The Molecular Switch of Telomere Phages: High Binding Specificity of the PY54 Cro Lytic Repressor to a Single Operator Site
Abstract
:1. Introduction
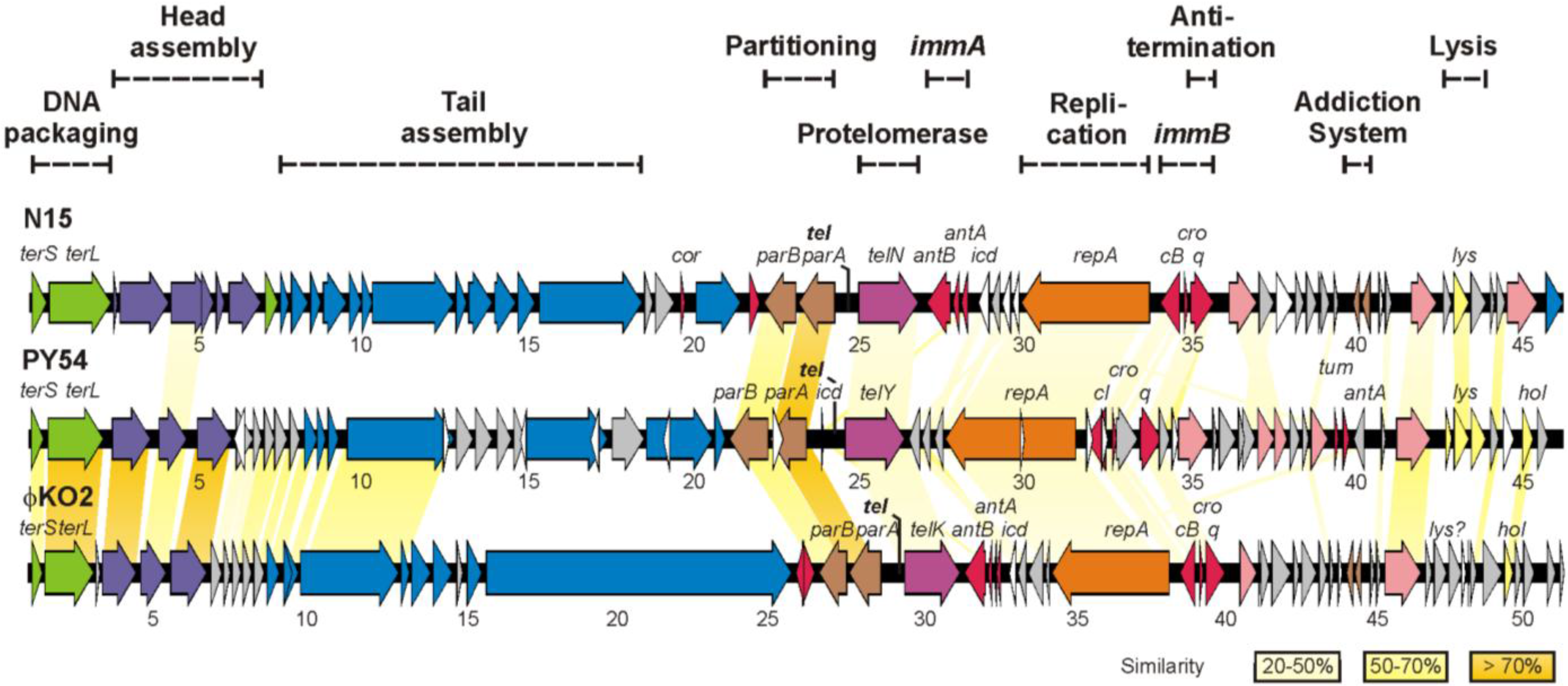
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Growth Conditions
2.2. Bacteriophages and Construction of Phage Mutants
2.3. In Vivo Assay for the PY54 Cro Repressor Activity
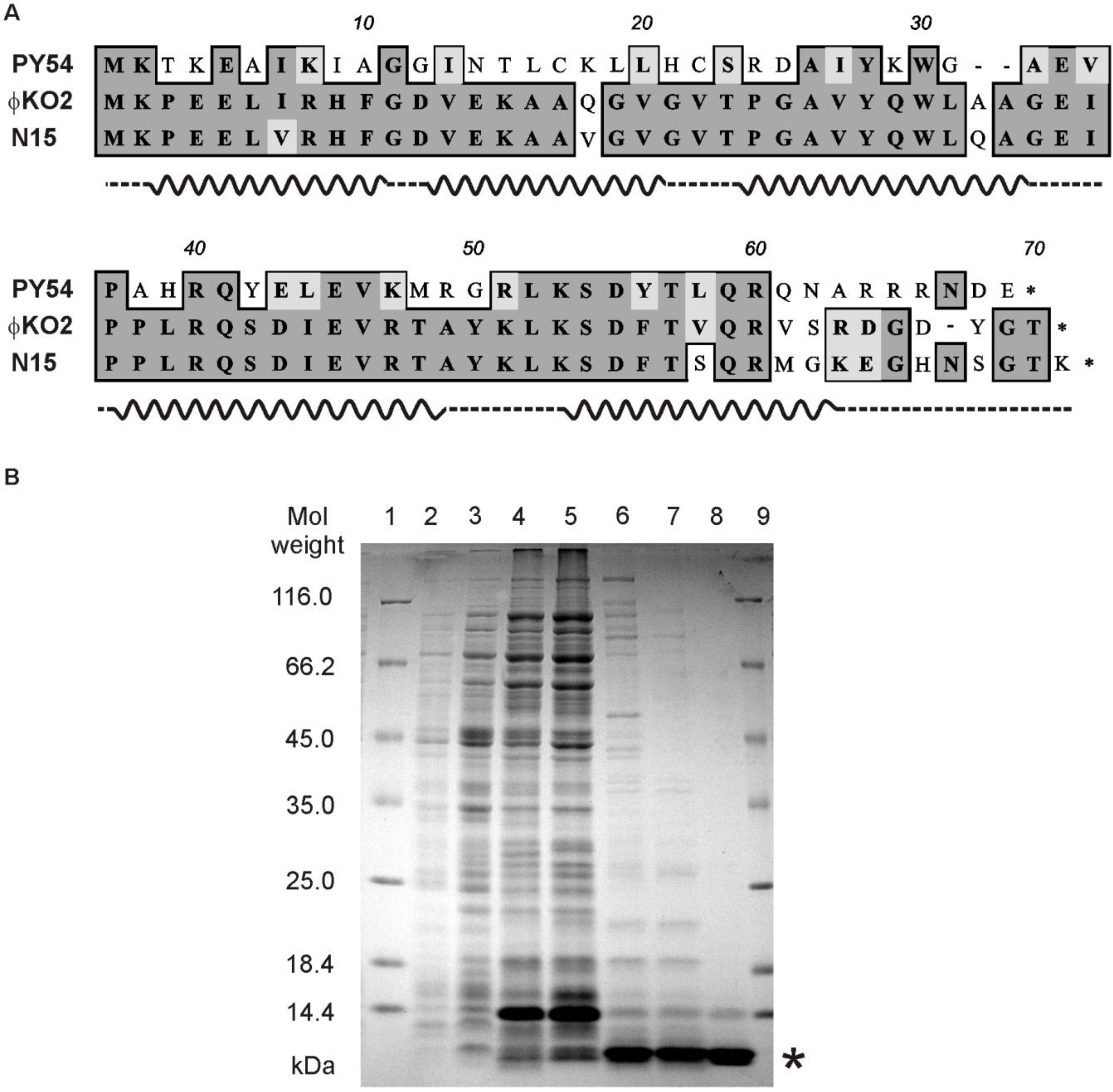
2.4. Overproduction and Purification of the Repressor Protein
2.5. SDS-PAGE and Mass Spectrometric Analysis
2.6. Electrophoretic Mobility Shift Assays (EMSA)
2.7. In Silico Analyses
2.8. Analysis of Promoter Activity
2.9. Determination of the cI and cro Transcription Start Sites
3. Results
3.1. croPY54 Triggers PY54 Induced Lysis but Does Not Affect N15 Propagation
3.2. The Purified cro Product of PY54 Shows Binding to a Specific Target Sequence Within immB
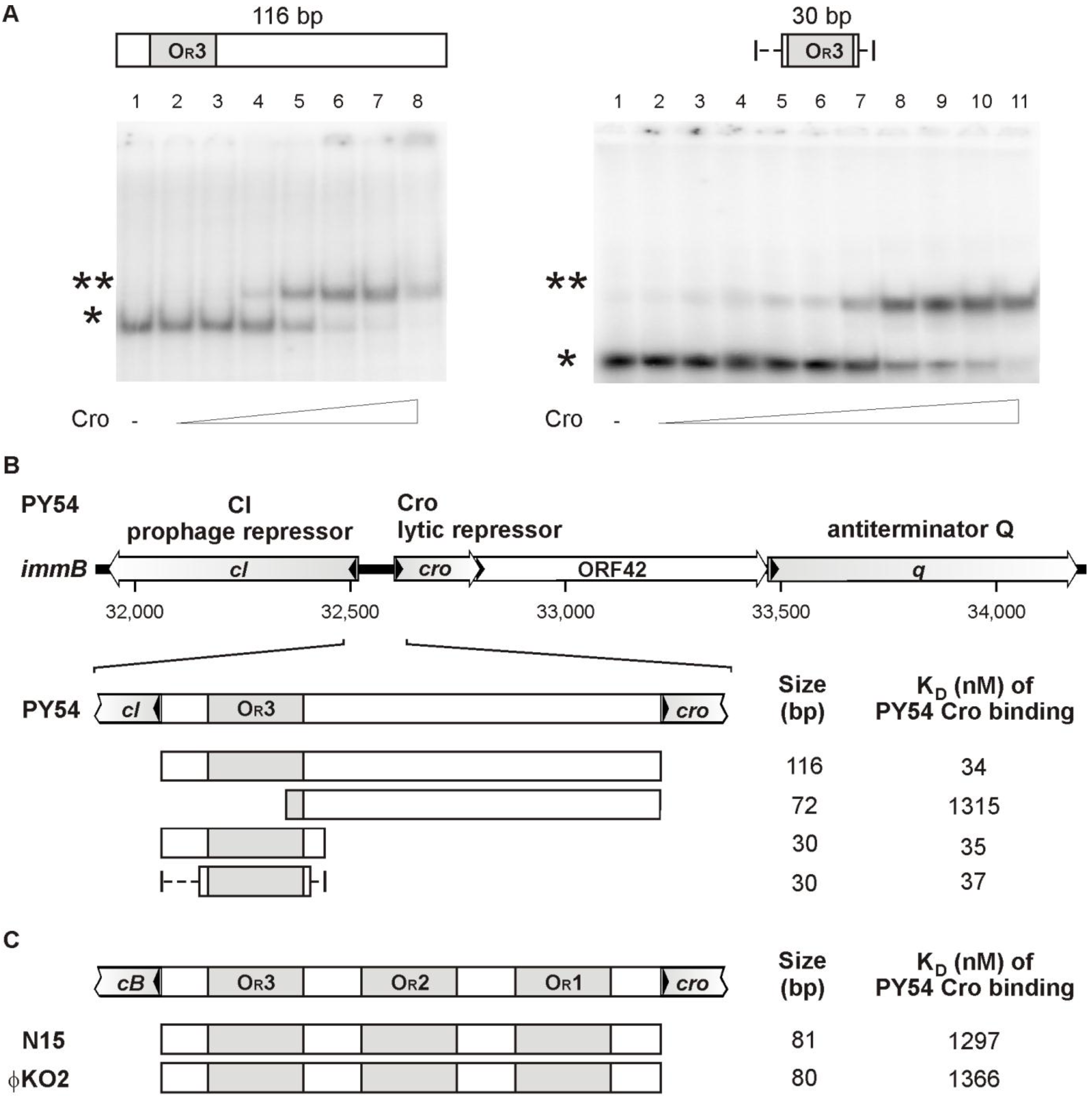
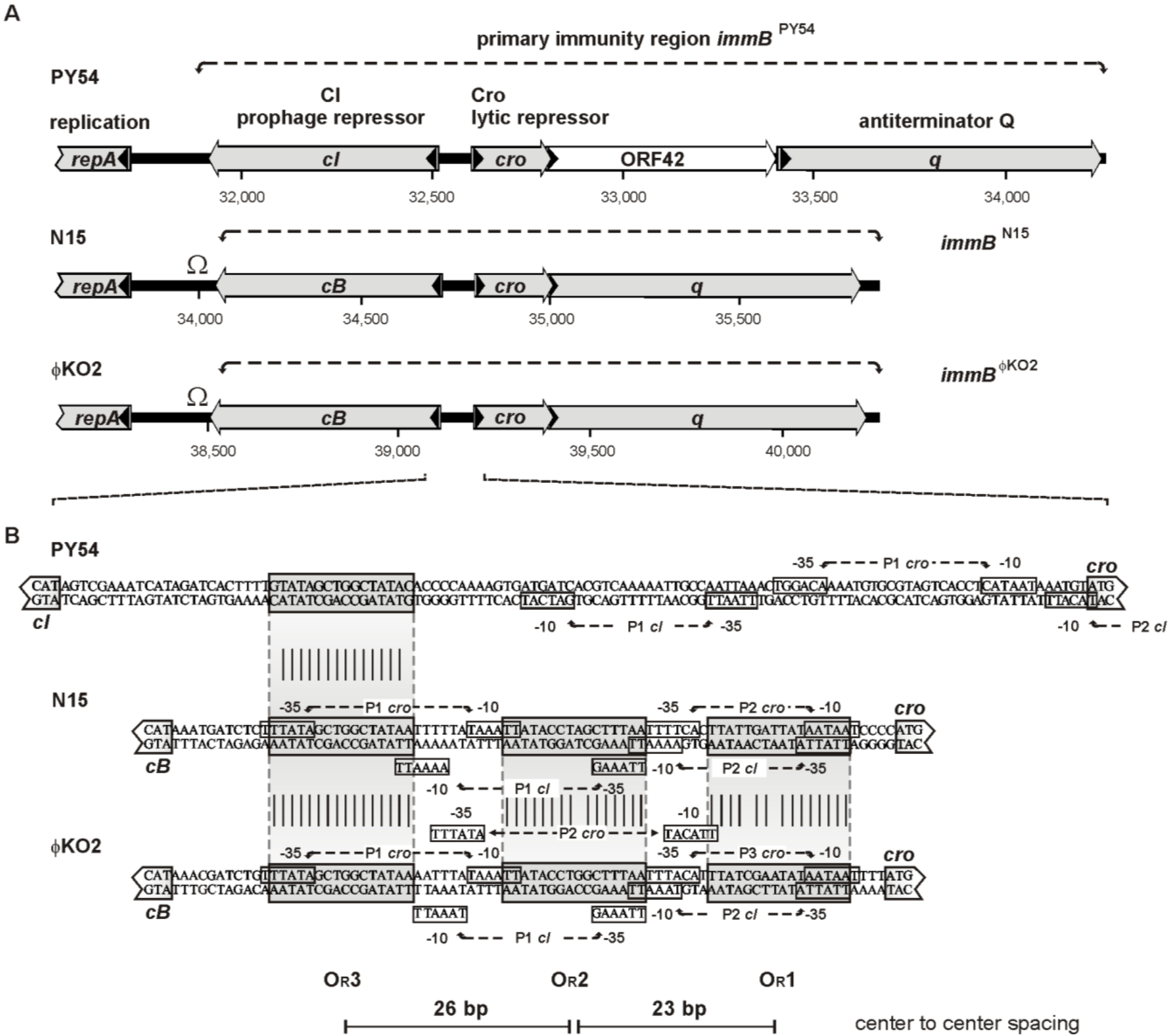
3.3. The Cro Repressor Binds to a Single Site Upstream of the Prophage Repressor Gene
3.4. The Central 16 bp of OR3 Are the Target of Cro
3.5. Both Peripheral Base Pairs of OR3 Determine Phage Specificity

3.6. Promoters of the Repressor Genes Center on the cro Region
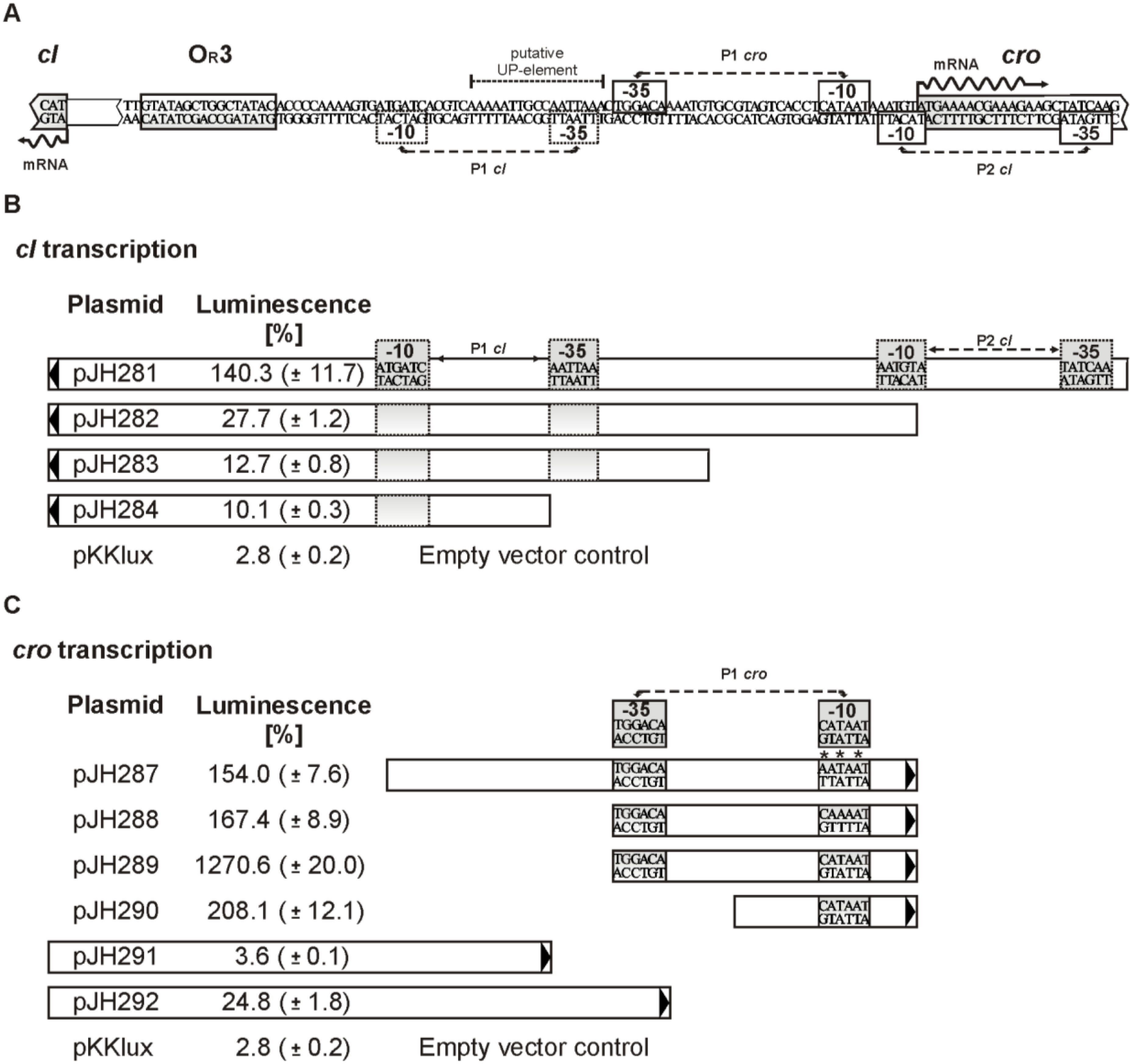
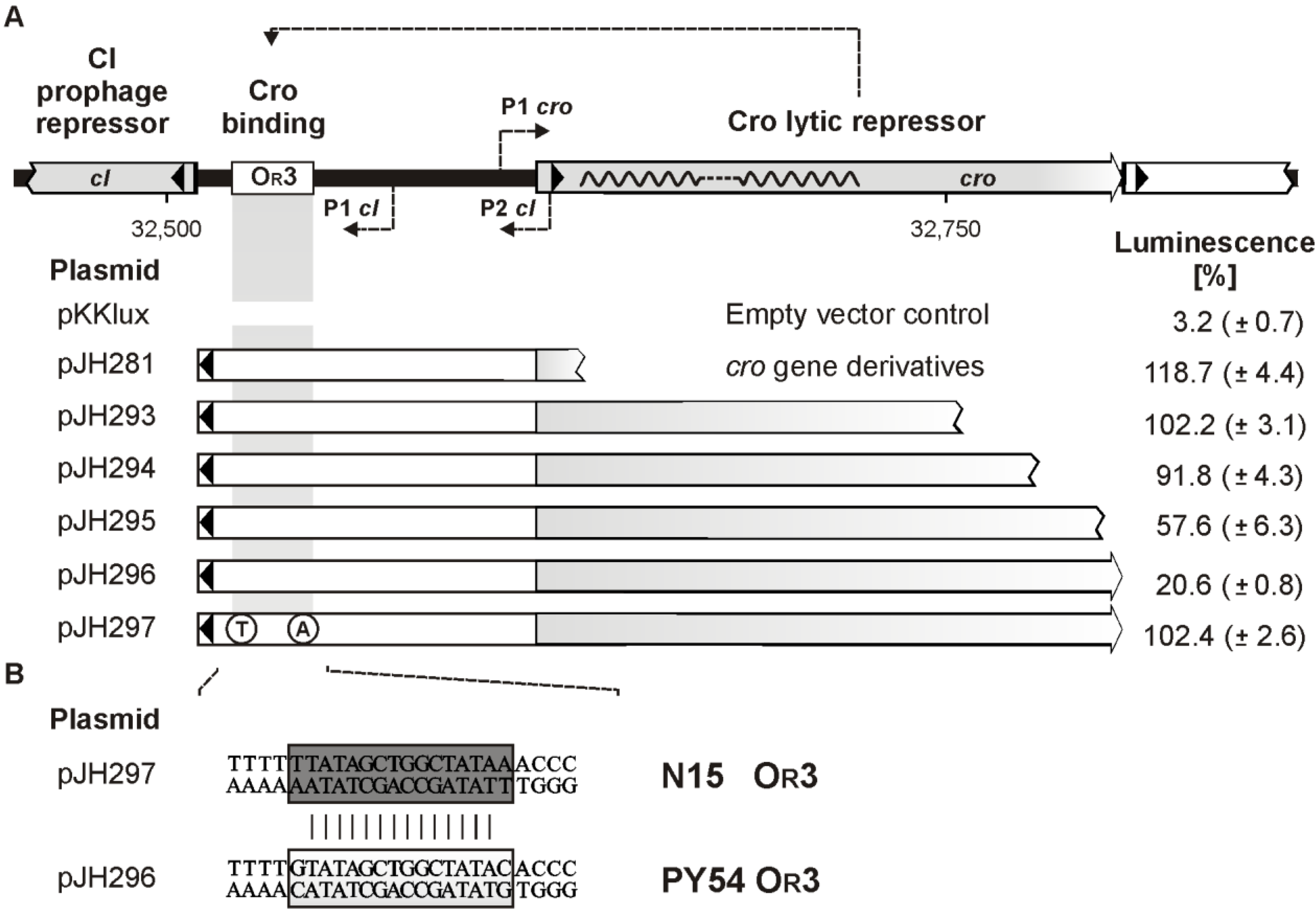
