suggesting recruitment of the entire eIF4F complex, possibly for translation of viral RNAs. Interestingly however, resistance to TEV in A. thaliana eIFiso4E deficient plants is not correlated with defects in viral RNA translation []. Thus, although the VPg-eIF4E/iso4E interaction may support viral RNA translation in some potyvirus-host combinations, it is likely that it also facilitates other steps of the infection cycle, notably viral RNA replication and/or cell-to-cell movement [,]. This is discussed below (Section 3.2 and Section 3.3). The VPg protein of rice yellow mottle virus (RYMV, a sobemovirus) was found to bind directly with eIFiso4G rather than eIF4E isoforms []. As will be discussed below (Section 4.2), natural recessive resistance to RYMV in rice is correlated with mutation of eIFiso4G. It is not known whether the VPg-eIFiso4G interaction plays a role in the translation of the viral RNA or in other aspects of the viral infection cycle. The role played by the VPg protein of secovirids and its possible interactions with translation factors is less clear. The VPg protein of secovirids (2–4 kDa) is smaller than that of potyviruses or sobemoviruses (10–26 kDa). The VPg-protease polyprotein of tomato ringspot virus (a nepovirus), was shown to bind eIFiso4E in vitro, and although the eIFiso4E-binding domain was within the protease region, binding to eIFiso4E was enhanced by the presence of the VPg []. However, the biological function of the interaction is not known.
3.1.2. eIF4A-Like DEAD-Box Helicases Assisting the Translation of Bromovirus and Tombusvirus RNAs
Using a yeast replication model, DED1, a DEAD-box helicase related to eIF4A and implicated in yeast mRNA translation, was shown to facilitate the translation of the RNA2-encoded viral RNA-dependent RNA polymerase (RdRp) from brome mosaic virus (BMV, a bromovirus) []. DED1 interacts with a unique cis-acting sequence present only in the 5′ UTR of RNA2. Thus the translation of RNA1 and RNA3 was not affected by DED1. Of interest, mutation of DED1 (ded1-18 mutant with two amino acid substitutions) hinders the accumulation of the 2a RdRp and reduces virus replication but does not affect yeast mRNAs translation or yeast growth. Although this is an attractive candidate for bromovirus resistance, it is not known whether mutation of an equivalent helicase in plant would have a similar effect on BMV accumulation. Knock-down of the yeast DED1 also impacted the translation of tomato bushy stunt virus (TBSV, a tombusvirus) RNA []. Interestingly and as described below, DED1 and other DEAD-box RNA helicases also play important roles in the regulation of viral RNA replication in yeast and in plants (Section 3.2).
3.1.3. Recruitment of eEF1A by tRNA-Like Structure at the 3′ End of Viral RNAs
The RNAs of several plant viruses (e.g., tobamoviruses, tymoviruses and bromoviruses) have a tRNA-like structure at their 3′ end rather than a polyA tail []. These structures are recognized by aminoacyl-tRNA synthetases and become aminoacylated. The aminoacylated tRNA-like structure of turnip yellow mosaic virus (TYMV, a tymovirus) interacts with eEF1A and with ribosomes [,] and has been shown to enhance translation of the viral RNA []. As will be discussed below, the interaction of eEF1A with the TYMV tRNA-like structure also regulates its replication
3.1.4. Recruitment of an eIF3 Subunit and Associated Factors by Cauliflower Mosaic Virus for Translation Re-Initiation
A distinctive feature of some viral RNAs is that they are polycistronic or contain small ORFs in their 5′ UTRs. Leaky scanning, ribosome shunting or reinitiation mechanisms allow tight regulation of the translation rate of downstream ORFs []. The CaMV transactivator p6 protein facilitates translation re-initiation downstream of the 35S RNA large and highly structured 5′ UTR that also contains multiple small ORFs []. P6 interacts with a subunit of eIF3, various ribosomal proteins and a new plant translation factor, RISP, which itself interacts with the 60S ribosome [,,,]. The CaMV p6 protein also binds and activates the serine/threonine protein kinase TOR (target-of-rapamycin), which phosphorylates and activates RISP []. These interactions were shown to facilitate the translation re-initiation mechanism in vitro and in vivo. An A. thaliana T-DNA insertion line that prevent the expression of RISPa, the gene encoding RISP, shows only modest resistance to CaMV, probably due to the presence of a second highly homologous gene (RISPb) with overlapping function []. Unfortunately, double knock-out of RISPa and RISPb is not viable. The A. thaliana 35-7 line, which is silenced for TOR expression by RNA interference (RNAi), seemed more promising as it displayed strong resistance to CaMV []. Interestingly, two A. thaliana lines silenced for TOR expression (including the 35-7 line) are partially resistant to watermelon mosaic virus (a potyvirus) but fully susceptible to TuMV []. Treatment of wild-type plants with AZD-8055, an ATP-competitive inhibitor of TOR, also resulted in partial resistance to watermelon mosaic virus but not to TuMV []. Thus, manipulation of TOR expression or activity may provide a useful source of resistance not only to CaMV (and possibly other caulimoviruses) but also to some potyviruses. Whether such resistance is transferable to crops of economic importance and whether it can be stable in a field situation will require further study.
3.2. Translation factors Regulating Viral RNA Replication
In addition to their obvious role in viral RNA translation, cellular translation factors are recruited to membrane-associated viral replication complexes (VRCs) and regulate viral RNA replication. In many viruses, translation and replication are tightly linked and viral RNAs or proteins are brought to the VRCs co-translationally, along with translation factors [,,]. Some translation factors interact with viral RdRps and are recruited to VRCs; e.g., eIF3 subunits for BMV and tobacco mosaic virus (TMV) [,] and eEF1A for TMV and TuMV [,]. Recruitment of translation factors to VRCs can also require interactions with cis-acting sequences on viral RNAs (see below) or with viral proteins other than the RdRp, for example the potyvirus VPg protein. The TuMV VPg, or rather the VPg domain in the larger membrane-targeted 6K-VPg-protease polyprotein, was initially shown to recruit eIFiso4E to the VRC suggesting an as yet-undefined role for this factor in viral RNA replication []. Later, other translation factors were also found in association with TuMV VRCs, including PABP, a DEAD-box helicase and eEF1A [,,]. DEAD-box helicases and eEF1A are discussed in detail below. The biological function of PABP association with potyvirus VRCs is not known, although it may enhance replication by allowing circularization of the viral RNA (jointly with eIF4E and eIF4G isoforms). A. thaliana T-DNA insertion lines that knock-out the expression of PABP show increased resistance to TuMV without visibly affecting plant growth or health [], providing a possible new source of resistance to potyviruses.
