Abstract
Klebsiella pneumoniae phages vB_KpnP_SU503 (SU503) and vB_KpnP_SU552A (SU552A) are virulent viruses belonging to the Autographivirinae subfamily of Podoviridae that infect and kill multi-resistant K. pneumoniae isolates. Phages SU503 and SU552A show high pairwise nucleotide identity to Klebsiella phages KP34 (NC_013649), F19 (NC_023567) and NTUH-K2044-K1-1 (NC_025418). Bioinformatic analysis of these phage genomes show high conservation of gene arrangement and gene content, conserved catalytically active residues of their RNA polymerase, a common and specific lysis cassette, and form a joint cluster in phylogenetic analysis of their conserved genes. Also, we have performed biological characterization of the burst size, latent period, host specificity (together with KP34 and NTUH-K2044-K1-1), morphology, and structural genes as well as sensitivity testing to various conditions. Based on the analyses of these phages, the creation of a new phage genus is suggested within the Autographivirinae, called “Kp34likevirus” after their type phage, KP34. This genus should encompass the recently genome sequenced Klebsiella phages KP34, SU503, SU552A, F19 and NTUH-K2044-K1-1.
1. Introduction
In the post-genomic era there is a substantial number of phage genomes being deposited in NCBI GenBank, and the understanding of phages and their relationship to each other is growing. Early taxonomical classifications mainly focused on morphological similarities and the composition of the nucleic acid of the phage, be it double stranded or single stranded RNA or DNA. With the increased availability of fast and cheap next generation sequencing, the number of sequenced phages has increased drastically, which has also increased the need of having accurate taxonomical groups that are easily classified and understood. This process has begun and has resulted in the classification of the subfamilies of Autographivirinae and Picovirinae in the Podoviridae family, based on comparison of conserved genes [1]. New phage genera are established and generally named after the first fully sequenced phage within each classification, as with the genera within the Autographivirinae subfamily, the T7likevirus [2], the Sp6likevirus [3] and the Phikmvlikevirus [4]. These genera share the defining Autographivirinae ability to self-transcribe its genes, an ability acquired with the single-subunit RNA polymerase, common gene arrangement and common transcriptional scheme.
Phages KP34 [5] and NTUH-K2044-K1-1 [6] are Podoviridae within the subfamily of Autographivirinae and have been loosely associated with the Phikmvlikevirus genus. Both infect the Gram-negative, rod shaped, encapsulated and facultative anaerobic organism Klebsiella pneumoniae. This bacterium is an opportunistic pathogen which is often the cause of nosocomial infections and community acquired pneumonia [7]. In this study, two new phages have been isolated that both belong to the subfamily of Autographivirinae, and bear close resemblance to Klebsiella phages KP34 [5], NTUH-K2044-K1-1 [6] and F19 (NC_023567). These phages show significant nucleotide identity, a conserved recognition and specificity loop of the single-subunit RNA polymerase and a highly similar lysis cassette. In the traditional taxonomic classifications, organisms are grouped into natural groups based on easily observed characteristics, which has moved from the earlier observations purely based on morphological features, into more precise groups based on features of their genomes, transcriptional regime and gene content. Based on these findings, the creation of a new bacteriophage genus is suggested, called “Kp34likevirus”, named after the type phage KP34.
2. Materials and Methods
2.1. Biological Materials
Two phages, vB_KpnP_SU503 (SU503) and vB_KpnP_SU552A (SU552A), were isolated from the bioactive and aerobic step in the waste water treatment process of the Henriksdal waste water treatment plant situated in Stockholm, Sweden. Phages were isolated according to Pieroni et al. with some modifications [8]. One hundred mL of pre-centrifuged effluent was mixed with 100 mL double strength LB-broth and 10 mL of a 2 h, mid-logarithmic K. pneumoniae culture. Phage SU503 was isolated and propagated on K. pneumoniae 07RAFM-KPN-503 and SU552A on 07RAFM-KPN-552. Both are extended spectrum β-lactamse (ESBL) positive clinical isolates collected by The Public Health Agency of Sweden (Folkhälsomyndigheten) in 2007 [9]. Phage KP34 was propagated on Klebsiella isolate KPN77 and NTUH-K2044-K1-1 on NTUH-K2044 (Table S1).
Phage titer and plaque morphology was investigated using the standard soft agar overlay method [10] and performed simultaneously using the same conditions (medium, temperature and time of incubation). KP34 was isolated in Wroclaw, Poland [5], while Klebsiella phage NTUH-K2044-K1-1 was isolated in Taipei City, Taiwan (Table 1) [6].

Table 1.
Bacteriophages used in this study.
| Phage name | Reference | Country of origin | NCBI accession no. |
|---|---|---|---|
| vB_KpnP_SU503 | this work | Sweden | KP708985 |
| vB_KpnP_SU552A | this work | Sweden | KP708986 |
| Klebsiella phage KP34 | [5] | Poland | GQ413938 |
| Klebsiella phage F19 | [11] | China | KF765493 |
| NTUH-K2044-K1-1 | [6] | Taiwan | AB716666 |
2.2. Electron Microscopy
A filtered high-titer phage lysate was centrifuged at 25,000 g for 60 min. The pellet was washed twice in ammonium acetate (0.1 M, pH 7.0). Phages were deposited on copper grids with carbon-coated Formvar films (Sigma-Aldrich Co., St. Louis, MO, USA) and stained for 10 sec with uranyl acetate (2%, pH 4.5). Excess liquid was blotted off and phages were examined using a Zeiss EM 900 electron microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) in Laboratory of Microscopy Techniques, University of Wroclaw, Poland. The magnification was calibrated using T4 phage tail length (114 nm) as a standard.
2.3. Burst Size Experiments
A one-step growth curve of isolated Klebsiella phages was performed according to the method of Pajunen et al. [12] with some modifications previously described [13].
2.4. Sensitivity of Phage Particles to Temperature, Chloroform and pH
Sensitivity of phage particles to temperature, chloroform and pH was evaluated as previously described [13]. Viruses were suspended in phosphate-buffered saline (PBS) (NaCl 137 mmol/L, KCl 2.7 mmol/L, Na2HPO4 10 mmol/L, KH2PO4 1.8 mmol/L, the pH adjusted to 7.4 with HCl and NaOH). Phage particles in PBS pH 7.4 were used as control. An equal volume of filter-sterilized bacteriophage of 108 plaque-forming units per mL (PFU/mL) was mixed with chloroform and incubated for 2 h at room temperature (RT) with intermittent shaking. Further preparations of phages were incubated at pH 2, 4, 5, 6, 8 and 10 for 1 h and 5 h, at RT and 37 °C. A phage preparation was also incubated at 60 °C for 10 min. Phage titers were then assessed using the soft agar overlay method [10].
2.5. Determination of Phage Bacterial Host Range
The host range of the bacteriophages was determined on the 26 clinical K. pneumoniae strains listed in Table S2 [14]. Bacteria were stored at −70 °C in Trypticase Soy Broth (TSB) (Becton Dickinson and Company, Cockeysville, MD, USA) supplemented with 20% glycerol (Avantor Performance Materials Poland S.A., Gliwice, Poland). Spot testing was used as a rapid and efficient method for determining the host range in large collections of bacteria [14]. The phage titer used for spot testing was 105 PFU/mL with final 5 × 102 PFU per spot. A majority of the clinical isolates used in the host range analysis were selected from ESBL-positive K. pneumoniae strains isolated in Sweden during the year of 2007, individually picked for the largest variation of PFGE patterns (using XbaI) by the Public Health Agency of Sweden (Folkhälsomyndigheten) (Solna, Sweden).
