Incomplete LPS Core-Specific Felix01-Like Virus vB_EcoM_VpaE1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Phage Techniques
2.3. Transmission Electron Microscopy (TEM)
2.4. Host Range Analysis
2.5. Adsorption Assays
2.5.1. Assay of Adsorbtion Kinetics
2.5.2. Periodate and Proteinase K Treatments
2.6. Phage DNA Isolation and Analysis
2.7. Genome Sequencing, Assembly and Annotation
2.8. Phylogenetic Analysis
2.9. Analysis of VpaE1 Virion Structural Proteins
2.9.1. SDS-PAGE
2.9.2. Filter-Aided Protein Sample Preparation (FASP) for Mass Spectrometry Analysis
2.9.3. Liquid Chromatography and Mass Spectrometry
2.9.4. Data Processing, Search and Analysis
3. Results
3.1. Phage Isolation and Analysis of VpaE1 Plaque and Virion Morphology
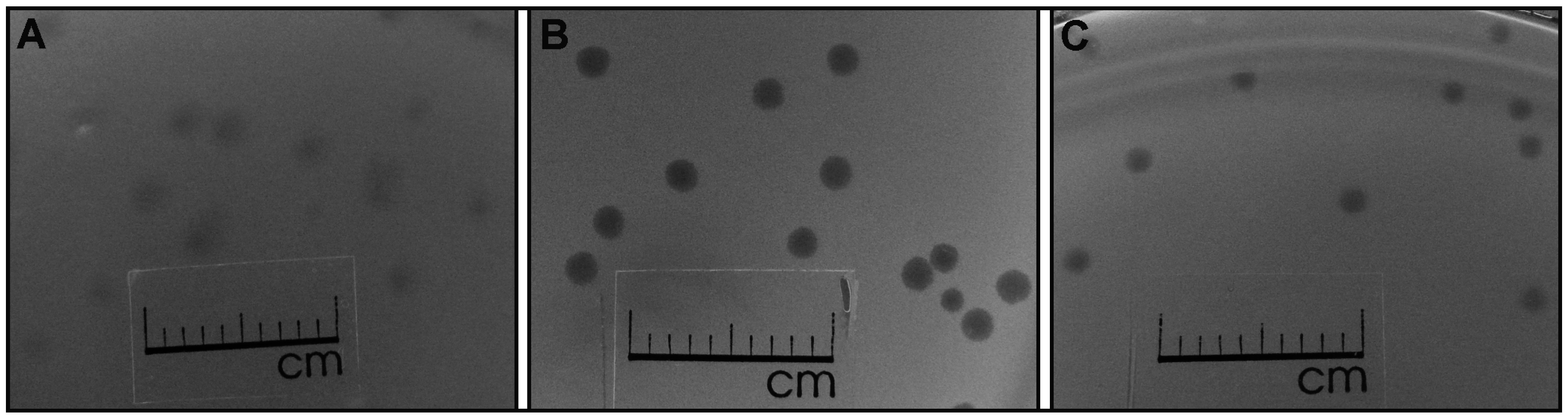

3.2. The Host Range of VpaE1
| Strain | Spot Test | Source/Reference |
|---|---|---|
| E. coli B strains | ||
| BE | + | Lindsay W. Black |
| BL21 | + | Novagen |
| BL21(DE3) | + | Novagen |
| B40 | + | Lindsay W. Black |
| BE-BS | + | Kenneth N. Kreuzer/[52] |
| E. coli K12 strains | ||
| MG1655 | – | Edita Sužiedėlienė |
| K803 | – | Lindsay W. Black |
| XL1blue | – | Stratagene |
| CR63 | – | Kenneth N. Kreuzer/[52] |
| BW25113 | – | [53] |
| E. coli STEC representatives | ||
| O157:H7 | – | Edita Sužiedėlienė |
| O103:H2 | – | Edita Sužiedėlienė |
| O111:H8 | – | Edita Sužiedėlienė |
| O145:H28 | – | Edita Sužiedėlienė |
| O26:H11 | – | Edita Sužiedėlienė |
| Other enterobacteria | ||
| Salmonella Typhimurium LT2 | – | Jaunius Urbonavičius |
| Klebsiella pneumoniae KV-3 | – | [54] |
3.3. Annotation and Overview of VpaE1 Genome

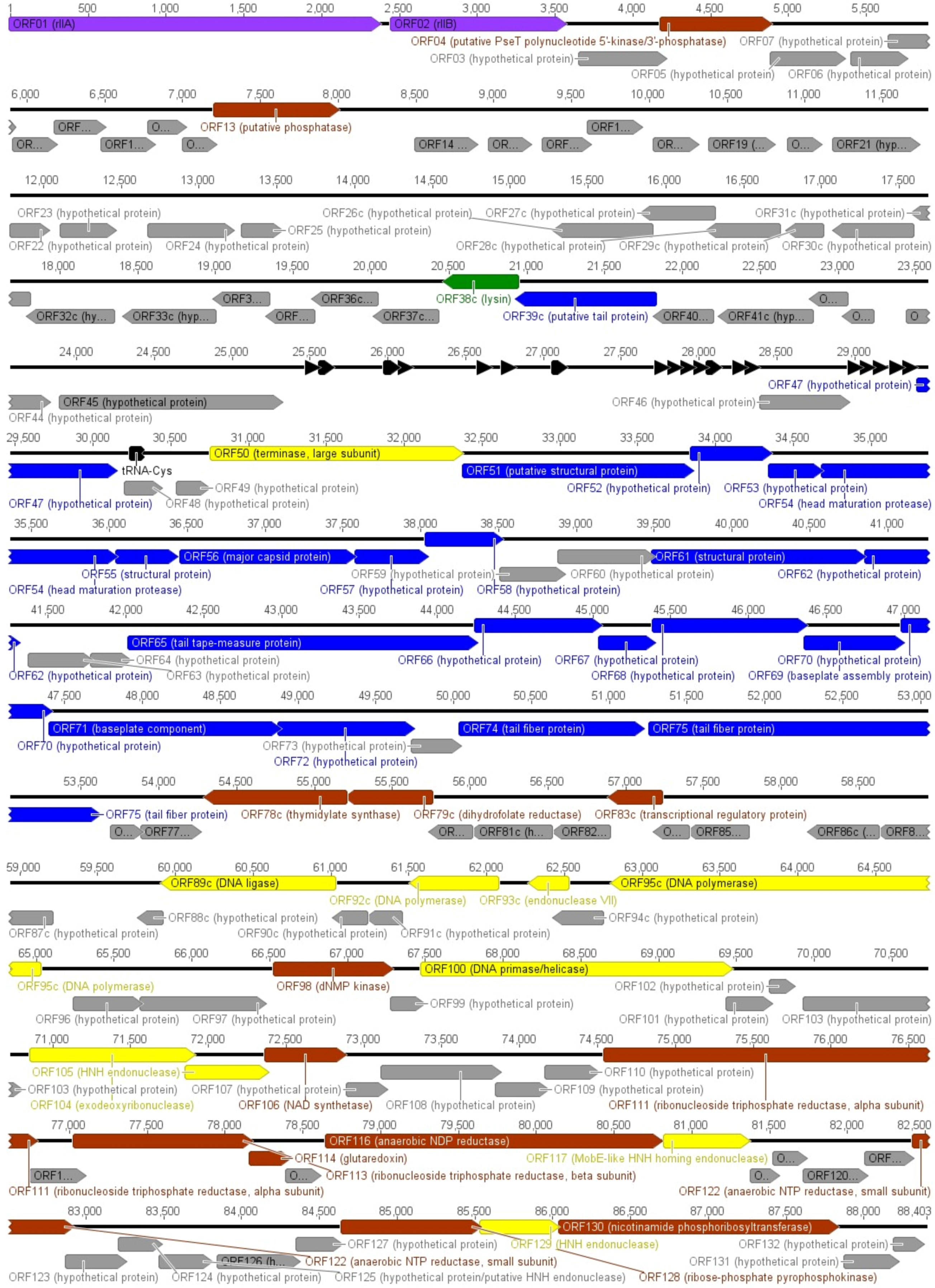
3.4. Structural Proteins and Virion Proteome

3.5. Phylogenetic Relatedness

3.6. Identification of VpaE1 Receptor
| E. coli K-12 BW25113 Mutants | Spot Test | Source/Reference |
|---|---|---|
| ΔwaaC | – | Keio collection/[36] |
| ΔwaaF | – | Keio collection/[36] |
| ΔwaaG | + | Keio collection/[36] |
| ΔwaaP | + | Keio collection/[36] |
| ΔwaaY | – | Keio collection/[36] |
| ΔwaaQ | – | Keio collection/[36] |
| ΔwaaO | – | Keio collection/[36] |
| ΔwaaR | + | Keio collection/[36] |
| ΔwaaB | ± | Keio collection/[36] |
| ΔwaaZ | ± | Keio collection/[36] |


4. Discussion

5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rakhuba, D.V.; Kolomiets, E.I.; Dey, E.S.; Novik, G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [PubMed]
- Shin, H.; Lee, J.H.; Kim, H.; Choi, Y.; Heu, S.; Ryu, S. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. PLoS ONE 2012, 7, e43392. [Google Scholar] [CrossRef] [PubMed]