3.2.1 Recruitment of DEAD-Box RNA Helicases to Tombusvirus and Potyvirus VRCs
DEAD-box RNA helicases, related to eIF4A, are recruited to tombusvirus VRCs []. Contrary to larger RNA viruses, tombusviruses do not encode a helicase to facilitate their replication. In yeast, DED1 binds to the 3′ end of the TBSV (−) strand RNA, is recruited to the VRCs and enhances (+) strand RNA synthesis by facilitating the unwinding of the (−) strand RNA []. The selective binding of DED1 to (−) strand RNA allows assymetric RNA synthesis and accumulation of excess (+) strand RNA for new rounds of translation/replication or for encapsidation. Overlapping functions were assigned to the yeast DBP2 and A. thaliana RH20 DEAD-box helicases that, similarly to DED1, bind to the 3′ end of TBSV (−) strand RNA to stimulate (+) strand RNA synthesis []. A second group of DEAD-box helicases including the yeast eIF4AIII-like Fal1p and DDX5-like Dbp3p helicases and the orthologous plant AtRH2 and AtRH5 helicases (from A. thaliana) also participate in the selective enhancement of TBSV (+) strand RNA synthesis by binding to another cis-acting replication enhancing element in the 5′ terminal region of the (−) strand RNA []. This allows unwinding of this highly structured region of the RNA. Overexpression of AtRH2 and AtRH5 in N. benthamiana results in enhanced viral accumulation and symptom severity []. Although down-regulation or mutation of these plant helicases may provide resistance to tombusviruses, this has not been tested.
Using a yeast-two hybrid screen, the VPg protein from PPV was shown to interact with a peach DEAD-box helicase (PpDXL) closely related to A. thaliana eIF4A genes []. Homozygous A. thaliana lines with T-DNA insertion in eIF4A genes could not be generated, likely because of pleiotropic effects of these mutations. However, knock-out of another related DEAD-box helicases, AtRH8, resulted in increased resistance to two potyviruses (PPV and TuMV) without visibly impacting the plant []. Interaction between AtRH8 and VPg was confirmed and AtRH8 was found in association with VRCs in TuMV-infected cells. The exact function of the cellular helicase in potyvirus VRCs is not known, as contrary to tombusviruses, potyviruses do encode a helicase, the CI protein, which is also recruited to the VRC []. Of practical interest, ectopic overexpression of a truncated AtRH8 protein, corresponding to the VPg-binding domain, also suppresses virus accumulation, probably by outcompeting the wild-type protein for VPg interaction []. This dominant-negative approach may be applicable to other helicases that interact with tombusviruses or other viruses.
3.2.2. Regulation of Tymovirus, Tobamovirus and Tombusvirus Replication by eEF1A
Potyviruses, tobamoviruses and tombusviruses recruit eEF1A to their VRCs [,,]. I have already discussed the role of eEF1A in promoting the translation of the TYMV RNA by binding to the 3′ terminal tRNA-like structure. Interestingly, the TYMV tRNA-like structure also acts as a promoter for (−) strand RNA synthesis. eEF1A was shown to down-regulate (−) strand RNA synthesis, probably by competing with the RdRp for binding to the tRNA-like structure []. It was suggested that conformation changes in the TYMV tRNA-like structure may regulate binding to eEF1A or RdRp at different stages of the infection cycle.
The multiple functions of eEF1A in tombusvirus RNA replication have been studied in details although primarily in the yeast model system [,]. eEF1A interacts with the TBSV RdRp and also binds to a cis-acting element in the 3′ UTR of the RNA. These interactions are thought to stimulate the recruitment of viral RNAs to the replication complex. eEF1A also helps stabilizing the p33 accessory protein and enhances (−) strand RNA synthesis. Screening of eEF1A mutants in yeast identified several mutants that could not stimulate viral replication, thereby resulting in partial resistance to TBSV [,]. However, the utility of these mutants in establishing virus resistance is tempered by their pleiotropic effects on host translation accuracy or efficiency. Perhaps of greater practical interest, another elongation factor eEF1Bγ, a member of the eEF1B complex and a non-essential cellular protein, is also associated with the TBSV VRCs []. eEF1Bγ was shown to interact with a loop in a tight stem-loop structure of the TBSV RNA 3′ UTR. The binding helps loosening the structure and stimulates (−) strand RNA synthesis synergistically with eEF1A. Silencing of eEF1Bγ expression in N. benthamiana plants resulted in severe reduction of TBSV RNA accumulation and symptom amelioration. The silenced plants also displayed partial resistance to TMV, suggesting a potential for the development of broad-spectrum resistance [].
In the case of tobamoviruses such as TMV, eEF1A also interacts with both the RdRp and with a cis-acting element in the viral RNA (a pseudoknot in the upstream region of the 3′ UTR) [,]. In N. benthamiana, eEF1A is encoded by at least six genes corresponding to two groups of related sequences []. Using the 3′ UTR sequence of representative eEF1A genes to specifically silence genes belonging to only one of the two groups, partial resistance to TMV was achieved with only moderate impairment of plant growth []. Silencing of all forms of eEF1A simultaneously, which improves virus resistance, also results in severe plant growth defects. The resistance is apparently due to reduced viral replication and/or cell-to-cell movement. It was suggested that the known interaction of eEF1A with the cytoskeleton may contribute to the assembly of VRCs or to the intracellular and/or cell-to-cell movement of these complexes. In a separate study, a virus-induced gene silencing approach using a tobacco rattle virus vector was used to down-regulate the expression of eEF1A and eEF1B(α/β) []. Partial resistance to TMV was achieved as evidenced by reduced virus accumulation. eEF1A and eEF1B were both shown to interact with the RdRp. Although it is likely that eEF1B accumulates in the VRCs, its exact biological function in viral replication or another step of the infection cycle is not known.
3.3. Other Roles for Translation Factors in Enhancing the Virus Infection Cycle
Most interactions described above regulate the translation or replication of viral RNAs. However, in some cases, and as already discussed for the TMV-eEF1A interaction, other steps of the infection cycle are apparently regulated by translation factors. Plants deficient for eIF4E show restricted cell-to-cell movement of some potyviruses, possibly related to the microtubule-binding affinity of the eIF4G binding partner of eIF4E [,]. In addition to binding the VPg, eIFiso4E was also found to bind the TEV coat protein and facilitate systemic movement of the virus in the plant []. Finally, eIF4E/iso4E is also found in the nucleus/nucleolus in infected cells, together with the nuclear VPg-protease polyprotein []. Possible roles for the nuclear localization of eIF4E/iso4E in mRNA transport, host gene regulation and RNA silencing have been suggested [].
5. Concluding Remarks
Characterization of natural recessive virus resistance has provided new insights on the interactions between plant viruses and translation factors and on the selection pressures controlling the emergence of virulent isolates. In addition, the systematic characterization of interactions between viral RNAs or proteins and translation factors has identified new potential targets for antiviral resistance. As discussed in Section 4.1, it is hoped that this knowledge can be used in the future to identify new sources of resistance in wild plant species that could be transferred to cultivated crops. In addition, specific mutations likely to provide broad-spectrum resistance can be selected and engineered in characterized translation factors. In that respect, the new CRISPR-Cas9 genome editing technology should allow targeted introduction of specific mutations in eIF4E isoform genes in plants for which natural resistance is not available []. It will be interesting to follow the evolving interactions between plant viruses and translation factors as new resistant varieties are implemented in the field.