2.6. Phage Structural Protein Analysis
Phage particles were purified by cesium chloride density gradient centrifugation method [15]. Phage structural protein analysis was performed by SDS-PAGE gel electrophoresis using Mini-Protean®TGX Stain-Free Precast Gels (Bio-Rad Laboratories, Inc., Hercules, CA, USA) [15]. Analysis of protein bands was performed using Molecular Imager® Gel Doc™ XR System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and Quantity One® software (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Precision Plus Protein™ Unstained Standard (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used as a size marker for the molecular analysis of the phage structural proteins.
2.7. DNA Purification
Phage stocks (~109 PFU/mL) were sterile filtered (0.45 µm Whatman Puradiscs, GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) and DNase I and RNase A treated (Thermo Fischer Scientific, Waltham, MA, USA) for one hour at 37 °C to remove bacterial DNA and RNA contamination prior to phage genome recovery. Subsequent Proteinase K treatment (Thermo Fischer Scientific, Waltham, MA, USA) released phage DNA for purification using phase lock gel Heavy (5 Prime Inc., Gaithersburg, MD, USA) with phenol:chloroform:isoamyl alcohol (Sigma-Aldrich Co., St. Louis, MO, USA) as organic phase in accordance to manufacturer protocols. DNA was then ethanol precipitated using standard protocols [10].
2.8. DNA Sequencing
Phages SU503 and SU552A were sequenced using Roche 454 FLX+ titanium pyrosequencing technology (454 Life Sciences, Branford, CT, USA). The sequencing run resulted in 10506 reads from SU503 and 57583 reads from SU552A. Ends were trimmed prior to de novo assembly. Roche Newbler (2.6, 454 Life Sciences, Branford, CT, USA, 2011) was used to de novo assemble the reads produced by the sequencing run, and contigs smaller than 1000 nucleotides were discarded. From the SU503 sequencing run, 92.09% of the reads assembled into one singular large contig of 43,809 nucleotides with an average coverage of 86 reads per nucleotide. From SU552A, 97.7% of the reads assembled into a singular large contig of 43,594 nucleotides with an average coverage of 509 reads per nucleotide. Genome ends were identified by sequence homology to phage KP34 (GQ413938) and separated.
2.9. In Silico Analysis and Annotation of Genomes
The genomes of SU503 and SU552A were annotated using the RAST engine in Glimmer 3.0 mode [16] and with MetaGeneAnnotator [17] in parallel. Overlapping open reading frames (ORF) were detected by manually investigating each ORF for upstream Shine-Dalgarno sequence, start codon usage and ORF overlap length [18].
All open reading frames were compared to the nucleotide collection database (nr/nt) using BLASTN and translated amino acid sequences were searched for homologous proteins using PSI-BLAST in the non-redundant protein sequences database (nr) [19,20]. To investigate possible transfer-RNAs in the phage genomes, tRNAscan-SE was employed [21]. Putative Rho-independent terminators were identified using ARNold (http://rna.igmors.u-psud.fr/toolbox/arnold/index.php) with minimal energy threshold value set to −15.0 [22,23].
Novel phage promoters were discovered by extracting 100 nucleotides before each putative ORF using ExtractUpstreamDNA (https://lfz.corefacility.ca/extractUpStreamDNA/) and running it through the Multiple RM for Motif Elicitation (MEME) [24]. The same procedure was then performed by only extracting 100 nucleotides before each putative operon. Motifs were then manually investigated in each phage genome and conserved nucleotides marked.
The same annotation procedure was performed on phages KP34, F19 and NTUH-K2044-K1-1 (Table 1) to establish a common annotation framework. Ambiguous and conflicting ORF lengths were harmonized for overlaps, length and ribosomal binding sites.
2.10. Comparative Genomics
To establish a set of conserved genes between the genera within the subfamily of Autographivirinae, CoreGenes 3.5 was utilized on the sequenced genomes of type phages from each phage genus [25]. These phages were φKMV (NC_005045), SP6 (NC_004831), T7 (NC_001604) and KP34 (NC_013649). Determination of conserved genes between the Phikmvlikevirus and “Kp34likevirus” members were made through comparison of φKMV, LKA1 (NC_009936), KP34 and NTUH-K2044-K1-1. Linear genome visualization and tblastx [19] comparison was performed using Easyfig 2.1 [26] with a minimum length of BLAST hits set to be drawn to 30.
A local BLAST database was setup in Geneious (6.1.8, Biomatters Ltd., Auckland, New Zealand, 2013) containing the amino acid sequences inferred from the nucleotide sequences of the predicted open reading frames of phages SU503, SU552A, KP34, NTUH-K2044, F19 for pairwise identity comparison [27]. The datasets used in the phylogenetic analyses were assembled by comparison of the amino acid sequences of the genes, common to these five phage genomes, to the annotated proteins of other phages from the Autographivirinae subfamily. The amino acid sequences of three proteins corresponding to phage KP34 RNA polymerase (KP-KP34p37), head-tail connector protein (KP-KP34p40) and DNA maturase B (KP-KP34p50) were selected for phylogenetic analysis in MEGA6 [28]. In all, 33 phages were shown to harbor homologs to these genes, and the corresponding annotated protein sequences (28) were downloaded from GenBank. The alignments of the amino acid sequences were performed with MUSCLE [29]. The datasets contained 693 positions in the analysis of the RNA polymerase, and 342 in both the head-tail connector protein and DNA maturase B protein sequences datasets.
Phylogenetic analyses were performed in MEGA6 [28] using the maximum likelihood (ML) method based on the Whelan and Goldman substitution model [30]. Gaps or missing data were completely deleted and uniform substitution rates among sites selected in the analyses. Initial starting trees for the ML searches were autogenerated by the maximum parsimony (MP) method. The ML searches were carried out with the Nearest-Neighbor-Interchange (NNI) heuristic method, and the robustness of the resulting trees was evaluated by 500 bootstrap replicates.
2.11. Analysis of Lysis Cassette
Identification and in silico analysis of the lysis proteins and lysis cassette has been done using protein and nucleotide BLAST analysis [19], biosequence HMMER analysis (using profile hidden Markov models) [31], TOPCONS (identifying the presence of transmembrane domains) [32] and ExPASy SIB Bioinformatics Resource Portal [33].
3. Results
3.1. Phage Isolation and Morphological Characteristics
Phages SU503 and SU552A were isolated from the aerated bioactive stage in the Henriksdal waste water treatment plant. This waste water treatment plant receives effluent from both domestic and commercial sources but also from a major hospital in the Stockholm region, the Stockholm South General Hospital (Södersjukhuset). SU503 was isolated on clinical K. pneumoniae 07RAFM-KPN503 and SU552A on 07RAFM-KPN-552 (Table S1).
The bacteriophages SU503 and SU552A were examined simultaneously with NTUH-K2044-K1-1 and KP34 by transmission electron microscopy (TEM) and classified on the basis of their morphological features to be members of the Podoviridae family (Figure 1). The icosahedral heads of these four tested phages were the same size and estimated to be approximately 63 nm between opposite apices and the head was connected to a short 15 nm tail. Twenty six K. pneumoniae strains (Table S1) were used to determine the specificity of the four tested bacteriophages: SU503, SU552A, NTUH-K2044-K1-1 and KP34. The host range of NTUH-K2044-K1-1 was limited to the host strain. Both phages SU503 and SU552A were found to infect two out of 26 clinical isolates (Table S2). By comparison, phage KP34 showed lytic activity against eight of these strains (31%).
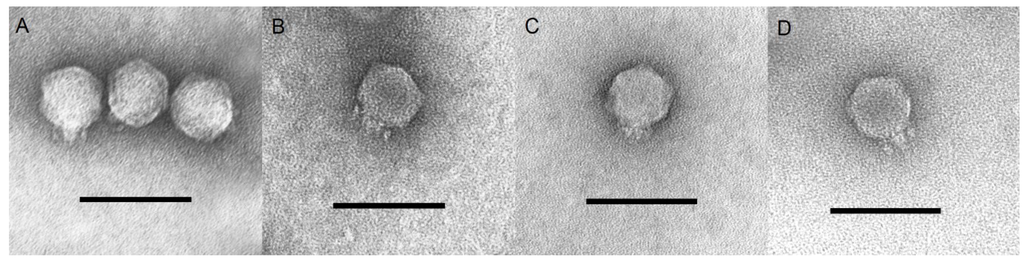
Figure 1.