3.7. The C-Terminal Region of Cro is Essential for OR3 Binding
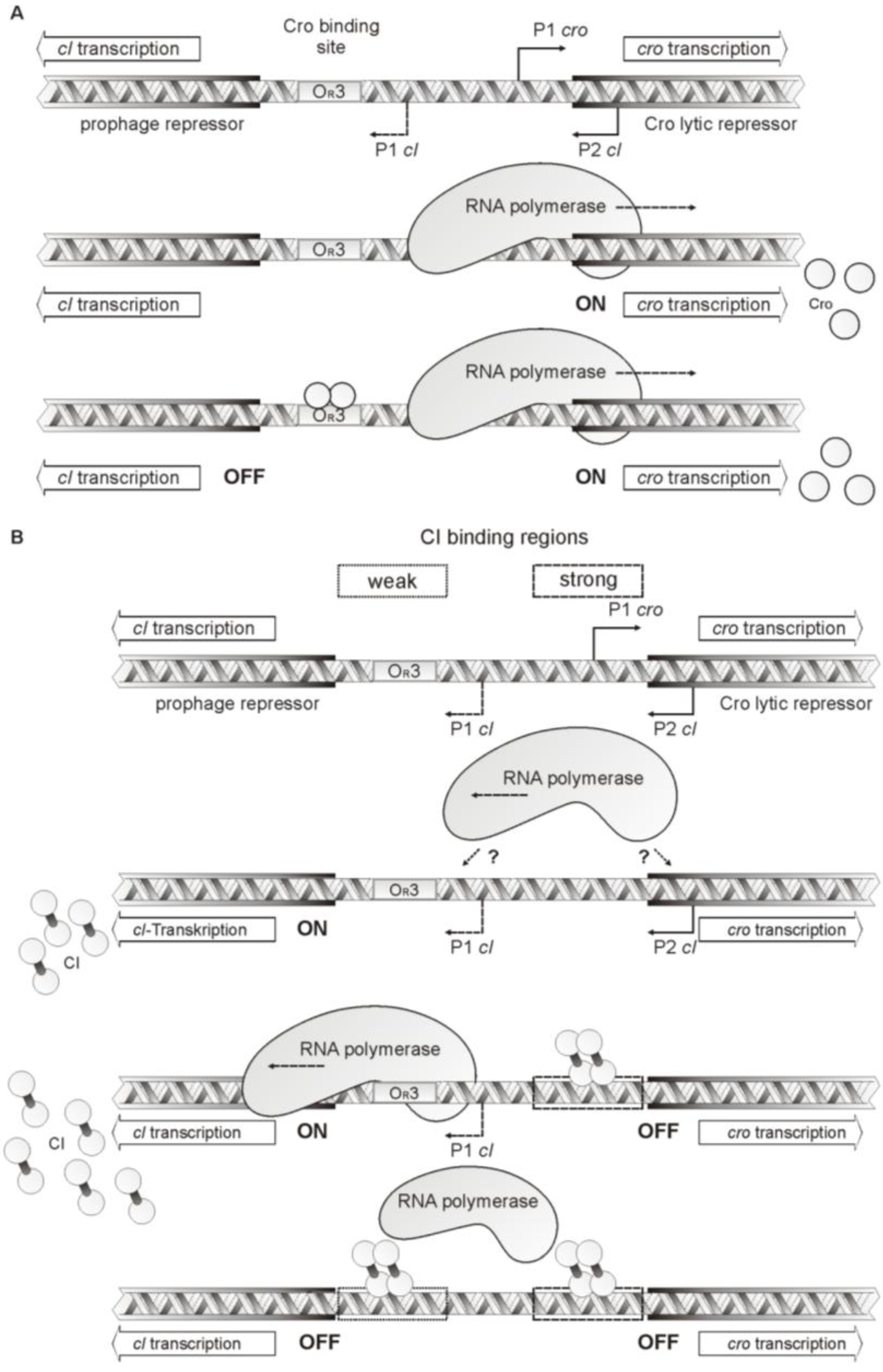
4. Discussion
Acknowledgments
Author Contributions
Supplementary Materials
Conflicts of Interest
References
- Casjens, S.R.; Gilcrease, E.B.; Huang, W.M.; Bunny, K.L.; Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Hatfull, G.F.; Hendrix, R.W. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 2004, 186, 1818–1832. [Google Scholar] [CrossRef] [PubMed]
- Hertwig, S.; Klein, I.; Lurz, R.; Lanka, E.; Appel, B. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol. Microbiol. 2003, 48, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Rybchin, V.N.; Svarchevsky, A.N. The plasmid prophage N15: A linear DNA with covalently closed ends. Mol. Microbiol. 1999, 33, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Klein, I.; Appel, B.; Hertwig, S. Interplay between the temperate phages PY54 and N15, linear plasmid prophages with covalently closed ends. J. Bacteriol. 2007, 189, 8366–8370. [Google Scholar] [CrossRef] [PubMed]
- Mardanov, A.V.; Ravin, N.V. The antirepressor needed for induction of linear plasmid-prophage N15 belongs to the SOS regulon. J. Bacteriol. 2007, 189, 6333–6338. [Google Scholar] [CrossRef] [PubMed]
