Abstract
Development of a vaccine against congenital infection with human cytomegalovirus is complicated by the issue of re-infection, with subsequent vertical transmission, in women with pre-conception immunity to the virus. The study of experimental therapeutic prevention of re-infection would ideally be undertaken in a small animal model, such as the guinea pig cytomegalovirus (GPCMV) model, prior to human clinical trials. However, the ability to model re-infection in the GPCMV model has been limited by availability of only one strain of virus, the 22122 strain, isolated in 1957. In this report, we describe the isolation of a new GPCMV strain, the CIDMTR strain. This strain demonstrated morphological characteristics of a typical Herpesvirinae by electron microscopy. Illumina and PacBio sequencing demonstrated a genome of 232,778 nt. Novel open reading frames ORFs not found in reference strain 22122 included an additional MHC Class I homolog near the right genome terminus. The CIDMTR strain was capable of dissemination in immune compromised guinea pigs, and was found to be capable of congenital transmission in GPCMV-immune dams previously infected with salivary gland‑adapted strain 22122 virus. The availability of a new GPCMV strain should facilitate study of re-infection in this small animal model.
1. Introduction
Development of a vaccine against human cytomegalovirus (HCMV) is a major public health priority [1]. The suggestion that passively transferred antibody protects the fetus against infection and injury [2] has driven efforts to develop recombinant subunit vaccines targeting major envelope glycoproteins, such as glycoprotein B (gB) [3,4]. Although clinical trials of recombinant gB vaccines have shown some degree of effectiveness in preventing HCMV infection and disease in high risk populations [5,6], vaccine mediated protection with vaccines targeting this single envelope glycoprotein appears to be incomplete. Moreover, the effectiveness of natural immunity in preventing congenital HCMV infection and its attendant sequelae is itself a matter of some controversy. A number of recent studies have described fetal HCMV transmission in women with preconception immunity, due to re-infection with new strains of HCMV [7,8,9,10,11,12]. Such infections can produce sequelae identical to those observed in congenitally infected infants born to women with primary HCMV infection in pregnancy [13,14]. These observations certainly complicate HCMV vaccine design, and suggest that: (1) for full protection, an HCMV vaccine may need to elicit responses superior to those conferred by natural immunity; (2) there may be a strong rationale for vaccinating women of childbearing age who are already HCMV seropositive, in addition to targeting and immunizing seronegative women, toward the goal of preventing re-infection with subsequent transmission of the “new” strain.
Several clinical studies have documented the phenomena of re-infection in women of childbearing age. In one prospective study performed at the University of Alabama-Birmingham, serum specimens from 46 women with preconceptional immunity against HCMV obtained during a previous pregnancy and a new pregnancy were analyzed for antibodies against the strain-specific epitopes of HCMV glycoprotein H (gH), and the nucleotide sequences of the gH gene from seven HCMV isolates were determined. Ten of the 16 mothers with infected children (62%) acquired new antibody specificities against gH, as compared with only 4 of the 30 mothers of uninfected infants (13%), suggesting that acquisition of an infection with a virus expressing a novel strain-specific gH genotype during pregnancy was associated with congenital transmission [9]. In another study in Brazil that followed 7,848 women prospectively, sera from 40 mothers of congenitally infected infants and 109 mothers of uninfected control newborns were analyzed for strain-specific anti-HCMV antibodies, based not only on polymorphisms within gH binding sites, but also a second antibody reactivity site on gB [14,15]. Seven of 40 (17.5%) study women, but only 5 of 109 (4.6%) controls acquired antibodies reactive with new HCMV strains during pregnancy (p = 0.002), suggesting that maternal reinfection by new strains of HCMV is a major source of congenital infection in this population. In a study of re-infection (based on acquisition of new gB and/or gH antibody specificities) in 205 seropositive women performed by Ross and colleagues at UAB, approximately one-third of the study participants (59 of 205) were noted to have reinfection, using this definition, during follow-up [10]. The molecular and immunological correlates of re-infection are unclear. There is some evidence that gB polymorphisms in clinical isolates may be less important for re-infection than polymorphisms in gH and other envelope glycoproteins. In a study in Brazil, infections in immunocompetent women with strains corresponding to more than one gB genotype were not common [16]. Additionally, in a study of the HCMV strains acquired longitudinally in women who developed infection in spite of being enrolled in the recombinant gB vaccine trial at UAB [5], there was no selection for or against any non-vaccine gB subtype, in spite of women being immunized only with gB protein corresponding to the Towne (gB1 subtype) strain [17]. Other evidence suggests that the response (or lack thereof) to the envelope glycoprotein N (gN) may play a role in predisposing to re-infection with new HCMV strains expressing heterologous gB and/or gH genotypes [18,19,20].