Transmission electron micrographs of (A) SU503; (B) SU552A; (C) KP34; (D) NTUH-K2044-K1-1. Length bar is 100 nm.
Using the soft agar overlay method, it was shown that phages SU552A and NTUH-K2044-K1-1 form small, clear plaques (about 1–2 mm diameter), whereas the remaining two phages (SU503 and KP34) produced turbid plaques with a diameter of 5 mm, on the strain used for their isolation. All plaques were surrounded by a hazy halo zone, indicating the ability of these phages to produce virion-associated polysaccharide-degrading enzymes [34]. The presence of a gene encoding pectate lyase has been confirmed in NTUH-K2044-K1-1 genome (Orf34) [6]. Homologous depolymerase genes have been identified in the other phages assessed in this study using in silico analysis, at locus tags KP-KP34p57, SU503_53, SU552A_54 and F19_51. The proteins encoded by mentioned genes are relatively large in size (55-70 kDa) and have dual functions, as a structural component of the tail fiber and enzymatic as polysaccharide-degrading enzyme. Their enzymatic activity according to software algorithms are probable pectate lyase. According to the one-step growth curves, all phages have a comparable latent period (15–20 min), but differ in respective average burst size. Phages KP34 and NTUH-K2044-K1-1 have approximately 50 virions per cell while SU503 and SU552A has less than half of that, approximately 20 virions per cell. These phage life cycle characteristics are in accord with the values observed for phage KP34 [5].
3.2. Physicochemical Properties
All phages were shown to be relatively sensitive to high temperature, with a 2-log (KP34 particles) or 3-log (remaining phages) decrease in titer observed after 10 min at 60 °C. While phage KP34 retained almost 100% infectivity after chloroform treatment, the activity reduction of other phages were by two orders (NTUH-K2044-K1-1) or three orders (SU503 and SU552A) of magnitude. Similar variation in chloroform sensitivity has been observed for Klebsiella phages KP15 and KP27, where closely related Myoviridae representatives showed different stability in chloroform treatment [13]. Such phenomenon could be explained by the observation that despite the general absence of lipids, about one third of tailed phages are chloroform-sensitive [35]. The optimal pH was determined by testing the stability of phages at different pH after 1 h and 5 h of incubation at RT and 37 °C. All tested phages lost their infective ability completely at pH 2.0 after 5 h incubation at both temperatures. One-hour incubation at the same acidity caused three and seven log decrease in phages SU503 and NTUH-K2044-K1-1, respectively. Phages SU552A and KP34 were found to be more sensitive to acidic conditions and completely lost its activity after 60 min at pH 2.0. Exposure to pH 4 at 37° C caused ten-fold PFU reduction of SU552A, SU503 and NTUH-K2044-K1-1, after 5 h incubation. In contrast, KP34 was less resistant to this pH value, where three and seven log decrease of titer was noticed after 1 and 5 h, respectively. All tested phages were stable within a pH range of 5–10 at both temperatures.
3.3. Analysis of Structural Proteins
Comparative analysis of the genome sequences of phages SU503 and SU552A reveals the presence of seven genes that encode putative structural proteins which share high amino acid sequence identity to the other “Kp34likeviruses”: KP34, F19 and NTUH-K2044-K1-1, ranging from 86%–99%. By contrast, pairwise BLAST comparison of these same structural proteins with their φKMV equivalent, only shows an average of 30% (between 24%–38%) in pairwise identity. To further characterize these structural genes, an SDS-PAGE was performed. A major protein band and several minor protein bands were observed on the gel, with molecular weights ranging from approximately 20 to 135 kDa (Figure 2).
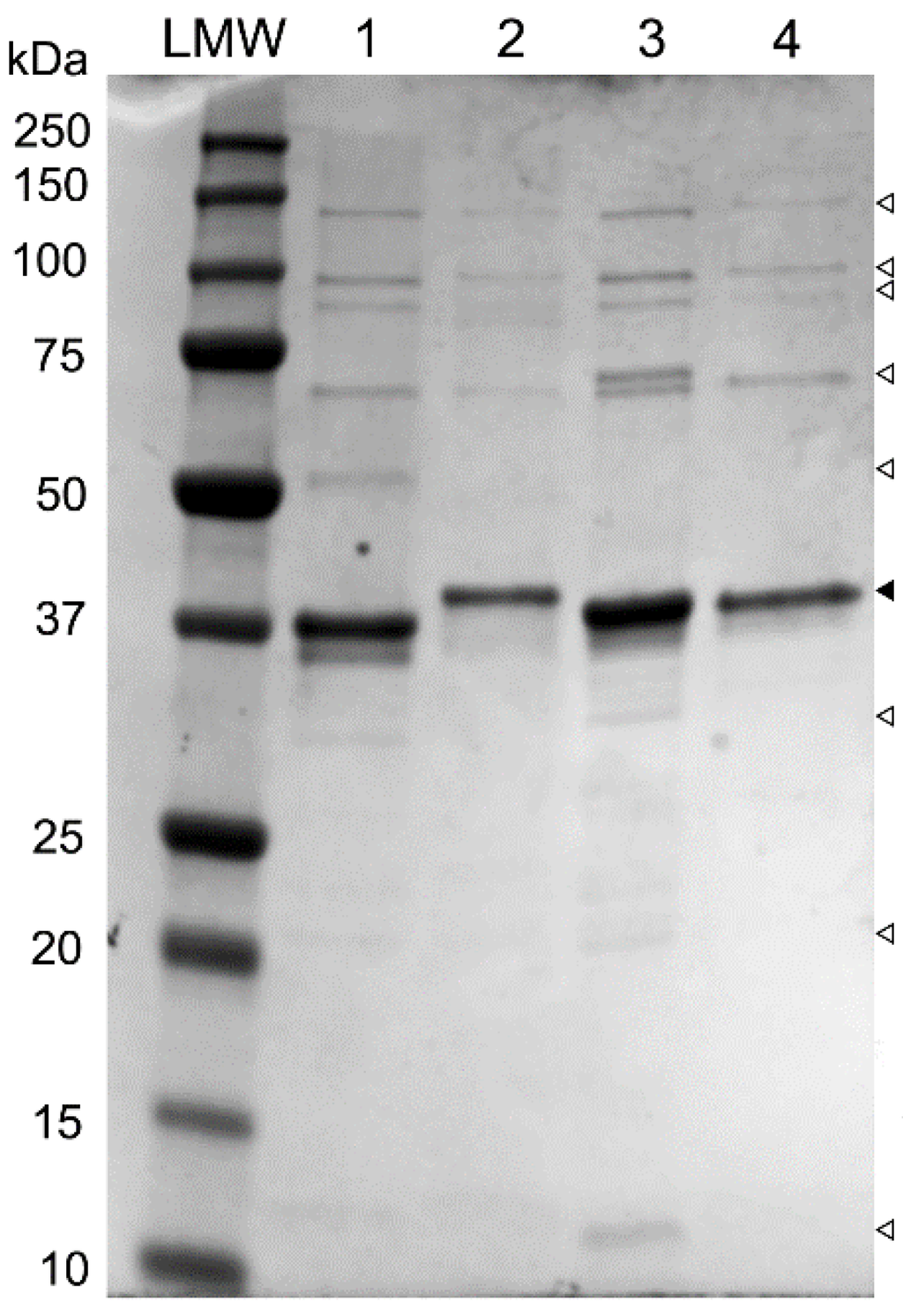
Figure 2.