- Michel, A.; Clermont, O.; Denamur, E.; Tenaillon, O. Bacteriophage PhiX174‘s ecological niche and the flexibility of its Escherichia coli lipopolysaccharide receptor. Appl. Environ. Microbiol. 2010, 76, 7310–7313. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Hull, S. Escherichia coli lipopolysaccharide in pathogenesis and virulence. In Escherichia coli: Mechanisms of Virulence; Sussman, M., Ed.; Cambridge University Press: Cambridge, UK, 1997; pp. 145–167. [Google Scholar]
- Amor, K.; Heinrichs, D.E.; Frirdich, E.; Ziebell, K.; Johnson, R.P.; Whitfield, C. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect. Immun. 2000, 68, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Ryu, S. Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2012, 86, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Perry, E.B.; Barrick, J.E.; Bohannan, B.J. The Molecular and genetic basis of repeatable coevolution between Escherichia coli and bacteriophage T3 in a laboratory microcosm. PLoS ONE 2015, 10, e0130639. [Google Scholar] [CrossRef] [PubMed]
- Brockhurst, M.A.; Rainey, P.B.; Buckling, A. The effect of spatial heterogeneity and parasites on the evolution of host diversity. Proc. Biol. Sci. 2004, 271, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Buckling, A.; Wei, Y.; Massey, R.C.; Brockhurst, M.A.; Hochberg, M.E. Antagonistic coevolution with parasites increases the cost of host deleterious mutations. Proc. Biol. Sci. 2006, 273, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Yethon, J.A.; Heinrichs, D.E.; Monteiro, M.A.; Perry, M.B.; Whitfield, C. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 1998, 273, 26310–26316. [Google Scholar] [CrossRef] [PubMed]
- Yethon, J.A.; Vinogradov, E.; Perry, M.B.; Whitfield, C. Mutation of the lipopolysaccharide core glycosyltransferase encoded by waaG destabilizes the outer membrane of Escherichia coli by interfering with core phosphorylation. J. Bacteriol. 2000, 182, 5620–5623. [Google Scholar] [CrossRef] [PubMed]
- Heinrichs, D.E.; Yethon, J.A.; Whitfield, C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 1998, 30, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Marti, R.; Zurfluh, K.; Hagens, S.; Pianezzi, J.; Klumpp, J.; Loessner, M.J. Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol. Microbiol. 2013, 87, 818–834. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, S.; Park, B.; Ryu, S. Core lipopolysaccharide-specific phage SSU5 as an auxiliary component of a phage cocktail for Salmonella biocontrol. Appl. Environ. Microbiol. 2014, 80, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Zhang, P.; Zhang, H.; Zhou, Y.; Zhang, L.; Wang, R. Bio-control of Salmonella enteritidis in foods using bacteriophages. Viruses 2015, 7, 4836–4853. [Google Scholar] [CrossRef] [PubMed]