Acknowledgments
This work was funded by grants from Agriculture and Agri-Food Canada (AAFC) and from the Natural Sciences and Engineering Research Council of Canada (NSERC). I would like to thank members of my lab for helpful discussions, in particular Dinesh Babu Paudel and Basudev Ghoshal.
Conflicts of Interest
The author declares no conflict of interest.
References
- Sanfacon, H.; Jovel, J. Interactions Between plant and Virus Proteomes in Susceptible Hosts: Identification of New Targets for Antiviral Strategies. In Biotechnology and Plant Disease Management; Punja, Z.K., De Boer, S.H., Sanfacon, H., Eds.; CAB International: Wallingford, UK, 2007; pp. 87–108. [Google Scholar]
- Nagy, P.D.; Pogany, J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012, 10, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, A. Dissecting the Molecular Network of Virus-Plant Interactions: The Complex Roles of Host Factors. Annu. Rev. Phytopathol. 2015, 53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Roberts, R.; Rakotondrafara, A.M. The role of the 5′ untranslated regions of Potyviridae in translation. Virus Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.E.; Miller, W.A. 3′ cap-independent translation enhancers of plant viruses. Annu. Rev. Microbiol. 2013, 67, 21–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Krishnaswamy, S. Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 2012, 13, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, O.; Aranda, M.A.; Caranta, C. Plant resistance to viruses mediated by translation initiation factors. In Recent Advances in Plant Virology; Caranta, C., Aranda, M.A., Tepfer, M., Lopez-Moya, J., Eds.; Caister Academic Press: Wymondham, Norfolk, VA, USA, 2011; pp. 177–194. [Google Scholar]
- Robaglia, C.; Caranta, C. Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 2006, 11, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Maule, A.J.; Caranta, C.; Boulton, M.I. Sources of natural resistance to plant viruses: Status and prospects. Mol. Plant Pathol. 2007, 8, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Calvo, M.; Martinez-Turino, S.; Garcia, J.A. Resistance to Plum pox virus Strain C in Arabidopsis thaliana and Chenopodium foetidum involves genome-linked viral protein and other viral determinants and might depend on compatibility with host translation initiation factors. Mol. Plant Microbe Interact. 2014, 27, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; Rodriguez-Moreno, L.; Rodriguez-Hernandez, A.M.; Aranda, M.A.; Truniger, V. Nicotiana benthamiana resistance to non-adapted Melon necrotic spot virus results from an incompatible interaction between virus RNA and translation initiation factor 4E. The Plant J. 2011, 66, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Sanfacon, H. Symptom recovery in virus-infected plants: Revisiting the role of RNA silencing mechanisms. Virology 2015, 479–480, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Valasek, L.S. “Ribozoomin”—Translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr. Protein Pept. Sci. 2012, 13, 305–330. [Google Scholar] [CrossRef] [PubMed]
- Browning, K.S. Plant translation initiation factors: it is not easy to be green. Biochem. Soc. Trans. 2004, 32, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, R.; Bailey-Serres, J. Regulation of translational initiation in plants. Curr. Opin. Plant Biol. 2002, 5, 460–465. [Google Scholar] [CrossRef]
- Patrick, R.M.; Browning, K.S. The eIF4F and eIFiso4F Complexes of Plants: An Evolutionary Perspective. Comp. Funct. Genomics 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; von Arnim, A.G. Translational regulation of cytoplasmic mRNAs. Arabidopsis Book 2013, 11. [Google Scholar] [CrossRef] [PubMed]
- Bush, M.S.; Hutchins, A.P.; Jones, A.M.; Naldrett, M.J.; Jarmolowski, A.; Lloyd, C.W.; Doonan, J.H. Selective recruitment of proteins to 5′ cap complexes during the growth cycle in Arabidopsis. Plant J. 2009, 59, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Browning, K.S.; Humphreys, J.; Hobbs, W.; Smith, G.B.; Ravel, J.M. Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. J. Biol. Chem. 1990, 265, 17967–17973. [Google Scholar] [PubMed]
- Duprat, A.; Caranta, C.; Revers, F.; Menand, B.; Browning, K.S.; Robaglia, C. The Arabidopsis eukaryotic initiation factor ISO4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 2002, 32, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Combe, J.P.; Petracek, M.E.; van Eldik, G.; Meulewaeter, F.; Twell, D. Translation initiation factors eIF4E and eIFiso4E are required for polysome formation and regulate plant growth in tobacco. Plant Mol. Biol. 2005, 57, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Mayberry, L.K.; Allen, M.L.; Nitka, K.R.; Campbell, L.; Murphy, P.A.; Browning, K.S. Plant cap-binding complexes eukaryotic initiation factors eIF4F and eIFISO4F: molecular specificity of subunit binding. J. Biol. Chem. 2011, 286, 42566–42574. [Google Scholar] [CrossRef] [PubMed]
- Mayberry, L.K.; Allen, M.L.; Dennis, M.D.; Browning, K.S. Evidence for Variation in the Optimal Translation Initiation Complex: Plant eIF4B, eIF4F, and eIFISO4F Differentially Promote Translation of mRNAs. Plant Physiol. 2009, 150, 1844–1854. [Google Scholar] [CrossRef] [PubMed]
- Andersen, G.R.; Nissen, P.; Nyborg, J. Elongation factors in protein biosynthesis. Trends Biochem. Sci. 2003, 28, 434–441. [Google Scholar] [CrossRef]
- Goodfellow, I.G.; Roberts, L.O. Eukaryotic initiation factor 4E. Int. J. Biochem. Cell. Biol. 2008, 40, 2675–2680. [Google Scholar] [CrossRef] [PubMed]
- Culjkovic, B.; Topisirovic, I.; Borden, K.L. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell. Cycle 2007, 6, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Bokros, C.L.; Hugdahl, J.D.; Kim, H.H.; Hanesworth, V.R.; van Heerden, A.; Browning, K.S.; Morejohn, L.C. Function of the p86 subunit of eukaryotic initiation factor ISO4F as a microtubule-associated protein in plant cells. Proc. Natl. Acad. Sci. USA 1995, 92, 7120–7124. [Google Scholar] [CrossRef] [PubMed]
- Hugdahl, J.D.; Bokros, C.L.; Morejohn, L.C. End-to-end annealing of plant microtubules by the p86 subunit of eukaryotic initiation factor-ISO4F. Plant Cell. 1995, 7, 2129–2138. [Google Scholar] [CrossRef] [PubMed]
- Mateyak, M.K.; Kinzy, T.G. eEF1A: solid thinking outside the ribosome. The Journal of biological chemistry 2010, 285, 21209–21213. [Google Scholar] [CrossRef] [PubMed]
- Au, H.H.; Jan, E. Novel viral translation strategies. Wiley Interdiscip. Rev. RNA 2014, 5, 779–801. [Google Scholar] [CrossRef] [PubMed]