- Lobocka, M.B.; Svarchevsky, A.N.; Rybchin, V.N.; Yarmolinsky, M.B. Characterization of the primary immunity region of the Escherichia coli linear plasmid prophage N15. J. Bacteriol. 1996, 178, 2902–2910. [Google Scholar] [PubMed]
- Ravin, N.V.; Svarchevsky, A.N.; Deho, G. The anti-immunity system of phage-plasmid N15: Identification of the antirepressor gene and its control by a small processed RNA. Mol. Microbiol. 1999, 34, 980–994. [Google Scholar] [CrossRef] [PubMed]
- Hochschild, A.; Douhan, J.; Ptashne, M. How lambda repressor and lambda Cro distinguish between OR1 and OR3. Cell 1986, 47, 807–816. [Google Scholar] [CrossRef]
- Ziegelin, G.; Tegtmeyer, N.; Lurz, R.; Hertwig, S.; Hammerl, J.A.; Appel, B.; Lanka, E. The repA gene of the linear Yersinia enterocolitica prophage PY54 functions as a circular minimal replicon in Escherichia coli. J. Bacteriol. 2005, 187, 3445–3454. [Google Scholar] [CrossRef] [PubMed]
- Ravin, N.V. N15: The linear phage-plasmid. Plasmid 2011, 65, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Dubrava, M.S.; Ingram, W.M.; Roberts, S.A.; Weichsel, A.; Montfort, W.R.; Cordes, M.H. N15 Cro and lambda Cro: orthologous DNA-binding domains with completely different but equally effective homodimer interfaces. Protein Sci. 2008, 17, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.M.; Lefevre, K.R.; Cordes, M.H. Sequence correlations between Cro recognition helices and cognate