Irrespective of the mechanism(s) involved, the issue of re-infection is a major challenge in vaccine design. There is increasing evidence that congenital HCMV infections after nonprimary maternal infections can lead to symptomatic disease and substantial long-term sequelae. Notably, recent evidence from a study at UAB indicated that the incidence of hearing loss in infants infected after nonprimary maternal infection was similar to that in infected infants born to women with primary infection [13], although in this study, infants in the primary infection group were more likely to demonstrate progressive and severe or profound hearing loss, compared to infants in the non-primary group.
Since the consequences of re-infection and congenital transmission with a newly acquired strain in pregnant seropositive women can be similar to those that occur after primary infection and transmission in CMV-seronegative women [10,11,13,14], the study of re-infection in a small animal model of congenital transmission would be very useful for the modeling of vaccine strategies to prevent maternal re-infection [21,22]. Ideally, HCMV reinfection would be studied in an animal model prior to clinical vaccine trials. Unfortunately, the strict species-specificity of cytomegaloviruses precludes preclinical testing of HCMV vaccines in animals. However, a number of rodent and primate cytomegaloviruses are useful in modeling HCMV vaccines and therapies, given the conservation of many immunogenic structural proteins amongst the various viruses [23,24,25].
Among the small animal models, the guinea pig cytomegalovirus (GPCMV) is uniquely useful, since, in contrast to other rodent models, transplacental infection of the fetus occurs following viral challenge during pregnancy [24,26]. Hence, the GPCMV model is particularly well-suited to the study of vaccines against congenital infection. However, until now this model has relied exclusively on a single strain of GPCMV, 22122, isolated by Hartley in 1957 [27]. While it has been feasible to study re-infection by experimentally inoculating strain 22122 virus into naturally seropositive guinea pigs [28], the lack of defined genetic information on other GPCMV strains (e.g., those presumably latent in the seropositive animals used in the above mentioned studies) has made it impossible to study of the role of strain variation in fetal infection in the setting of preconception immunity.
In this report, we describe the isolation of a novel strain of GPCMV, the CIDMTR strain. This communication represents the first report of detailed characterization of a GPCMV isolate since the original publication of isolation of the 22122 strain [27]. Although sequence and ORF structure were generally well conserved with the 22122 strain of GPCMV, the CIDMTR strain demonstrated some differences in genome structure, particularly in the right-hand end of the viral genome. There are also substantive differences in some protein coding sequences between the two strains, including sequences in envelope glycoproteins, suggesting that these proteins may have been the targets of immune selection during the evolution of GPCMV in the guinea pig host. We describe in this report the morphology and DNA sequence of this newly isolated strain, and report preliminary experiments regarding its pathogenesis in vivo. The availability of a second strain of GPCMV should enable the study of re-infection and, potentially, the development of vaccine strategies designed to protect against maternal re-infection in the guinea pig model of congenital cytomegalovirus infection.
2. Results and Discussion
2.1. Isolation of the CIDMTR Strain
In the course of ongoing vaccine and pathogenesis studies, guinea pigs were screened at the time of purchase for GPCMV antibodies, using an ELISA based on GPCMV strain 22122 [29]. Within a group of 24 guinea pigs purchased from a commercial source, 5 (21%) were found to be GPCMV-seropositive by ELISA. Western blot analysis was performed using sera from commercially purchased, ELISA-positive animals using purified strain 22122 virions as the source of target antigens (Figure 1). These studies confirmed that sera from these animals were broadly cross-reactive with GPCMV strain 22122 virion-associated polypeptides, suggesting that these animals were infected with GPCMV strains that were closely related antigenically to strain 22122.
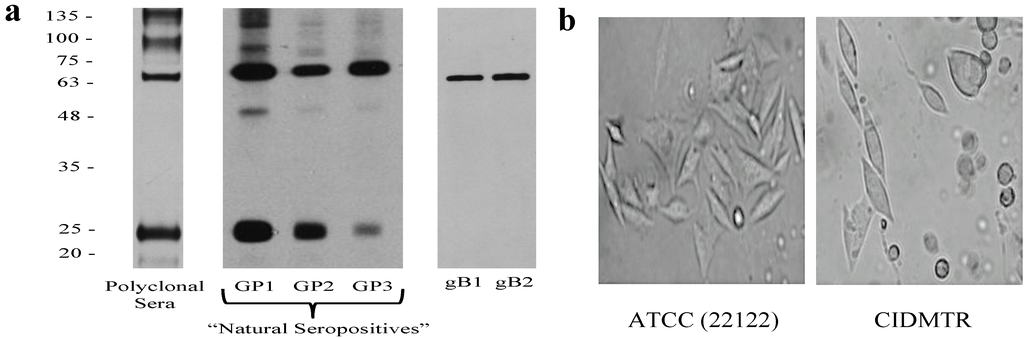
Figure 1.