- Firth, A.E.; Brierley, I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012, 93, 1385–1409. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.R. Tricks an IRES uses to enslave ribosomes. Trends Microbiol. 2012, 20, 558–66. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.A.; Jackson, J.; Feng, Y. Cis- and trans-regulation of luteovirus gene expression by the 3′ end of the viral genome. Virus Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.E. 3′UTRs of carmoviruses. Virus Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Ishibashi, K.; Miyashita, S.; Ishikawa, M. Functions of the 5′- and 3′-untranslated regions of tobamovirus RNA. Virus Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Nieto, C.; Gonzalez-Ibeas, D.; Aranda, M. Mechanism of plant eIF4E-mediated resistance against a Carmovirus (Tombusviridae): Cap-independent translation of a viral RNA controlled in cis by an (a)virulence determinant. Plant J. 2008, 56, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Laliberte, J.F. The genome-linked protein VPg of plant viruses-A protein with many partners. Curr. Opin. Virol. 2011, 1, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Rantalainen, K.I.; Eskelin, K.; Tompa, P.; Makinen, K. Structural flexibility allows the functional diversity of potyvirus genome-linked protein VPg. J. Virol. 2011, 85, 2449–2457. [Google Scholar] [CrossRef] [PubMed]
- Hebrard, E.; Bessin, Y.; Michon, T.; Longhi, S.; Uversky, V.N.; Delalande, F.; Van Dorsselaer, A.; Romero, P.; Walter, J.; Declerck, N.; Fargette, D. Intrinsic disorder in Viral Proteins Genome-Linked: Experimental and predictive analyses. Virology J. 2009, 6. [Google Scholar] [CrossRef] [PubMed]
- Grzela, R.; Szolajska, E.; Ebel, C.; Madern, D.; Favier, A.; Wojtal, I.; Zagorski, W.; Chroboczek, J. Virulence factor of potato virus Y, genome-attached terminal protein VPg, is a highly disordered protein. J. Biol. Chem. 2008, 283, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Rantalainen, K.I.; Uversky, V.N.; Permi, P.; Kalkkinen, N.; Dunker, A.K.; Makinen, K. Potato virus A genome-linked protein VPg is an intrinsically disordered molten globule-like protein with a hydrophobic core. Virology 2008, 377, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, S.; Chatel, H.; Fortin, M.G.; Laliberte, J.F. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor ISO4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 1997, 234, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.; Plante, D.; Wittmann, S.; Daigneault, N.; Fortin, M.G.; Laliberte, J.F. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 2000, 74, 7730–7737. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 2001, 75, 12141–12152. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Miyoshi, H.; Gallie, D.R.; Goss, D.J. Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: interactions with translation initiation factors eIF4F and eIFiso4F. J. Biol. Chem. 2008, 283, 1340–1349. [Google Scholar] [CrossRef] [PubMed]
- Plante, D.; Viel, C.; Leonard, S.; Tampo, H.; Laliberte, J.F.; Fortin, M.G. Turnip mosaic virus VPg does not disrupt the translation initiation complex but interfere with cap binding. Physiol. Molec. Plant Pathol. 2004, 64, 219–226. [Google Scholar] [CrossRef]
- Eskelin, K.; Hafren, A.; Rantalainen, K.I.; Makinen, K. Potyviral VPg enhances viral RNA Translation and inhibits reporter mRNA translation in planta. J. Virol. 2011, 85, 9210–9221. [Google Scholar] [CrossRef] [PubMed]
- Hafren, A.; Eskelin, K.; Makinen, K. Ribosomal protein P0 promotes Potato virus A infection and functions in viral translation together with VPg and eIFISO4E. J. Virol. 2013, 87, 4302–4312. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, V.; Gallois, J.L.; Chafiai, F.; Allen, L.M.; Schurdi-Levraud, V.; Browning, K.S.; Candresse, T.; Caranta, C.; Le Gall, O.; German-Retana, S. Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana. FEBS Lett. 2007, 581, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Paredes, C.A.; Silva-Rosales, L.; Daros, J.A.; Alejandri-Ramirez, N.D.; Dinkova, T.D. The absence of eukaryotic initiation factor eIFISO4E affects the systemic spread of a Tobacco etch virus isolate in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2013, 26, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Hebrard, E.; Poulicard, N.; Gerard, C.; Traore, O.; Wu, H.C.; Albar, L.; Fargette, D.; Bessin, Y.; Vignols, F. Direct interaction between the Rice yellow mottle virus (RYMV) VPg and the central domain of the rice eIFISO4G1 factor correlates with rice susceptibility and RYMV virulence. Mol. Plant Microbe Interact. 2010, 23, 1506–1513. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.; Chisholm, J.; Laliberte, J.F.; Sanfacon, H. Interaction in vitro between the proteinase of Tomato ringspot virus (genus Nepovirus) and the eukaryotic translation initiation factor iso4E from Arabidopsis thaliana. J. Gen. Virol. 2002, 83, 2085–2089. [Google Scholar] [PubMed]
- Noueiry, A.O.; Chen, J.; Ahlquist, P. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc. Natl. Acad. Sci. USA 2000, 97, 12985–12990. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, N.; Pogany, J.; Nagy, P.D. A Co-Opted DEAD-Box RNA helicase enhances tombusvirus plus-strand synthesis. PLoS Pathog. 2012, 8, e1002537. [Google Scholar] [CrossRef] [PubMed]
- Dreher, T.W. Role of tRNA-like structures in controlling plant virus replication. Virus Res. 2009, 139, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Dreher, T.W.; Uhlenbeck, O.C.; Browning, K.S. Quantitative assessment of EF-1α.GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J. Biol. Chem. 1999, 274, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Colussi, T.M.; Costantino, D.A.; Hammond, J.A.; Ruehle, G.M.; Nix, J.C.; Kieft, J.S. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature 2014, 511, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, D.; Dreher, T.W. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology 2004, 321, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Ryabova, L.A.; Pooggin, M.M.; Hohn, T. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 2006, 119, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Bonneville, J.M.; Sanfacon, H.; Futterer, J.; Hohn, T. Posttranscriptional trans-activation in cauliflower mosaic virus. Cell. 1989, 59, 1135–1143. [Google Scholar] [CrossRef]
- Park, H.S.; Himmelbach, A.; Browning, K.S.; Hohn, T.; Ryabova, L.A. A plant viral "reinitiation" factor interacts with the host translational machinery. Cell. 2001, 106, 723–733. [Google Scholar] [CrossRef]
- Thiebeauld, O.; Schepetilnikov, M.; Park, H.S.; Geldreich, A.; Kobayashi, K.; Keller, M.; Hohn, T.; Ryabova, L.A. A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation. EMBO J. 2009, 28, 3171–3184. [Google Scholar] [CrossRef] [PubMed]
- Bureau, M.; Leh, V.; Haas, M.; Geldreich, A.; Ryabova, L.; Yot, P.; Keller, M. P6 protein of Cauliflower mosaic virus, a translation reinitiator, interacts with ribosomal protein L13 from Arabidopsis thaliana. J. Gen. Virol. 2004, 85, 3765–3775. [Google Scholar] [CrossRef] [PubMed]