O(R) consensus half-sites suggest conserved rules of protein-DNA recognition. J. Mol. Biol. 2005, 350, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Hertwig, S.; Klein, I.; Schmidt, V.; Beck, S.; Hammerl, J.A.; Appel, B. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 2003, 331, 605–622. [Google Scholar] [CrossRef]
- Ravin, V.; Ravin, N.; Casjens, S.; Ford, M.E.; Hatfull, G.F.; Hendrix, R.W. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 2000, 299, 53–73. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Klein, I.; Lanka, E.; Appel, B.; Hertwig, S. Genetic and functional properties of the self-transmissible Yersinia enterocolitica plasmid pYE854, which mobilizes the virulence plasmid pYV. J. Bacteriol. 2008, 190, 991–1010. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Freytag, B.; Lanka, E.; Appel, B.; Hertwig, S. The pYV virulence plasmids of Yersinia pseudotuberculosis and Y. pestis contain a conserved DNA region responsible for the mobilization by the self-transmissible plasmid pYE854. Environ. Microbiol. Rep. 2012, 4, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Hertwig, S.; Klein, I.; Hammerl, J.A.; Appel, B. Characterization of two conjugative Yersinia plasmids mobilizing pYV. Adv. Exp. Med. Biol. 2003, 529, 35–38. [Google Scholar] [PubMed]