- Felix, A.; Callow, B.R. Typing of paratyphoid B bacilli by Vi bacteriophage. Brit. Med. J. 1943, 2, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Whichard, J.M.; Weigt, L.A.; Borris, D.J.; Li, L.L.; Zhang, Q.; Kapur, V.; Pierson, F.W.; Lingohr, E.J.; She, Y.M.; Kropinski, A.M.; et al. Complete genomic sequence of bacteriophage Felix O1. Viruses 2010, 2, 710–730. [Google Scholar] [CrossRef] [PubMed]
- Hirsh, D.C.; Martin, L.D. Rapid detection of Salmonella spp. by using Felix-O1 bacteriophage and high-performance liquid chromatography. Appl. Environ. Microbiol 1983, 45, 260–264. [Google Scholar] [PubMed]
- Whichard, J.M.; Sriranganathan, N.; Pierson, F.W. Suppression of Salmonella growth by wild-type and large-plaque variants of bacteriophage Felix O1 in liquid culture and on chicken frankfurters. J. Food Protect. 2003, 66, 220–225. [Google Scholar]
- Lavigne, R.; Darius, P.; Summer, E.J.; Seto, D.; Mahadevan, P.; Nilsson, A.S.; Ackermann, H.W.; Kropinski, A.M. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 2009, 9. [Google Scholar] [CrossRef] [PubMed]
- Villegas, A.; She, Y.M.; Kropinski, A.M.; Lingohr, E.J.; Mazzocco, A.; Ojha, S.; Waddell, T.E.; Ackermann, H.W.; Moyles, D.M.; Ahmed, R.; et al. The genome and proteome of a virulent Escherichia coli O157:H7 bacteriophage closely resembling Salmonella phage Felix O1. Virol. J. 2009, 6. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, B.R.; Kim, J. Complete genome sequence of bacteriophage EC6, capable of lysing Escherichia coli O157:H7. Genome Announc. 2013, 1, e00085-12. [Google Scholar] [CrossRef] [PubMed]
- Hoa, N.X.; Tang, F.; Bai, Q.; Shen, C.; Vo, T.T.; Lu, C. Isolation and characterization of bacteriophages against enterotoxigenic Escherichia coli strains causing swine diseases. Afr. J. Microbiol. Res. 2013, 7, 2787–2793. [Google Scholar]
- Cowley, L.A.; Beckett, S.J.; Chase-Topping, M.; Perry, N.; Dallman, T.J.; Gally, D.L.; Jenkins, C. Analysis of whole genome sequencing for the Escherichia coli O157:H7 typing phages. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Niu, Y.D.; Chen, J.; Anany, H.; Ackermann, H.W.; Johnson, R.P.; Ateba, C.N.; Stanford, K.; McAllister, T.A. Feces of feedlot cattle contain a diversity of bacteriophages that lyse non-O157 Shiga toxin-producing Escherichia coli. Can. J. Microbiol. 2015, 61, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Bardina, C.; Spricigo, D.A.; Cortes, P.; Llagostera, M. Significance of the bacteriophage treatment schedule in reducing Salmonella colonization of poultry. Appl. Environ. Microbiol. 2012, 78, 6600–6607. [Google Scholar] [CrossRef] [PubMed]