SDS-PAGE analysis of phage structural proteins. Lane LMW, reference ladder (kDa; BioRad), lane 1, phage SU503; lane 2, phage SU552A; lane 3, phage NTUH-K2044-K1-1; lane 4, phage KP34. ▼: solid arrow indicate major protein band; ∇: blank arrows show minor protein bands.
The predominant protein band at 37 kDa on the SDS gel are suggestive of major capsid protein, as it was shown earlier for phage KP34 [5]. The major capsid protein has at least 93% of sequence homology in phages KP34, F19 and NTUH-K2044-K1-1, which are all members of the proposed “Kp34likevirus” genus. The amino acid similarities of these capsid proteins, corresponding to the 37.7 kDa bands, compared to the homologous protein of phage φKMV are all less than 37%. In addition, other observed proteins bands could be correlated with KP34 structural proteins: the head-tail connector protein, the tail tubular proteins A and B, the internal virion protein B, the internal core protein, and the tail fiber protein.
3.4. Genome Analysis
Comparison of the phages within the suggested “Kp34likevirus” genus show high pairwise nucleotide identity, similar G+C content and genome size. Coding sequences (CDSs) in the described phages vary from 52 CDS in phage F19 to 57 CDS in phage KP34 (Table 2). SU503 has a genome size of 43,809 nucleotides and a G+C content of 53.7%. SU503 has a total of 55 predicted CDSs where 50 start with ATG start codons, two with TTG and three with GTG. SU552A has 56 predicted CDSs, with 51 ATG, three TTG and two GTG start codons. All phages within the proposed “Kp34likevirus” genus show large pairwise identity to each other (Table 2).

Table 2.
Genome information and pairwise identity of the members of “Kp34likevirus”.
| Phage | Size (bp) | G+C content, % | CDS | Nucleotide pairwise identity, % | ||||
|---|---|---|---|---|---|---|---|---|
| KP34 | SU503 | SU552A | NTUH-K2044-K1-1 | F19 | ||||
| KP34 | 43,809 | 54.1 | 57 | - | 78.6 | 79.3 | 77.5 | 77.5 |
| SU503 | 43,809 | 53.7 | 55 | 78.6 | - | 75.1 | 78.1 | 76.8 |
| SU552A | 43,594 | 54.2 | 56 | 79.3 | 75.1 | - | 76.4 | 76.6 |
| NTUH-K2044-K1-1 | 43,871 | 54.2 | 54 | 77.5 | 78.1 | 76.4 | - | 76.1 |
| F19 | 43,766 | 53.8 | 52 | 77.5 | 76.8 | 76.6 | 76.1 | - |
Host transcription of the phage genomes starts at predicted sigma 70 promoter sequences, located upstream from the first predicted operon (Table 3). A putative phage-specific promoter was discovered on two locations in the phage genomes, corresponding to the early-middle genes in the phage genomes. This conserved motif shows high conservation across all “Kp34likevirus” member phages (Table 3), and is placed in intra genic regions of all phages. Since no promoter sequence have been identified before the late genes, this conserved motif cannot be solely responsible for phage specific transcription initiation. An alternative function of this sequence motif could be the location of single-stranded nicks that have been found in Phikmvlikevirus relative phage φkF77 [36]. An Rho-independent transcription terminator is located downstream of the conserved motif, located in between the genes corresponding to the structural genes of KP34, genes KP-KP34p43 and KP-KP34p44. It shares this terminator with the other phages of “Kp34likevirus” (Table 3). Sequences encoding tRNAs were not found in the genomes.

Table 3.
Regulatory sequences of the phages in the suggested “Kp34likevirus” genus. Conserved motifs are presented in bold while underlined nucleotides depict loops.
| Host Promoter | ||
|---|---|---|
| Phage | location | nucleotide sequence |
| KP34 | 931..959 | TTGACACCGCGAAGAACATAAG |
| SU503 | 1040..1068 | TTGACACCGCGAAGGACATAAGCTAGATT |
| 1169..1196 | TTAAAATAAACGCTTGACAAGTTATGAT | |
| 1182..1210 | TTGACAAGTTATGATTCACTGAGTAACTT | |
| SU552A | 683..710 | TTGCCCTGCTTACCATTTTTGCTATAAG |
| 874..902 | TTGACACCGCGAAGAACATAAGCTAGATT | |
| 943..971 | TTGACACCGCGAAGAACATAAGCTAGATT | |
| NTUH-K2044-K1-1 | 953..981 | TTGACACCGCGAAGGACATAAGCTAGATT |
| 1022..1050 | TTGACAAGTTCTGATTCACTGAGTAACTT | |
| 1251..1280 | CTCACAGGTTAGCAGTCCTGAGCCGATAAG | |
| F19 | 968..996 | TTGACACCGCGAAGGACATAAGCTAGATT |
| 1051..1079 | TTGACAAGTTCCGATTCACTGAGTAACTT | |
| 1192..1220 | ACGACAAACGGCGGGTGCGCTTAGATGAT | |
| E. coli consensus sequence | TTGACA-(N15-18)-TATAAT | |
| Putative phage promoters | ||
| Phage | location | nucleotide sequence |
| KP34 | 1487..1538 | CACTAATTACAGCCTATAGCATCCTACGGGGTGCTATGTGAAGTAATTACCT |
| 2508..2559 | TTTAGTAGCAAGCCTATAGCGTCCTATGGGGCGCTATGTGAATGCAACTGGC | |
| SU503 | 2010..2061 | CGTTAATTACAGCCTATAGCATCCTACGGGGTGCTATGTGAAGTAATTACCT |
| 3192..3243 | TCCAGTAGCAAGCCTATAGCGTCCTACGGGGCGCTATGTGAATGCAACTGGC | |
| SU552A | 1449..1500 | CGTTAATTACAGCCTATAGCATCCTACGGGGTGCTATGTGAAGTAATTACCT |
| 2225..2276 | TATAGTAGCAAGCCTATAGCGTCCTACGGGGCGCTATGTGAATGCAACTAGC | |
| NTUH-K2044-K1-1 | 1752..1803 | TACTAATTACAGCCTATAGCATCCTATGGGGTGCTATGTGAAGTAATTACCT |
| 2618..2669 | TCTAGTAGCAAGCCTATAGCGTCCTACGGGGCGCTATGTGAATGCAACCGGC | |
| F19 | 1795..1846 | CGTTAATTACAGCCTATAGCATCCTATGGGGTGCTATGTGAAGTAATTACAT |
| 2571..2622 | TATAGTAGCAAGCCTATAGCGTCCGACTGGGCGCTATGTGAATGCAACTAGC | |
| Phage promoter consensus | AGCCTATAGCGTCCTACGGGGCGCTATGTGAA | |
| Rho-independent terminators | ||
| Phage | location | nucleotide sequence |
| KP34 | 26623..26660 | GCCCCTGGTGCCTTCGGGTGCCAGGGGCTTTTTTTTTT |
| SU503 | 26936..26972 | GCCCCTGGTGCCTTCGGGTGCCAGGGGCTTTTTTTTT |
| SU552A | 25384..25420 | GCCCCTGGTGCCTTCGGGTGCCAGGGGCTTTTTTTTT |
| NTUH-K2044-K1-1 | 25925..25961 | GCCCCTGGTGCCTTCTGGTGCCGGGGGCTTTTTTTTT |
| F19 | 26363..26400 | GCCCCTGGTGCCTTCGGGTGCCAGGGGCTTTTTTTTTT |
3.5. Comparative Genomics
A total of seven conserved proteins were identified, using CoreGenes 3.5 (Figure 3) [25], among phages within the subfamily of Autographivirinae. These included the DNA-dependent RNA polymerase, DNA maturase A and B, head-tail connector protein, the tail tubular protein A and B and the major capsid protein gene.