- Schepetilnikov, M.; Kobayashi, K.; Geldreich, A.; Caranta, C.; Robaglia, C.; Keller, M.; Ryabova, L.A. Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J. 2011, 30, 1343–1356. [Google Scholar] [CrossRef] [PubMed]
- Ouibrahim, L.; Giner Rubio, A.; Moretti, A.; Montane, M.H.; Menand, B.; Meyer, C.; Robaglia, C.; Caranta, C. Potyviruses differ in their requirement for TOR signaling. J. Gen. Virol. 2015, in press. [Google Scholar] [CrossRef] [PubMed]
- Cotton, S.; Grangeon, R.; Thivierge, K.; Mathieu, I.; Ide, C.; Wei, T.; Wang, A.; Laliberte, J.F. Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 2009, 83, 10460–10471. [Google Scholar] [CrossRef] [PubMed]
- Kawamura-Nagaya, K.; Ishibashi, K.; Huang, Y.P.; Miyashita, S.; Ishikawa, M. Replication protein of tobacco mosaic virus cotranslationally binds the 5′ untranslated region of genomic RNA to enable viral replication. Proc. Natl. Acad. Sci. USA 2014, 111, E1620–E1628. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Noueiry, A.; Ahlquist, P. An alternate pathway for recruiting template RNA to the brome mosaic virus RNA replication complex. J. Virol. 2003, 77, 2568–2577. [Google Scholar] [CrossRef] [PubMed]
- Quadt, R.; Kao, C.C.; Browning, K.S.; Hershberger, R.P.; Ahlquist, P. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 1993, 90, 1498–1502. [Google Scholar] [CrossRef] [PubMed]
- Osman, T.A.; Buck, K.W. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J. Virol. 1997, 71, 6075–6082. [Google Scholar] [PubMed]
- Thivierge, K.; Cotton, S.; Dufresne, P.J.; Mathieu, I.; Beauchemin, C.; Ide, C.; Fortin, M.G.; Laliberte, J.F. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 2008, 377, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, Y.; Kobayashi, T.; Hamada, K.; Sakurai, K.; Yoshii, A.; Suzuki, M.; Namba, S.; Hibi, T. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 2006, 347, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, C.; Boutet, N.; Laliberte, J.F. Visualisation of the interaction between the precursors of the viral protein linked to the genome (VPg) of Turnip mosaic virus and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 2007, 81, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.S.; Wei, T.; Laliberte, J.F.; Wang, A. A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol. 2010, 152, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, C.; Laliberte, J.F. The poly(A) binding protein is internalized in virus-induced vesicles or redistributed to the nucleolus during Turnip mosaic virus infection. J. Virol. 2007, 81, 10905–10913. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, P.J.; Ubalijoro, E.; Fortin, M.G.; Laliberte, J.F. Arabidopsis thaliana class II poly(A)-binding proteins are required for efficient multiplication of turnip mosaic virus. J. Gen. Virol. 2008, 89, 2339–2348. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, N.; Barajas, D.; Nagy, P.D. Similar roles for yeast Dbp2 and Arabidopsis RH20 DEAD-box RNA helicases to Ded1 helicase in tombusvirus plus-strand synthesis. Virology 2012, 432, 470–484. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, N.; Nagy, P.D. The expanding functions of cellular helicases: the tombusvirus RNA replication enhancer co-opts the plant eIF4AIII-like AtRH2 and the DDX5-like AtRH5 DEAD-box RNA helicases to promote viral asymmetric RNA replication. PLoS Pathog. 2014, 10, e1004051. [Google Scholar] [CrossRef] [PubMed]
- Nishikiori, M.; Dohi, K.; Mori, M.; Meshi, T.; Naito, S.; Ishikawa, M. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 2006, 80, 8459–8468. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pogany, J.; Panavas, T.; Xu, K.; Esposito, A.M.; Kinzy, T.G.; Nagy, P.D. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 2009, 385, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, D.; Yoshinari, S.; Dreher, T.W. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology 2004, 321, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pogany, J.; Tupman, S.; Esposito, A.M.; Kinzy, T.G.; Nagy, P.D. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLOS Pathog. 2010, 6, e1001175. [Google Scholar] [CrossRef] [PubMed]
- Sasvari, Z.; Izotova, L.; Kinzy, T.G.; Nagy, P.D. Synergistic roles of eukaryotic translation elongation factors 1Bgamma and 1A in stimulation of tombusvirus minus-strand synthesis. PLoS Pathog. 2011, 7, e1002438. [Google Scholar] [CrossRef] [PubMed]
- Zeenko, V.V.; Ryabova, L.A.; Spirin, A.S.; Rothnie, H.M.; Hess, D.; Browning, K.S.; Hohn, T. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 2002, 76, 5678–5691. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, Y.; Sakurai, K.; Hamada, K.; Komatsu, K.; Ozeki, J.; Yoshida, A.; Yoshii, A.; Shimizu, T.; Namba, S.; Hibi, T. Significance of eukaryotic translation elongation factor 1A in tobacco mosaic virus infection. Arch. Virol. 2010, 155, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Oh, C.S.; Kang, B.C. Translation elongation factor 1B (eEF1B) is an essential host factor for Tobacco mosaic virus infection in plants. Virology 2013, 439, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Johansen, E.; Eyers, S.; Thomas, C.L.; Noel Ellis, T.H.; Maule, A.J. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 2004, 40, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, R.; Soto, M.J.; Martinez-Zapater, J.; Ponz, F. Impaired cell-to-cell movement of Potato virus Y in pepper plants carrying the ya (pr21) resistance gene. Mol. Plant Microbe Interact. 1996, 9, 314–318. [Google Scholar] [CrossRef]
- Csorba, T.; Kontra, L.; Burgyan, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, in press. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Lanet, E.; Delannoy, E.; Sormani, R.; Floris, M.; Brodersen, P.; Crete, P.; Voinnet, O.; Robaglia, C. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell. 2009, 21, 1762–1768. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread translational inhibition by plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.O.; Tomari, Y. Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell. 2013, 52, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.S.; Hart-Smith, G.; Eamens, A.L.; Wilkins, M.R.; Waterhouse, P.M. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat. Plants 2015, 1. [Google Scholar] [CrossRef]