- Moreno Switt, A.I.; Orsi, R.H.; den Bakker, H.C.; Vongkamjan, K.; Altier, C.; Wiedmann, M. Genomic characterization provides new insight into Salmonella phage diversity. BMC Genom. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Spricigo, D.A.; Bardina, C.; Cortes, P.; Llagostera, M. Use of a bacteriophage cocktail to control Salmonella in food and the food industry. Int. J. Food Microbiol. 2013, 165, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Colom, J.; Cano-Sarabia, M.; Otero, J.; Cortes, P.; Maspoch, D.; Llagostera, M. Liposome-encapsulated bacteriophages for enhanced oral phage therapy against Salmonella spp. Appl. Environ. Microbiol. 2015, 81, 4841–4849. [Google Scholar] [CrossRef] [PubMed]
- Tolen, T.N.; Xie, Y.; Hernandez, A.C.; Kuty Everett, G.F. Complete genome sequence of Salmonella enterica serovar Typhimurium myophage mushroom. Genome Announc. 2015, 3, e00154-15. [Google Scholar] [CrossRef] [PubMed]
- Lehman, S.M.; Kropinski, A.M.; Castle, A.J.; Svircev, A.M. Complete genome of the broad-host-range Erwinia amylovora phage phiEa21-4 and its relationship to Salmonella phage felix O1. Appl. Environ. Microbiol. 2009, 75, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.T.; Luna, A.J.; Hernandez, A.C.; Kuty Everett, G.F. Complete genome sequence of Citrobacter freundii myophage Moogle. Genome Announc. 2015, 3, e01426-14. [Google Scholar] [CrossRef] [PubMed]
- Henry, M.; Bobay, L.M.; Chevallereau, A.; Saussereau, E.; Ceyssens, P.J.; Debarbieux, L. The search for therapeutic bacteriophages uncovers one new subfamily and two new genera of Pseudomonas-infecting Myoviridae. PLoS ONE 2015, 10, e0117163. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.J.; Sacher, J.C.; Szymanski, C.M. Exploring the interactions between bacteriophage-encoded glycan binding proteins and carbohydrates. Curr. Opin. Struct. Biol. 2015, 34, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E. Phage host range and efficiency of plating. In Bacteriophages: Methods in Molecular Biology; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: New York, NY, USA, 2009; Volume 1, pp. 141–149. [Google Scholar]
- Kropinski, A.M. Measurement of the bacteriophage inactivation kinetics with purified receptors. In Bacteriophages: Methods in Molecular Biology; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: New York, NY, USA, 2009; Volume 1, pp. 157–160. [Google Scholar]