- Sambrook, J.; Russel, D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Hertwig, S.; Klein, I.; Appel, B. Properties of the temperate Yersinia enterocolitica bacteriophage PY54. Adv. Exp. Med. Biol. 2003, 529, 241–243. [Google Scholar] [PubMed]
- Covarrubias, L.; Bolivar, F. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene 1982, 17, 79–89. [Google Scholar] [CrossRef]
- Schiemann, D.A. Synthesis of a selective agar medium for Yersinia enterocolitica. Can. J. Microbiol. 1979, 25, 1298–1304. [Google Scholar] [CrossRef] [PubMed]
- Strauch, E.; Voigt, I.; Broll, H.; Appel, B. Use of a plasmid of a Yersinia enterocolitica biogroup 1A strain for the construction of cloning vectors. J. Biotechnol. 2000, 79, 63–72. [Google Scholar] [CrossRef]
- Balzer, D.; Ziegelin, G.; Pansegrau, W.; Kruft, V.; Lanka, E. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucl. Acids Res. 1992, 20, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Di Jeso, F. Ammonium sulfate concentration conversion nomograph for 0 degrees. J. Biol. Chem. 1968, 243, 2022–2023. [Google Scholar] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, J.; Capdevielle, J.; Guillemot, J.C.; Ferrara, P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 1992, 203, 173–179. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl. Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993, 233, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Hawley, D.K.; McClure, W.R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucl. Acids Res. 1983, 11, 2237–2255. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.B.; Reynolds, R.P. Analysis of E. coli promoter sequences. Nucl. Acids Res. 1987, 15, 2343–2361. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Malloch, R.A.; Fujita, N.; Smillie, D.A.; Ishihama, A.; Hayward, R.S. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J. Mol. Biol. 1993, 232, 406–418. [Google Scholar] [CrossRef] [PubMed]
- Jacob, D.; Lewin, A.; Meister, B.; Appel, B. Plant-specific promoter sequences carry elements that are recognised by the eubacterial transcription machinery. Transgenic Res. 2002, 11, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Degnan, P.H.; Michalowski, C.B.; Babic, A.C.; Cordes, M.H.; Little, J.W. Conservation and diversity in the immunity regions of wild phages with the immunity specificity of phage lambda. Mol. Microbiol. 2007, 64, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.; Meyer, B.J.; Ptashne, M. Mechanism of action of the cro protein of bacteriophage lambda. Proc. Natl. Acad. Sci. USA 1978, 75, 1783–1787. [Google Scholar] [CrossRef] [PubMed]
- Estrem, S.T.; Gaal, T.; Ross, W.; Gourse, R.L. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA 1998, 95, 9761–9766. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.M.; Roberts, S.A.; Heroux, A.; Cordes, M.H. Two structures of a lambda Cro variant highlight dimer flexibility but disfavor major dimer distortions upon specific binding of cognate DNA. J. Mol. Biol. 2008, 375, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Resch, A.; Tedin, K.; Graschopf, A.; Haggard-Ljungquist, E.; Blasi, U. Ternary complex formation on leaderless phage mRNA. FEMS Microb. Rev. 1995, 17, 151–157. [Google Scholar] [CrossRef]
- Alanis, V.A.; Kropinski, A.M.; Abbasifar, R.; Griffiths, M.W. Complete genome sequence of Vibrio parahaemolyticus bacteriophage vB_VpaM_MAR. J. Virol. 2012, 86, 13138–13139. [Google Scholar] [CrossRef] [PubMed]
- Lan, S.F.; Huang, C.H.; Chang, C.H.; Liao, W.C.; Lin, I.H.; Jian, W.N.; Wu, Y.G.; Chen, S.Y.; Wong, H.C. Characterization of a new plasmid-like prophage in a pandemic Vibrio parahaemolyticus O3:K6 strain. Appl. Environ. Microb. 2009, 75, 2659–2667. [Google Scholar] [CrossRef] [PubMed]
- Mobberley, J.M.; Authement, R.N.; Segall, A.M.; Paul, J.H. The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J. Virol. 2008, 82, 6618–6630. [Google Scholar] [CrossRef] [PubMed]