- Yang, L.; Wu, G.; Poethig, R.S. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, L.; Zhuang, X.; Yu, Y.; Liu, X.; Cui, X.; Ji, L.; Pan, Z.; Cao, X.; Mo, B.; Zhang, F.; Raikhel, N.; Jiang, L.; Chen, X. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013, 153, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Nicole, M.C.; Meteignier, L.V.; Hong, N.; Wang, G.; Moffett, P. Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. J. Exp. Bot. 2015, in press. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Zamora, A.; Azhar, M.T.; Sacco, M.A.; Lambert, L.H.; Moffett, P. Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J. 2009, 58, 940–951. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Sanfacon, H. Temperature-dependent symptom recovery in Nicotiana benthamiana plants infected with tomato ringspot virus is associated with reduced translation of viral RNA2 and requires ARGONAUTE 1. Virology 2014, 456–457, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Karran, R.A.; Sanfacon, H. Tomato ringspot virus Coat Protein Binds to ARGONAUTE 1 and Suppresses the Translation Repression of a Reporter Gene. Mol. Plant Microbe Interact. 2014, 27, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Bushell, M. The complexity of miRNA-mediated repression. Cell. Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Fukaya, T.; Tomari, Y. MicroRNAs mediate gene silencing via multiple different pathways in Drosophila. Mol. Cell. 2012, 48, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Meijer, H.A.; Kong, Y.W.; Lu, W.T.; Wilczynska, A.; Spriggs, R.V.; Robinson, S.W.; Godfrey, J.D.; Willis, A.E.; Bushell, M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 2013, 340, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.; Ohshima, K. Potyviruses and the digital revolution. Annu. Rev. Phytopathol. 2010, 48, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Charron, C.; Nicolai, M.; Gallois, J.L.; Robaglia, C.; Moury, B.; Palloix, A.; Caranta, C. Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 2008, 54, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Moury, B.; Charron, C.; Janzac, B.; Simon, V.; Gallois, J.L.; Palloix, A.; Caranta, C. Evolution of plant eukaryotic initiation factor 4E (eIF4E) and potyvirus genome-linked protein (VPg): A game of mirrors impacting resistance spectrum and durability. Infect. Genet. Evol. 2014, 27, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Aranda, M.A. Recessive resistance to plant viruses. Adv. Vir. Res. 2009, 75, 119–159. [Google Scholar]
- Kang, B.C.; Yeam, I.; Frantz, J.D.; Murphy, J.F.; Jahn, M.M. The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 2005, 42, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Ashby, J.A.; Stevenson, C.E.; Jarvis, G.E.; Lawson, D.M.; Maule, A.J. Structure-based mutational analysis of eIF4E in relation to sbm1 resistance to pea seed-borne mosaic virus in pea. PLoS ONE 2011, 6, e15873. [Google Scholar] [CrossRef] [PubMed]
- German-Retana, S.; Walter, J.; Doublet, B.; Roudet-Tavert, G.; Nicaise, V.; Lecampion, C.; Houvenaghel, M.C.; Robaglia, C.; Michon, T.; Le Gall, O. Mutational analysis of plant cap-binding protein eIF4E reveals key amino acids involved in biochemical functions and potyvirus infection. J. Virol. 2008, 82, 7601–7612. [Google Scholar] [CrossRef] [PubMed]
- Ibiza, V.P.; Canizares, J.; Nuez, F. EcoTILLING in Capsicum species: Searching for new virus resistances. BMC Genomics 2010, 11, e631. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; Piron, F.; Dalmais, M.; Marco, C.F.; Moriones, E.; Gomez-Guillamon, M.L.; Truniger, V.; Gomez, P.; Garcia-Mas, J.; Aranda, M.A.; Bendahmane, A. EcoTILLING for the identification of allelic variants of melon eIF4E, a factor that controls virus susceptibility. BMC Plant Biol. 2007, 7. [Google Scholar] [CrossRef] [PubMed]
- Konecna, E.; Safarova, D.; Navratil, M.; Hanacek, P.; Coyne, C.; Flavell, A.; Vishnyakova, M.; Ambrose, M.; Redden, R.; Smykal, P. Geographical gradient of the eIF4E alleles conferring resistance to potyviruses in pea (Pisum) germplasm. PLoS ONE 2014, 9, e90394. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.; Nicolai, M.; Caranta, C.; Palloix, A. Allele mining in the pepper gene pool provided new complementation effects between pvr2-eIF4E and pvr6-eIFISO4E alleles for resistance to pepper veinal mottle virus. J. Gen. Virol. 2009, 90, 2808–2814. [Google Scholar] [CrossRef] [PubMed]
- Piron, F.; Nicolai, M.; Minoia, S.; Piednoir, E.; Moretti, A.; Salgues, A.; Zamir, D.; Caranta, C.; Bendahmane, A. An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 2010, 5, e11313. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Hernandez, A.M.; Gosalvez, B.; Sempere, R.N.; Burgos, L.; Aranda, M.A.; Truniger, V. Melon RNA interference (RNAi) lines silenced for Cm-eIF4E show broad virus resistance. Mol. Plant Pathol. 2012, 3, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Mazier, M.; Flamain, F.; Nicolai, M.; Sarnette, V.; Caranta, C. Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato. PLoS ONE 2011, 6, e29595. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kohalmi, S.E.; Svircev, A.; Wang, A.; Sanfacon, H.; Tian, L. Silencing of the host factor eIFISO4E gene confers plum pox virus resistance in plum. PLoS ONE 2013, 8, e50627. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.C.; Yeam, I.; Li, H.; Perez, K.W.; Jahn, M.M. Ectopic expression of a recessive resistance gene generates dominant potyvirus resistance in plants. Plant Biotechnol. J. 2007, 4, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Cavatorta, J.; Perez, K.W.; Gray, S.M.; Van Eck, J.; Yeam, I.; Jahn, M. Engineering virus resistance using a modified potato gene. Plant Biotechnol. J. 2011, 9, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Richael, C.; Rommens, C.M. Overexpression of the wild potato eIF4E-1 variant Eva1 elicits Potato virus Y resistance in plants silenced for native eIF4E-1. Transgenic Res. 2012, 21, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kang, W.H.; Hwang, J.; Yang, H.B.; Dosun, K.; Oh, C.S.; Kang, B.C. Transgenic Brassica rapa plants over-expressing eIFISO4E variants show broad-spectrum Turnip mosaic virus (TuMV) resistance. Mol. Plant Pathol. 2014, 15, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Schaad, M.C.; Anderberg, R.J.; Carrington, J.C. Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 2000, 273, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Estevan, J.; Marena, A.; Callot, C.; Lacombe, S.; Moretti, A.; Caranta, C.; Gallois, J.L. Specific requirement for translation initiation factor 4E or its isoform drives plant host susceptibility to Tobacco etch virus. BMC Plant Biol. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Nakahara, K.; Yoshii, M.; Ishikawa, M.; Uyeda, I. Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 2005, 579, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Li, J.; Liu, W.Y.; An, S.J.; Cho, H.; Her, N.H.; Yeam, I.; Kim, D.; Kang, B.C. Double mutations in eIF4E and eIFiso4E confer recessive resistance to Chilli veinal mottle virus in pepper. Mol. Cells 2009, 27, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Gallois, J.L.; Moury, B.; Robaglia, C.; Palloix, A.; Caranta, C. Simultaneous mutations in translation initiation factors eIF4E and eIFISO4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 2006, 87, 2089–2098. [Google Scholar] [CrossRef] [PubMed]