Between phages of the genera Phikmvlikevirus and phages from the suggested “Kp34likevirus” there are 18 conserved proteins. A total of 29 genes are conserved between the phages within the “Kp34likevirus” genus (Table 4).
To investigate the evolutionary relationship between phages from the “Kp34likevirus”, Phikmvlikevirus, and the more distant Sp6likevirus and T7likeviruses within the Autographivirinae subfamily, phylogenetic analyses were performed on the amino acid residues of the RNA polymerase, head-tail connector protein and the DNA maturase B protein (Figure 4). In all three phylograms the phages of the proposed “K34likevirus” cluster into a single monophyletic clade. This division is also conserved when analyzing the other conserved proteins from phages of the Autographivirinae subfamily, the major capsid protein, tail tubular protein A and B and DNA maturase A. The whole genome nucleotide identity against the two phages in the sister group, consisting of the Vibrio phage VP93 and the Pantoea phage LIMElight, was shown to be only 9% and 15% respectively.
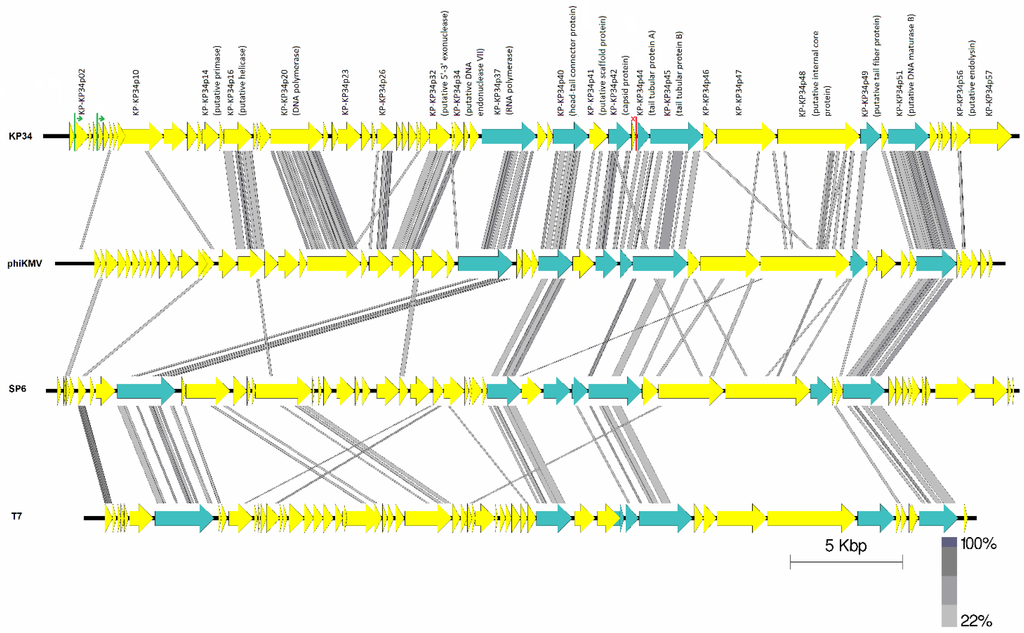
Figure 3.
Pairwise comparison using tblastx of the four type phages KP34, φKMV, SP6 and T7. Lines between genomes represent high amino acid similarity between the phages using tblastx analysis. Blue arrows denotes conserved genes across the Autographivirinae subfamily. Green lines show putative phage specific promoter sites and red rho-independent terminator in phage KP34.

Table 4.
Conserved genes among phages within the “Kp34likevirus” and their amino acid pairwise identity to phage KP34. Conserved genes between KP34 and φKMV shown in bold. a: a.a. query coverage range from 84.7%–85.04%. b: a.a. query coverage range from 91.31%–99.06%. c: a.a. query coverage range from 50.0%–76.43%.
| No. | Description | KP34 Locus_Tag | Accession no. | % Pairwise AA identity to KP34 | |||
|---|---|---|---|---|---|---|---|
| SU503 | SU552A | K2044 | F19 | ||||
| 1 | hypothetical protein | KP-KP34p02 | GI:282554636 | 96.8 | 97.9 | 94.1 | 97.9 |
| 2 | hypothetical protein | KP-KP34p05 | GI:294661414 | 63.0 | 65.8 | 64.4 | 65.8 |
| 3 | hypothetical protein | KP-KP34p10 | GI:282554643 | 84.0 a | 89.0 a | 88.3 a | 87.9 a |
| 4 | putative peptidase | KP-KP34p11 | GI:282554645 | 87.4 | 97.1 | 98.0 | 98.0 |
| 5 | hypothetical protein | KP-KP34p12 | GI:282554646 | 68.8 | 70.5 | 62.7 | 67.1 |
| 6 | putative DNA primase | KP-KP34p14 | GI:282554648 | 93.6 | 95.8 | 95.0 | 96.5 |
| 7 | putative DNA helicase | KP-KP34p16 | GI:294661415 | 96.0 b | 98.6 | 97.7 | 97.7 b |
| 8 | DNA polymerase | KP-KP34p20 | GI:282554654 | 81.5 | 84.1 | 81.8 | 80.8 |
| 9 | hypothetical protein | KP-KP34p23 | GI:282554657 | 88.1 | 81.9 | 85.8 | 72.9 |
| 10 | hypothetical protein | KP-KP34p26 | GI:282554660 | 96.5 | 98.7 | 96.5 | 95.8 |
| 11 | hypothetical protein | KP-KP34p29 | GI:282554662 | 78.5 | 83.5 | 91.8 | 86.0 |
| 12 | putative 5'-3' exonuclease | KP-KP34p32 | GI:282554665 | 93.5 | 92.2 | 91.9 | 92.5 |
| 13 | putative DNA endo- nuclease VII | KP-KP34p34 | GI:294661421 | 99.3 | 94.3 c | 51.4 c | 98.6 |
| 14 | DNA-dependent RNA polymerase | KP-KP34p37 | GI:282554612 | 96.8 | 96.4 | 98.4 | 94.6 |
| 15 | hypothetical protein | KP-KP34p38 | GI:282554613 | 97.9 | 90.4 | 89.7 | 91.1 |
| 16 | head-tail connector protein | KP-KP34p40 | GI:294661422 | 98.9 | 98.9 | 99.2 | 98.3 |
| 17 | putative scaffolding protein | KP-KP34p41 | GI:282554617 | 98.2 | 98.2 | 97.9 | 97.5 |
| 18 | capsid protein | KP-KP34p42 | GI:282554619 | 92.9 | 93 | 93.5 | 92.7 |
| 19 | tail tubular protein A | KP-KP34p44 | GI:282554621 | 87.9 | 91.9 | 97.7 | 92.2 |
| 20 | tail tubular protein B | KP-KP34p45 | GI:282554622 | 99.0 | 98.0 | 96.2 | 88.4 |
| 21 | putative internal virion protein B | KP-KP34p46 | GI:282554623 | 75.4 | 98.5 | 75.9 | 99.0 |
| 22 | hypothetical protein | KP-KP34p47 | GI:282554624 | 96.1 | 95.0 | 92.6 | 88.0 |
| 23 | putative internal core protein | KP-KP34p48 | GI:282554625 | 97.4 | 97.9 | 89.0 | 70.2 |
| 24 | putative tail fiber protein | KP-KP34p49 | GI:282554626 | 93.8 | 65.1 | 90.3 | 92.5 |
| 25 | putative DNA maturase A | KP-KP34p50 | GI:282554627 | 97.0 | 100 | 100 | 100 |
| 26 | putative DNA maturase B | KP-KP34p51 | GI:282554628 | 98.7 | 98.9 | 98.1 | 98.9 |
| 27 | hypothetical protein | KP-KP34p52 | GI:282554629 | 97.4 | 98.4 | 98.4 | 97.6 |
| 28 | hypothetical protein (spanin) | KP-KP34p54 | GI:282554631 | 98.5 | 97.8 | 97.0 | 96.3 |
| 29 | putative endolysin | KP-KP34p56 | GI:282554633 | 90.6 | 96.0 | 93.3 | 89.8 |
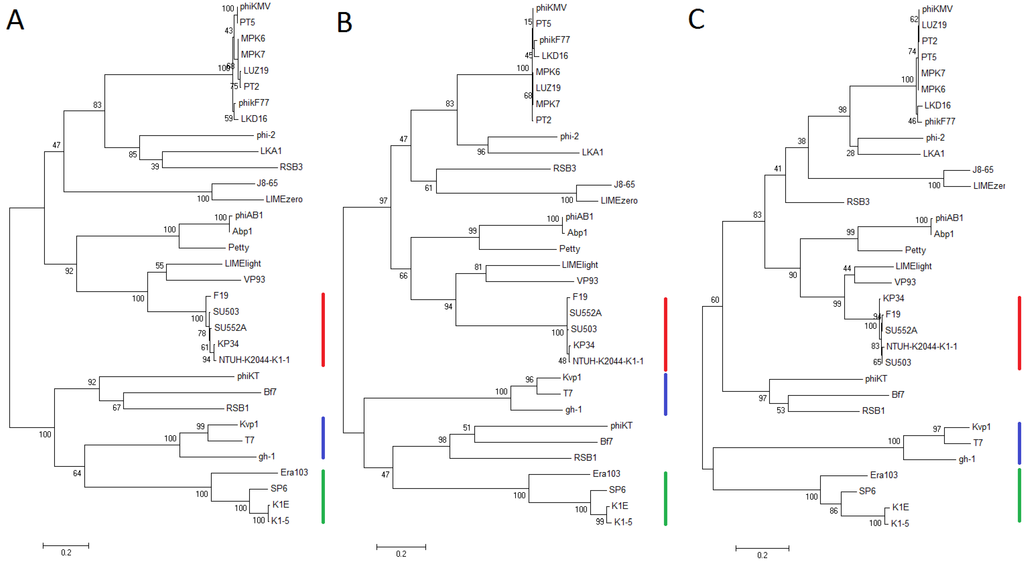
Figure 4.