- Zabala, B.; Hammerl, J.A.; Espejo, R.T.; Hertwig, S. The linear plasmid prophage Vp58.5 of Vibrio parahaemolyticus is closely related to the integrating phage VHML and constitutes a new incompatibility group of telomere phages. J. Virol. 2009, 83, 9313–9320. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammerl, J.A.; Roschanski, N.; Lurz, R.; Johne, R.; Lanka, E.; Hertwig, S. The Molecular Switch of Telomere Phages: High Binding Specificity of the PY54 Cro Lytic Repressor to a Single Operator Site. Viruses 2015, 7, 2771-2793. https://doi.org/10.3390/v7062746
Hammerl JA, Roschanski N, Lurz R, Johne R, Lanka E, Hertwig S. The Molecular Switch of Telomere Phages: High Binding Specificity of the PY54 Cro Lytic Repressor to a Single Operator Site. Viruses. 2015; 7(6):2771-2793. https://doi.org/10.3390/v7062746
Chicago/Turabian StyleHammerl, Jens Andre, Nicole Roschanski, Rudi Lurz, Reimar Johne, Erich Lanka, and Stefan Hertwig. 2015. "The Molecular Switch of Telomere Phages: High Binding Specificity of the PY54 Cro Lytic Repressor to a Single Operator Site" Viruses 7, no. 6: 2771-2793. https://doi.org/10.3390/v7062746
APA StyleHammerl, J. A., Roschanski, N., Lurz, R., Johne, R., Lanka, E., & Hertwig, S. (2015). The Molecular Switch of Telomere Phages: High Binding Specificity of the PY54 Cro Lytic Repressor to a Single Operator Site. Viruses, 7(6), 2771-2793. https://doi.org/10.3390/v7062746