- Jenner, C.E.; Nellist, C.F.; Barker, G.C.; Walsh, J.A. Turnip mosaic virus (TuMV) is able to use alleles of both eIF4E and eIFISO4E from multiple loci of the diploid Brassica rapa. Mol. Plant Microbe Interact. 2010, 23, 1498–1505. [Google Scholar] [CrossRef] [PubMed]
- Ayme, V.; Petit-Pierre, J.; Souche, S.; Palloix, A.; Moury, B. Molecular dissection of the potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 2007, 88, 1594–1601. [Google Scholar] [CrossRef] [PubMed]
- Ayme, V.; Souche, S.; Caranta, C.; Jacquemond, M.; Chadoeuf, J.; Palloix, A.; Moury, B. Different mutations in the genome-linked protein VPg of potato virus Y confer virulence on the pvr2(3) resistance in pepper. Mol. Plant Microbe Interact. 2006, 19, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Moury, B.; Morel, C.; Johansen, E.; Guilbaud, L.; Souche, S.; Ayme, V.; Caranta, C.; Palloix, A.; Jacquemond, M. Mutations in potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant Microbe Interact. 2004, 17, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Gallois, J.L.; Charron, C.; Sanchez, F.; Pagny, G.; Houvenaghel, M.C.; Moretti, A.; Ponz, F.; Revers, F.; Caranta, C.; German-Retana, S. Single amino acid changes in the turnip mosaic virus viral genome-linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIFISO4E and eIFISO4G. J. Gen. Virol. 2010, 91, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Razzak, A.; Guiraud, T.; Peypelut, M.; Walter, J.; Houvenaghel, M.C.; Candresse, T.; Le Gall, O.; German-Retana, S. Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E-mediated resistance against Lettuce mosaic potyvirus. Mol. Plant Pathol. 2009, 10, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Sorel, M.; Svanella-Dumas, L.; Candresse, T.; Acelin, G.; Pitarch, A.; Houvenaghel, M.C.; German-Retana, S. Key mutations in the cylindrical inclusion involved in lettuce mosaic virus adaptation to eIF4E-mediated resistance in lettuce. Mol. Plant Microbe Interact. 2014, 27, 1014–1024. [Google Scholar] [CrossRef] [PubMed]
- Tavert-Roudet, G.; Abdul-Razzak, A.; Doublet, B.; Walter, J.; Delaunay, T.; German-Retana, S.; Michon, T.; Le Gall, O.; Candresse, T. The C terminus of lettuce mosaic potyvirus cylindrical inclusion helicase interacts with the viral VPg and with lettuce translation eukaryotic initiation factor 4E. J. Gen. Virol. 2012, 93, 184–93. [Google Scholar] [CrossRef] [PubMed]
- Sorel, M.; Garcia, J.A.; German-Retana, S. The Potyviridae cylindrical inclusion helicase: a key multipartner and multifunctional protein. Mol. Plant Microbe Interact. 2014, 27, 215–26. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, K.S.; Shimada, R.; Choi, S.H.; Yamamoto, H.; Shao, J.; Uyeda, I. Involvement of the P1 cistron in overcoming eIF4E-mediated recessive resistance against Clover yellow vein virus in pea. Mol. Plant Microbe Interact. 2010, 23, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Moury, B.; Janzac, B.; Ruellan, Y.; Simon, V.; Ben Khalifa, M.; Fakhfakh, H.; Fabre, F.; Palloix, A. Interaction patterns between potato virus Y and eIF4E-mediated recessive resistance in the Solanaceae. J. Virol. 2014, 88, 9799–9807. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Caranta, C.; Palloix, A.; Lefebvre, V.; Caboche, M.; Bendahmane, A. Structural analysis of the eukaryotic initiation factor 4E gene controlling potyvirus resistance in pepper: Exploitation of a BAC library. Gene 2004, 338, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Dussault, M.H.; Palloix, A.; Moury, B.; Bendahmane, A.; Robaglia, C.; Caranta, C. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 2002, 32, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, V.; German-Retana, S.; Sanjuan, R.; Dubrana, M.P.; Mazier, M.; Maisonneuve, B.; Candresse, T.; Caranta, C.; LeGall, O. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus Lettuce mosaic virus. Plant Physiol. 2003, 132, 1272–1282. [Google Scholar] [CrossRef] [PubMed]
- Andrade, M.; Abe, Y.; Nakahara, k. S.; Uyeda, I. The cyv-2 resistance to clover yellow vein virus in pea is controlled by the eukaryotic initiation factor 4E. J. Gen. Plant Pathol. 2009, 75, 241–249. [Google Scholar] [CrossRef]
- Bruun-Rasmussen, M.; Moller, I.S.; Tulinius, G.; Hansen, J.K.; Lund, O.S.; Johansen, I.E. The same allele of translation initiation factor 4E mediates resistance against two Potyvirus spp. in Pisum sativum. Mol. Plant Microbe Interact. 2007, 20, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Naderpour, M.; Lund, O.S.; Larsen, R.; Johansen, E. Potyviral resistance derived from cultivars of Phaseolus vulgaris carrying bc-3 is associated with the homozygotic presence of a mutated eIF4E allele. Mol. Plant Pathol. 2010, 11, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Hart, J.P.; Griffiths, P.D. A series of eIF4E alleles at the Bc-3 locus are associated with recessive resistance to Clover yellow vein virus in common bean. Theor. Appl. Genet. 2013, 126, 2849–2863. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Gallois, J.L.; Lesage, M.L.; Caranta, C. The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genomics 2005, 274, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Hofinger, B.J.; Russell, J.R.; Bass, C.G.; Baldwin, T.; dos Reis, M.; Hedley, P.E.; Li, Y.; Macaulay, M.; Waugh, R.; Hammond-Kosack, K.E.; Kanyuka, K. An exceptionally high nucleotide and haplotype diversity and a signature of positive selection for the eIF4E resistance gene in barley are revealed by allele mining and phylogenetic analyses of natural populations. Mol. Ecol. 2011, 20, 3653–3668. [Google Scholar] [CrossRef] [PubMed]
- Kanyuka, K.; Druka, A.; Caldwell, D.G.; Tymon, A.; McCallum, N.; Waugh, R.; Adams, M.J. Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol. Plant Pathol. 2005, 6, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Kanyuka, K.; McGrann, G.; Alhudaib, K.; Hariri, D.; Adams, M.J. Biological and sequence analysis of a novel European isolate of Barley mild mosaic virus that overcomes the barley rym5 resistance gene. Arch. Virol. 2004, 149, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Kuhne, T.; Shi, N.; Proeseler, G.; Adams, M.J.; Kanyuka, K. The ability of a bymovirus to overcome the rym4-mediated resistance in barley correlates with a codon change in the VPg coding region on RNA1. J. Gen. Virol. 2003, 84, 2853–2859. [Google Scholar] [CrossRef] [PubMed]
- Stein, N.; Perovic, D.; Kumlehn, J.; Pellio, B.; Stracke, S.; Streng, S.; Ordon, F.; Graner, A. The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J. 2005, 42, 912–922. [Google Scholar] [CrossRef] [PubMed]