The phylogenetic analyses of amino acid sequences, inferred from three genes from 33 phages in the Autographivirinae subfamily, were performed by using the maximum likelihood (ML) method based on the Whelan and Goldman model [30], with 500 bootstrap replicates. The bootstrap percentages are shown next to each node. Initial trees for the searches were autogenerated by the maximum parsimony (MP) method. The following ML searches were performed using uniform substitution rates among sites and the Nearest-Neighbor-Interchange (NNI) heuristic search method. Positions containing gaps and missing data were excluded. The trees with the highest likelihoods are drawn to scale, with branch lengths measured in the number of substitutions per site. Red lines indicate the “Kp34likevirus” cluster, blue T7likevirus members and green SP6likevirus members. Phikmvlikevirus members’ φKMV, LKA1, LIMElight and LIMEzero are scattered in the upper clade of the phylograms. The analyses were conducted in MEGA6 [28]. (a) The RNA polymerase tree (log likelihood = −26580.7487); (b) The head-tail connector protein tree (log likelihood = −12447.9348); (c) The DNA maturase B (terminase) tree (log likelihood = −10492.0745).
3.6. RNA Polymerase
Three structurally important areas in the DNA-dependent RNA polymerase protein sequence were analyzed, the catalytically essential residues Asp537, Lys631, Tyr639 and Asp812, the recognition loop between amino acids 93 to 101 and the specificity loop between amino acids 739 to 770 [4,37,38]. All catalytically essential amino acid residues are conserved within all RNA polymerases, both in φKMV and KP34-like viruses, but there is a large divergence of the recognition loop, where the residues are identical amongst the KP34-likeviruses (Table 5). The specificity loop shows conservation within the “Kp34likevirus”, with substitutions in residues 751 to 755. Phages NTUH-K2044-K1-1 and KP34 have an identical specificity loop and phages F19, SU503 and SU552A are identical to each other.

Table 5.
Alignment of the recognition and specificity loops of the RNA polymerase in φKMV, “Kp34likevirus”. Phages VP93 (NC_012662) and LIMElight (NC_019454), which assembled close to the “Kp34likevirus” members in the phylogenetic analysis are added for comparison. Underlined peptides show sites of substitutions compared to KP34.
| Phage | Recognition loop | Specificity loop |
|---|---|---|
| φKMV | HQEAKAAKPAAKL | EEVRVRLRAEAVEYVTLYEAK-DEL |
| KP34 | MRNVKAPGIGGKY | EEVRVRIDCMNLSAVLVHNRDFKTC |
| K2044 | MRNVKAPGIGGKY | EEVRVRIDCMNLSAVLVHNRDFKTC |
| F19 | MRNVKAPGIGGKY | EEVRVRIDCMNLTIMRVHNRDFKTC |
| SU503 | MRNVKAPGIGGKY | EEVRVRIDCMNLTIMRVHNRDFKTC |
| SU552A | MRNVKAPGIGGKY | EEVRVRIDCMNLTIMRVHNRDFKTC |
| LIMElight | IKAEKAPGVGGKY | EEKRVNIRSMGLTQVVAYNRNYDLN |
| VP93 | LKASKTRGVGAKY | HETRVKVRSMGINQVVLYNFDYERN |
3.7. Lysis Cassette
Through genomic analysis of all five “Kp34likevirus” members: KP34, SU503, SU552A, NTUH-K2044-K1-1 and F19, a total of three proteins involved in the host lysis have been identified. These enzymes act jointly in the sequential order: holin, endolysin and spanin. The holins are small, hydrophobic proteins, able to form pores in the inner membrane allowing endolysins (peptidoglycan hydrolases) to gain access to the periplasm and attack the bonds in the murein structure [39,40,41,42]. Instead of a classic holin, bacteriophages may encode a variant, the pinholin, capable of forming small pores with a diameter too narrow to transfer endolysins but wide enough for ion movement and membrane depolarization, thereby for signal arrest release (SAR) or signal peptide (SP) endolysin activation [40,43]. The spanins acts at the end of the lysis process leading to the destruction of the outer membrane and phage progeny release.
The mentioned lysis proteins have been identified and characterized through in silico analysis of the “Kp34likevirus” genomes. At the outset, it is worth noting that the amino acid sequences of these enzymes are highly similar, over 95%. In silico analysis of each genome revealed holin genes with characteristic features; (i) a small size of about 83 residues; (ii) positively charged hydrophilic domain at the C-terminus; (iii) one transmembrane domain of class III holins, (iv) a holin gene adjacent to the endolysin gene. The analysis of the endolysins genes in the investigated phages indicates that these enzymes possess a SAR domain at the N-terminus. The SAR signal identification follows the presence of two consecutive sections; positively charged initial section, containing lysine or arginine (or both), and a hydrophobic H-region [40]. Both elements have been identified for the analyzed endolysins encoded by the KP34-like viruses. The catalytic domain of these endolysins with lysozyme activity is located directly behind the H-region. The presence of a SAR signal within the endolysin gene indicates that the described holins are in fact pinholins responsible for the endolysin activation. Analysed spanins belong to the u-spanin group and are represented by a single protein with two transmembrane domains arranged at distal parts of the enzyme. Spanins may exist as an i-spanin/o-spanin complex of two proteins such as Rz and Rz1, or as a single acting unimolecular protein called u-spanin [44,45].