- Kiljunen, S.; Datta, N.; Dentovskaya, S.V.; Anisimov, A.P.; Knirel, Y.A.; Bengoechea, J.A.; Holst, O.; Skurnik, M. Identification of the lipopolysaccharide core of Yersinia pestis and Yersinia pseudotuberculosis as the receptor for bacteriophage phiA1122. J. Bacteriol. 2011, 193, 4963–4972. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Harmon, D.; Kasif, S.; White, O.; Salzberg, S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999, 27, 4636–4641. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Feist, P.; Hummon, A.B. Proteomic challenges: Sample preparation techniques for microgram-quantity protein analysis from biological samples. Int. J. Mol. Sci. 2015, 16, 3537–3563. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Ant. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Kaliniene, L.; Klausa, V.; Truncaite, L. Low-temperature T4-like coliphages vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20. Arch. Virol. 2010, 155, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Kaliniene, L.; Klausa, V.; Zajanckauskaite, A.; Nivinskas, R.; Truncaite, L. Genome of low-temperature T4-related bacteriophage vB_EcoM-VR7. Arch. Virol. 2011, 156, 1913–1916. [Google Scholar] [CrossRef] [PubMed]
- Kaliniene, L.; Zajanckauskaite, A.; Simoliunas, E.; Truncaite, L.; Meskys, R. Low-temperature bacterial viruses VR—A small but diverse group of E. coli phages. Arch. Virol. 2015, 160, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Truncaite, L.; Simoliunas, E.; Zajanckauskaite, A.; Kaliniene, L.; Mankeviciute, R.; Staniulis, J.; Klausa, V.; Meskys, R. Bacteriophage vB_EcoM_FV3: A new member of “rV5-like viruses”. Arch. Virol. 2012, 157, 2431–2435. [Google Scholar] [CrossRef] [PubMed]
- Selick, H.E.; Kreuzer, K.N.; Alberts, B.M. The bacteriophage T4 insertion/substitution vector system. A method for introducing site-specific mutations into the virus chromosome. J. Biol. Chem. 1988, 263, 11336–11347. [Google Scholar] [PubMed]
- Grenier, F.; Matteau, D.; Baby, V.; Rodrigue, S. Complete genome sequence of Escherichia coli BW25113. Genome Announc. 2014, 2, e01038-14. [Google Scholar] [CrossRef] [PubMed]
- Simoliunas, E.; Kaliniene, L.; Truncaite, L.; Klausa, V.; Zajanckauskaite, A.; Meskys, R. Genome of Klebsiella sp.-infecting bacteriophage vB_KleM_RaK2. J. Virol. 2012, 86. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. In Advances in Applied Microbiology; Laskin, A.I., Sariaslani, S., Gadd, G.M., Eds.; Elsevier Inc. Academic Press: Burlington, MA, USA, 2010; Volume 70, pp. 217–248. [Google Scholar]