- Perovic, D.; Kramer, I.; Habekuss, A.; Perner, K.; Pickering, R.; Proeseler, G.; Kanyuka, K.; Ordon, F. Genetic analyses of BaMMV/BaYMV resistance in barley accession HOR4224 result in the identification of an allele of the translation initiation factor 4e (Hv-eIF4E) exclusively effective against Barley mild mosaic virus (BaMMV). Theor. Appl. Genet. 2014, 127, 1061–71. [Google Scholar] [CrossRef] [PubMed]
- Ling, K.S.; Harris, K.R.; Meyer, J.D.; Levi, A.; Guner, N.; Wehner, T.C.; Bendahmane, A.; Havey, M.J. Non-synonymous single nucleotide polymorphisms in the watermelon eIF4E gene are closely associated with resistance to zucchini yellow mosaic virus. Theor. Appl. Genet. 2009, 120, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Marandel, G.; Salava, J.; Abbott, A.; Candresse, T.; Decroocq, V. Quantitative trait loci meta-analysis of Plum pox virus resistance in apricot (Prunus armeniaca L.): new insights on the organization and the identification of genomic resistance factors. Mol. Plant Pathol. 2009, 10, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Rusholme, R.L.; Higgins, E.E.; Walsh, J.A.; Lydiate, D.J. Genetic control of broad-spectrum resistance to turnip mosaic virus in Brassica rapa (Chinese cabbage). J. Gen. Virol. 2007, 88, 3177–3186. [Google Scholar] [CrossRef] [PubMed]
- Nellist, C.F.; Qian, W.; Jenner, C.E.; Moore, J.D.; Zhang, S.; Wang, X.; Briggs, W.H.; Barker, G.C.; Sun, R.; Walsh, J.A. Multiple copies of eukaryotic translation initiation factors in Brassica rapa facilitate redundancy, enabling diversification through variation in splicing and broad-spectrum virus resistance. Plant J. 2014, 77, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.A.; Rusholme, R.L.; Hughes, S.L.; Jenner, C.E.; Bambridge, J.M.; Lydiate, D.J.; Green, S.K. Different classes of resistance to Turnip mosaic virus in Brassica rapa. Eur. J. Plant Pathol. 2002, 108, 15–20. [Google Scholar] [CrossRef]
- Lellis, A.D.; Kasschau, K.D.; Whitham, S.A.; Carrington, J.C. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIFISO4E during potyvirus infection. Curr. Biol. 2002, 12, 1046–1051. [Google Scholar] [CrossRef]
- Albar, L.; Bangratz-Reyser, M.; Hebrard, E.; Ndjiondjop, M.N.; Jones, M.; Ghesquiere, A. Mutations in the eIFISO4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus. Plant J. 2006, 47, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Thiemele, D.; Boisnard, A.; Ndjiondjop, M.N.; Cheron, S.; Sere, Y.; Ake, S.; Ghesquiere, A.; Albar, L. Identification of a second major resistance gene to Rice yellow mottle virus, RYMV2, in the African cultivated rice species, O. glaberrima. Theor. Appl. Genet. 2010, 121, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Poulicard, N.; Pinel-Galzi, A.; Traore, O.; Vignols, F.; Ghesquiere, A.; Konate, G.; Hebrard, E.; Fargette, D. Historical contingencies modulate the adaptability of Rice yellow mottle virus. PLoS Pathog. 2012, 8, e1002482. [Google Scholar] [CrossRef] [PubMed]
- Traore, O.; Pinel-Galzi, A.; Issaka, S.; Poulicard, N.; Aribi, J.; Ake, S.; Ghesquiere, A.; Sere, Y.; Konate, G.; Hebrard, E.; Fargette, D. The adaptation of Rice yellow mottle virus to the eIFISO4G-mediated rice resistance. Virology 2010, 408, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Hebrard, E.; Pinel-Galzi, A.; Fargette, D. Virulence domain of the RYMV genome-linked viral protein VPg towards rice rymv1–2-mediated resistance. Arch. Virol. 2008, 153, 1161–1164. [Google Scholar] [CrossRef] [PubMed]
- Hebrard, E.; Pinel-Galzi, A.; Bersoult, A.; Sire, C.; Fargette, D. Emergence of a resistance-breaking isolate of Rice yellow mottle virus during serial inoculations is due to a single substitution in the genome-linked viral protein VPg. J. Gen. Virol. 2006, 87, 1369–1373. [Google Scholar] [CrossRef] [PubMed]
- Poulicard, N.; Pinel-Galzi, A.; Hebrard, E.; Fargette, D. Why Rice yellow mottle virus, a rapidly evolving RNA plant virus, is not efficient at breaking rymv1–2 resistance. Mol. Plant Pathol. 2010, 11, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Poulicard, N.; Pinel-Galzi, A.; Fargette, D.; Hebrard, E. Alternative mutational pathways, outside the VPg, of rice yellow mottle virus to overcome eIFISO4G-mediated rice resistance under strong genetic constraints. J. Gen. Virol. 2014, 95, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; Morales, M.; Orjeda, G.; Clepet, C.; Monfort, A.; Sturbois, B.; Puigdomenech, P.; Pitrat, M.; Caboche, M.; Dogimont, C.; Garcia-Mas, J.; Aranda, M.A.; Bendahmane, A. An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J. 2006, 48, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Diaz, J.A.; Nieto, C.; Moriones, E.; Truniger, V.; Aranda, M.A. Molecular characterization of a Melon necrotic spot virus strain that overcomes the resistance in melon and nonhost plants. Mol. Plant Microbe Interact. 2004, 17, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Miras, M.; Sempere, R.N.; Kraft, J.J.; Miller, W.A.; Aranda, M.A.; Truniger, V. Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 2014, 202, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.L.; Zaslaver, O.; Mayberry, L.K.; Browning, K.S.; White, K.A. Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J. Virol. 2013, 87, 1872–1883. [Google Scholar] [CrossRef] [PubMed]
- Pinel-Galzi, A.; Traore, O.; Sere, Y.; Hebrard, E.; Fargette, D. The biogeography of viral emergence: Rice yellow mottle virus as a case study. Curr. Opin. Virol. 2015, 10, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Pinel-Galzi, A.; Rakotomalala, M.; Sangu, E.; Sorho, F.; Kanyeka, Z.; Traore, O.; Sereme, D.; Poulicard, N.; Rabenantoandro, Y.; Sere, Y.; Konate, G.; Ghesquiere, A.; Hebrard, E.; Fargette, D. Theme and variations in the evolutionary pathways to virulence of an rna plant virus species. PLoS Pathog. 2007, 3, e180. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Muhsin, M.; Atienza, G.A.; Kwak, D.Y.; Kim, S.M.; De Leon, T.B.; Angeles, E.R.; Coloquio, E.; Kondoh, H.; Satoh, K.; Cabunagan, R.C.; Cabauatan, P.Q.; Kikuchi, S.; Leung, H.; Choi, I.R. Single nucleotide polymorphisms in a gene for translation initiation factor (eIF4G) of rice (Oryza sativa) associated with resistance to Rice tungro spherical virus. Mol. Plant Microbe Interact. 2010, 23, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, M.; Nishikiori, M.; Tomita, K.; Yoshioka, N.; Kozuka, R.; Naito, S.; Ishikawa, M. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 2004, 78, 6102–6111. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2014, 32C, 76–84. [Google Scholar] [CrossRef] [PubMed]
© 2015 by Her Majesty the Queen in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