The genes involved in lysis of KP34-like viruses are located next to each other (successively: spanin –holin–endolysin) and thereby form a “lysis cassette” (Figure 5A). The presence and organization of a lysis cassette that encodes a holin, endolysin, Rz and Rz1, in this particular order, has been previously described for phage φKMV (Figure 5B) [40]. However, there are major differences between the lysis cassette of φKMV and “Kp34likevirus”. These differences relate to the amino acid sequences, length, gene organization, and spanin construction. In contrast to φKMV, the lysis cassette of KP34-like viruses is formed from a unimolecular spanin, holin and endolysin, respectively.
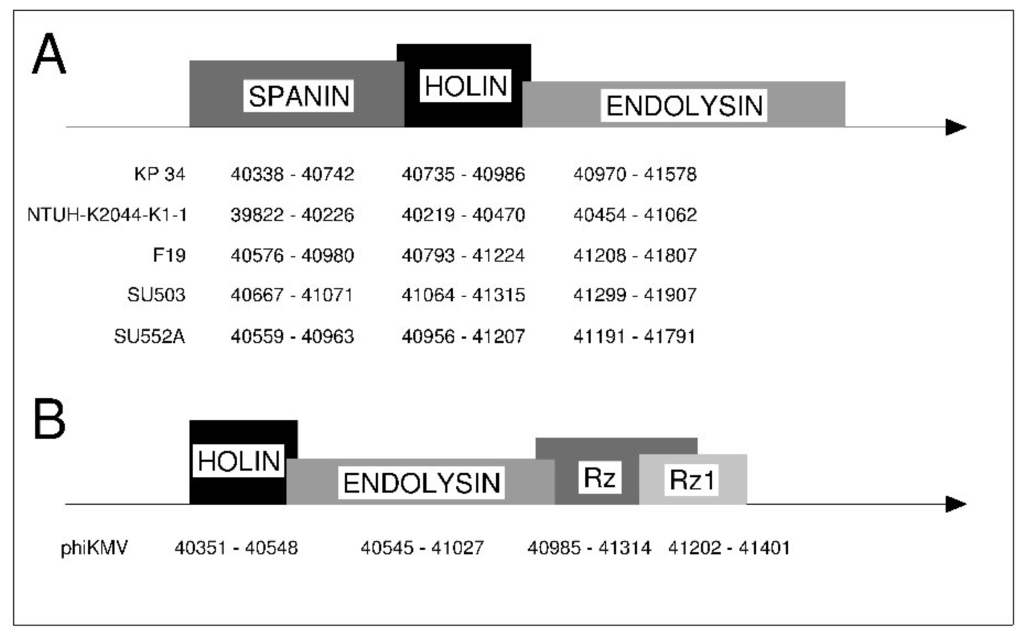
Figure 5.
Lysis cassette scheme in the analyzed (A) “Kp34likevirus” and (B) φKMV.
Compared to KP34, the amino acid sequence similarity of the lysis cassette is 96% (at a coverage 100%) for NTUH-K2044-K1-1, 93% (with 98% coverage) for F19, 93% (with 98% coverage) for SU503 and 97% (with 99% coverage) for SU552A, whereas the similarity of the φKMV to the KP34 lysis cassette is about 30% (22% coverage). The low similarity of lysis cassettes indicates a distant relationship between these genes in phage φKMV and the “Kp34likevirus” member phages.
4. Discussion
In this article we propose the creation of the new phage genus, “Kp34likevirus”, based on the high pairwise identity of the investigated phage genomes, the use of a single tail fiber gene, the highly conserved RNA polymerase recognition sites and specificity loops, as well as conserved lysis cassette genes. The “Kp34likevirus” genus is suggested to encompass Klebsiella phages KP34, SU503, SU552A, F19 and NTUH-K2044-K1-1.
At nucleotide level, the pairwise identity of phages KP34, SU503, SU552A, NTUH-K2044-K1-1 and F19 range from between 75.1 to 79.3% (Table 2). The similarity between the “Kp34likevirus” phages is even higher when comparing the pairwise identity of the amino acids of the gene products, with few exceptions (Table 4) and highly conserved order of genes between each other in comparison to φKMV. All phages within the “Kp34likevirus” genus have different host ranges, shown experimentally, which suggest that they utilize different host receptors (Table S2).
In contrast to the phages in the Phikmvlikevirus genus, the RNA polymerase responsible for transcribing the phage genes from phage specific promoters, show identical recognition loops between all the five phages, and a small variation of the specificity loop. The differences of the residues in the specificity loop divides them into two separate subgroups, with phage KP34 and NTUH-K2044-K1-1 in one group and phages SU503, SU552A and F19 in another (Table 5). The high identity of the specificity and recognition loops in their RNA polymerases suggest that the “Kp34likevirus” phages utilize a common promoter sequence, of which one has been predicted showing large conservation among the phages (Table 2).
Phylogenetic analysis of conserved genes places the “Kp34likevirus” phages in a monophyletic clade (Figure 4). In comparison to phages in the Sp6likevirus and T7likevirus genera, the division of phages into the Phikmvlikevirus and “Kp34likevirus” genera is a more recent occurrence in evolutionary terms from the Autographivirinae ancestor. Interestingly, the Phikmvlikevirus phage LIMElight and the unclassified phage VP93 closely affiliate to the suggested “Kp34likevirus” genus by phylogenetic analysis of the conserved genes, but while LIMElight shares the KP34 feature of a single tail fiber gene, it does not show any homology to the KP34 tail fiber genes, but shows instead larger homology to SP6 tail fiber genes. This has previously been suggested to be due to lateral acquisition of the tail fiber gene in this phage [46]. LIMElight has a slightly “Kp34likevirus”-like conserved recognition loop, and lesser conserved specificity loop of its RNA polymerase, and also a lysis cassette similar to the “Kp34likevirus” phages. The low nucleotide identity of LIMElight to KP34, 15%, also argues a distant relationship between the two phages.
Vibrio phage VP93, on the other hand, shows a clear divergence from the phages in “Kp34likevirus”, with dual tail fiber genes, no identifiable holin or endolysin, and no similarity in the RNA polymerase specificity and recognition loops (Table 5). The only commonality with the “Kp34likevirus” is the overall peptide similarity of the conserved genes across Autographivirinae (Figure 2). DNA pairwise identity of VP93 to KP34 is 9%. In depth analysis of the VP93 proteome using HHpred indicates that VPP93_gp41 encodes a protein structurally similar (E-value = 5.6 × 10−25) to a muramoyl-pentapeptide carboxypeptidase from Streptomyces albus (RCSB Protein Data Bank accession number: 1lbu). This protein shows weak similarity to gp46 (NP_853606) of Enterobacteria phage SP6.
The organization of the “Kp34likevirus” lysis cassette is based on spanin, holin and endolysin genes located sequentially in contrast to the phage φKMV lysis system (holin-endolysin-spanins sequence) (Figure 5). The differences were also detected in spanin structure where an i-spanin/o-spanin complex of two proteins is present in φKMV phage and u-spanin (unimolecular) exists in “Kp34likevirus” phages.
Taken together, the high nucleotide pairwise identity and amino acid similarity, promoter specificity and lysis cassette organization argues for the creation of the new Autographivirinae genus named “Kp34likevirus”, composed of phages KP34, SU503, SU552A, NTUH-K2044-K1-1 and F19. The phages belonging to this suggested genus have been collected from two different continents, and display a high degree of conservation. This genus will be proposed to the International Committee on Taxonomy of Viruses (ICTV) for ratification.
Supplementary Files
Supplementary File 1Acknowledgments
This work was partly funded by the foundation Olle Engqvist byggmästare (ASN). All 07RAFM Klebsiella pneumoniae isolates in Table S1 were kindly received by Barbro Olsson Liljequist from the Public Health Agency of Sweden (Folkhälsomyndigheten). BM project “Preparation of highly purified, recombinant endolysins encoded by Klebsiella phages and determination of their antibacterial activity and effect on human cells (research task: in silico analysis of the phage enzymes)” is carried-out within the VENTURES program of the Foundation for Polish Science, co-financed from the European Union under the European Regional Development Fund. Agnieszka Latka was co-financed by the European Union as part of the European Social Fund. The authors thank Sylwia Nowak (UWR) for excellent technical assistance.