- Dy, R.L.; Richter, C.; Salmond, G.P.; Fineran, P.C. Remarkable mechanisms in microbes to resist phage infections. Annu. Rev. Virol. 2014, 1, 307–331. [Google Scholar] [CrossRef]
- Lavigne, R.; Noben, J.P.; Hertveldt, K.; Ceyssens, P.J.; Briers, Y.; Dumont, D.; Roucourt, B.; Krylov, V.N.; Mesyanzhinov, V.V.; Robben, J.; et al. The structural proteome of Pseudomonas aeruginosa bacteriophage phiKMV. Microbiology 2006, 152, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Ceyssens, P.J.; Robben, J. Phage proteomics: Applications of mass spectrometry. In Bacteriophages: Methods in Molecular Biology; Clockie, M.R.J., Kropinski, A.M., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 2, pp. 239–251. [Google Scholar]
- Pupo, G.M.; Karaolis, D.K.; Lan, R.; Reeves, P.R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 1997, 65, 2685–2692. [Google Scholar] [PubMed]
- Reid, S.D.; Herbelin, C.J.; Bumbaugh, A.C.; Selander, R.K.; Whittam, T.S. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 2000, 406, 64–67. [Google Scholar] [PubMed]
- Hayashi, T.; Makino, K.; Ohnishi, M.; Kurokawa, K.; Ishii, K.; Yokoyama, K.; Han, C.G.; Ohtsubo, E.; Nakayama, K.; Murata, T.; et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001, 8, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, J.; Ren, G.; Li, Y.; Wang, X. Influence of core oligosaccharide of lipopolysaccharide to outer membrane behavior of Escherichia coli. Mar. Drugs 2015, 13, 3325–3339. [Google Scholar] [CrossRef] [PubMed]
- Frirdich, E.; Lindner, B.; Holst, O.; Whitfield, C. Overexpression of the waaZ gene leads to modification of the structure of the inner core region of Escherichia coli lipopolysaccharide, truncation of the outer core, and reduction of the amount of O polysaccharide on the cell surface. J. Bacteriol. 2003, 185, 1659–1671. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.M.; Goldrick, M.; Lord, L.; Roberts, I.S. Mutations in the waaR gene of Escherichia coli which disrupt lipopolysaccharide outer core biosynthesis affect cell surface retention of group 2 capsular polysaccharides. J. Bacteriol. 2006, 188, 1165–1168. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Barbe, V.; Lee, C.H.; Vallenet, D.; Yu, D.S.; Choi, S.H.; Couloux, A.; Lee, S.W.; Yoon, S.H.; Cattolico, L.; et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 2009, 394, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Garrett, T.A.; Raetz, C.R. In vitro assembly of the outer core of the lipopolysaccharide from Escherichia coli K-12 and Salmonella typhimurium. Biochemistry 2014, 53, 1250–1262. [Google Scholar] [CrossRef] [PubMed]