Author Contributions
Study conception and design: H.E., Z.D.-K., A.M.K.; Acquisition of data: H.E., J.-T.W., B.M., A.L., G.M.-S., M.H., O.M.; Drafting of manuscript: H.E., A.M.K., Z.D.-K., A.S.N.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lavigne, R.; Seto, D.; Mahadevan, P.; Ackermann, H.W.; Kropinski, A.M. Unifying classical and molecular taxonomic classification: Analysis of the Podoviridae using blastp-based tools. Res. Microbiol. 2008, 159, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.J.; Studier, F.W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 1983, 166, 477–535. [Google Scholar] [CrossRef] [PubMed]
- Dobbins, A.T.; George, M., Jr.; Basham, D.A.; Ford, M.E.; Houtz, J.M.; Pedulla, M.L.; Lawrence, J.G.; Hatfull, G.F.; Hendrix, R.W. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J. Bacteriol. 2004, 186, 1933–1944. [Google Scholar] [CrossRef] [PubMed]
- Ceyssens, P.J.; Lavigne, R.; Mattheus, W.; Chibeu, A.; Hertveldt, K.; Mast, J.; Robben, J.; Volckaert, G. Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: Establishment of the φKMV subgroup within the T7 supergroup. J. Bacteriol. 2006, 188, 6924–6931. [Google Scholar] [CrossRef] [PubMed]
- Drulis-Kawa, Z.; Mackiewicz, P.; Kesik-Szeloch, A.; Maciaszczyk-Dziubinska, E.; Weber-Dabrowska, B.; Dorotkiewicz-Jach, A.; Augustyniak, D.; Majkowska-Skrobek, G.; Bocer, T.; Empel, J.; et al. Isolation and characterisation of KP34—A novel φKMV-like bacteriophage for Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2011, 90, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.L.; Hsieh, P.F.; Huang, Y.T.; Lee, W.C.; Tsai, Y.T.; Su, P.A.; Pan, Y.J.; Hsu, C.R.; Wu, M.C.; Wang, J.T. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: Implication in typing and treatment. J. Infect. Dis. 2014, 210, 1734–1744. [Google Scholar] [CrossRef] [PubMed]
- Podschun, R.; Ullmann, U. Klebsiella spp. As nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [PubMed]
- Pieroni, P.; Rennie, R.P.; Ziola, B.; Deneer, H.G. The use of bacteriophages to differentiate serologically cross-reactive isolates of Klebsiella pneumoniae. J. Med. Microbiol. 1994, 41, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Brolund, A.; Haeggman, S.; Edquist, P.J.; Gezelius, L.; Olsson-Liljequist, B.; Wisell, K.T.; Giske, C.G. The diversilab system versus pulsed-field gel electrophoresis: Characterisation of extended spectrum beta-lactamase producing Escherichia coli and Klebsiella pneumoniae. J. Microbiol. Methods 2010, 83, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning—A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Klebsiella Phage F19, Complete Genome. Available online: http://www.ncbi.nlm.nih.gov/nuccore/NC_023567.2 (accessed on 2 April 2015).
- Pajunen, M.; Kiljunen, S.; Skurnik, M. Bacteriophage φyeo3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 2000, 182, 5114–5120. [Google Scholar] [CrossRef] [PubMed]
- Kesik-Szeloch, A.; Drulis-Kawa, Z.; Weber-Dabrowska, B.; Kassner, J.; Majkowska-Skrobek, G.; Augustyniak, D.; Lusiak-Szelachowska, M.; Zaczek, M.; Gorski, A.; Kropinski, A.M. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol. J. 2013, 10, e100. [Google Scholar] [CrossRef]
- Kutter, E. Phage host range and efficiency of plating. In Bacteriophages: Methods and Protocols Volume 1—Isolation, Characterization and Interactions, 1st ed.; Martha, R.J.C., Andrew, K., Eds.; Humana Press: New York, NY, USA, 2009; pp. 141–149. [Google Scholar]
- Boulanger, P. Purification of bacteriophages and SDS-PAGE analysis of phage structural proteins from ghost particles. Methods Mol. Biol. 2009, 502, 227–238. [Google Scholar] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, e75. [Google Scholar] [CrossRef]
- Noguchi, H.; Taniguchi, T.; Itoh, T. MetaGeneAnnotator: Detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008, 15, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Palleja, A.; Harrington, E.D.; Bork, P. Large gene overlaps in prokaryotic genomes: Result of functional constraints or mispredictions? BMC Genomics 2008, 9, e335. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-se: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Gautheret, D.; Lambert, A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 2001, 313, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Macke, T.J.; Ecker, D.J.; Gutell, R.R.; Gautheret, D.; Case, D.A.; Sampath, R. RNAmotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001, 29, 4724–4735. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Turner, D.; Reynolds, D.; Seto, D.; Mahadevan, P. CoreGenes3.5: A webserver for the determination of core genes from sets of viral and small bacterial genomes. BMC Res. Notes 2013, 6, 140. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Bernsel, A.; Viklund, H.; Hennerdal, A.; Elofsson, A. TOPCONS: Consensus prediction of membrane protein topology. Nucleic Acids Res. 2009, 37, W465–W468. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H.; Park, B.H. An enzyme produced by a phage-host cell system. Ii. The properties of the polysaccharide depolymerase. Virology 1956, 2, 719–736. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. Classification of bacteriophages. In The Bacteriophages, 2nd ed.; Calendar, R., Ed.; Oxford University Press: New York, NY, USA, 2006; pp. 8–16. [Google Scholar]
- Kulakov, L.A.; Ksenzenko, V.N.; Shlyapnikov, M.G.; Kochetkov, V.V.; del Casale, A.; Allen, C.C.; Larkin, M.J.; Ceyssens, P.J.; Lavigne, R. Genomes of “φKMV-like viruses” of Pseudomonas aeruginosa contain localized single-strand interruptions. Virology 2009, 391, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, G.M.; Steitz, T.A. Structure of a transcribing T7 RNA polymerase initiation complex. Science 1999, 286, 2305–2309. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, G.M.; Steitz, T.A. Insights into transcription: Structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2000, 10, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Peeters, L.M.; Volckaert, G.; Lavigne, R. The lysis cassette of bacteriophage φKMV encodes a signal-arrest-release endolysin and a pinholin. Bacteriophage 2011, 1, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B.; Delattre, A.S.; Lavigne, R. Learning from bacteriophages—Advantages and limitations of phage and phage-encoded protein applications. Curr. Protein Pept. Sci. 2012, 13, 699–722. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Azeredo, J.; Lavigne, R.; Kluskens, L.D. Bacteriophage endolysins as a response to emerging foodborne pathogens. Trends Food Sci. Technol. 2012, 28, 103–115. [Google Scholar] [CrossRef]
- Pang, T.; Fleming, T.C.; Pogliano, K.; Young, R. Visualization of pinholin lesions in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, E2054–E2063. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.D.; Rajaure, M.; Young, R. Spanin function requires subunit homodimerization through intermolecular disulfide bonds. Mol. Microbiol. 2013, 88, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Young, R. Phage lysis: Three steps, three choices, one outcome. J. Microbiol. 2014, 52, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Ceyssens, P.J.; Dunon, V.; Ackermann, H.W.; van Vaerenbergh, J.; Maes, M.; de Proft, M.; Lavigne, R. Bacteriophages LIMElight and LIMEzero of Pantoea agglomerans, belonging to the “φKMV-like viruses”. Appl. Environ. Microbiol. 2011, 77, 3443–3450. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).