- Kaszowska, M.; Niedziela, T.; Maciejewska, A.; Lukasiewicz, J.; Jachymek, W.; Lugowski, C. Core oligosaccharide of Escherichia coli B-the structure required for bacteriophage T4 recognition. Carbohydr. Res. 2015, 413, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Mizushima, S. Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J. Bacteriol. 1982, 151, 718–722. [Google Scholar] [PubMed]
- Studier, F.W.; Daegelen, P.; Lenski, R.E.; Maslov, S.; Kim, J.F. Understanding the differences between genome sequences of Escherichia coli B strains REL606 and BL21(DE3) and comparison of the E. coli B and K-12 genomes. J. Mol. Biol. 2009, 394, 653–680. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.; Duperchy, E.; Depeyrot, J.; Coursange, E.; Lenski, R.E.; Blot, M. Genomic comparisons among Escherichia coli strains B, K-12, and O157: H7 using IS elements as molecular markers. BMC Microbiol. 2002, 2. [Google Scholar] [CrossRef]
- Yoon, S.H.; Han, M.J.; Jeong, H.; Lee, C.H.; Xia, X.X.; Lee, D.H.; Shim, J.H.; Lee, S.Y.; Oh, T.K.; Kim, J.F. Comparative multi-omics systems analysis of Escherichia coli strains B and K-12. Genome Biol. 2012, 13. [Google Scholar] [CrossRef]
- Niu, Y.D.; Johnson, R.P.; Xu, Y.; McAllister, T.A.; Sharma, R.; Louie, M.; Stanford, K. Host range and lytic capability of four bacteriophages against bovine and clinical human isolates of Shiga toxin-producing Escherichia coli O157:H7. J. Appl. Microbiol. 2009, 107, 646–656. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šimoliūnas, E.; Vilkaitytė, M.; Kaliniene, L.; Zajančkauskaitė, A.; Kaupinis, A.; Staniulis, J.; Valius, M.; Meškys, R.; Truncaitė, L. Incomplete LPS Core-Specific Felix01-Like Virus vB_EcoM_VpaE1. Viruses 2015, 7, 6163-6181. https://doi.org/10.3390/v7122932
Šimoliūnas E, Vilkaitytė M, Kaliniene L, Zajančkauskaitė A, Kaupinis A, Staniulis J, Valius M, Meškys R, Truncaitė L. Incomplete LPS Core-Specific Felix01-Like Virus vB_EcoM_VpaE1. Viruses. 2015; 7(12):6163-6181. https://doi.org/10.3390/v7122932
Chicago/Turabian StyleŠimoliūnas, Eugenijus, Monika Vilkaitytė, Laura Kaliniene, Aurelija Zajančkauskaitė, Algirdas Kaupinis, Juozas Staniulis, Mindaugas Valius, Rolandas Meškys, and Lidija Truncaitė. 2015. "Incomplete LPS Core-Specific Felix01-Like Virus vB_EcoM_VpaE1" Viruses 7, no. 12: 6163-6181. https://doi.org/10.3390/v7122932
APA StyleŠimoliūnas, E., Vilkaitytė, M., Kaliniene, L., Zajančkauskaitė, A., Kaupinis, A., Staniulis, J., Valius, M., Meškys, R., & Truncaitė, L. (2015). Incomplete LPS Core-Specific Felix01-Like Virus vB_EcoM_VpaE1. Viruses, 7(12), 6163-6181. https://doi.org/10.3390/v7122932