Western blot analysis of sera from three “naturally seropositive” commercially purchased outbred Hartley guinea pigs. (a) Western analysis using a pooled, high-titer polyclonal anti-GPCMV antisera from guinea pigs immunized with adjuvanted viral particles [30] or sera from three “natural seropositive” guinea pigs (GP1, GP2, GP3) obtained from a commercial supplier, using sucrose gradient-purified virions from 22122 strain as target antigen. Two independently derived, monospecific anti-GPCMV gB antibodies (moab 29-29 [31] (gB1) and moab IE321 (gB2)) are included as controls. (b) Photomicrographs comparing CPE of low-passage (P2) CIDMTR virus (right panel) to GPCMV strain 22122 (left panel) following infection of GPL cells. CIDMTR plaques are more rounded, with elliptical appearance, compared to the more spindle-like morphology of 22122-infected fibroblasts. Plaques photographed at 60× magnification.
One of the seropositive animals identified in these preliminary experiments was immunosuppressed with cyclophosphamide, 100 mg/kg [32], and seven days later the animal was sacrificed and salivary gland (SG) homogenates harvested. Presumptive virus in the SG homogenate was passaged in vivo by inoculation of two GPCMV seronegative inbred strain 2 guinea pigs with 1 mL SG homogenate by subcutaneous route in the dorsal neck. Both of these animals demonstrated DNAemia (3.2 and 6.9 × 103 genomes/mL of blood, respectively) at 14 days post-inoculation. Three weeks following inoculation, these animals were also immunosuppressed with 100 mg/kg of cyclophosphamide. One week later, these animals were sacrificed and SG homogenates were cultured on guinea pig lung fibroblast cells (GPLs). Eleven days after inoculation plaques with characteristic cytopathic effect (CPE) were observed in one of the SG homogenate cultures (designated P0). Supernatant from this flask was used to inoculate GPLs to generate a P1 stock of virus. When CPE was extensive (approximately 2 weeks later), an aliquot was removed for electron microscopy (EM) studies (described below, Section 2.2) and the remainder of this flask was expanded for large-scale virus propagation (20 flasks). These flasks were then incubated for an additional week prior to harvest of P2 viral stock (Figure 1b). Some of these infected flasks were used for DNA purification for Illumina MiSeq and Pacific Biosciences PacBio RS sequencing, as described below. The remainder of this stock was used for in vivo challenge experiments in guinea pigs, as described in Section 2.4.
2.2. Morphological Analyses by EM
To confirm that the isolated virus had morphological characteristics of a cytomegalovirus, EM was performed. Analysis of P1 virus stock by conventional microscopy (Section 2.1) revealed 20% of the cells showed cytopathic effect. Cells were enlarged with large intranuclear and intracytoplasmic basophilic inclusions visible upon examination by light microscopy on thick section of plastic preparation stained with Toluidine blue (data not shown). Ultrastructurally, both intranuclear and intracytoplasmic inclusions were noted, interpreted as representative of nuclear and cytoplasmic viral factories (Figure 2A,B). Nuclear inclusions were typical for the center of replication and assembly characteristic of Herpesvirinae (Figure 2C). These nuclear factories showed a large amount of fibrillary electron dense material admixed with moderate to large numbers of empty A-capsids, scaffold-containing B-capsids, and DNA-containing C-capsids that are characteristic of herpesvirus capsid formation and genome packaging (Figure 2C; most capsids in this image are B-capsids). By comparison, a virtually identical appearance was noted in EMs from strain 22122-infected fibroblasts (Figure 2H). First envelopment was acquired at the inner nuclear membrane. The EM morphology supports a model whereby de-envelopment of nucleocapsids takes place at the outer nuclear membrane during egress, with the second envelopment and tegumentation occurring in the cytoplasm factory (Figure 2D). Rearrangement of Golgi cisterns, Golgi vesicles, multivesicular body and endoplasmic reticulum in conjunction with formation of nucleocapsids produced large aggregations that were often mixed with electron dense material. Numerous dense bodies were present in the intercellular space (Figure 2A,F). On negative contrast preparation, a large number of non-infectious enveloped particles (220.24 ± 48.15 nm) and dense bodies (357.83 ± 97.85 nm) were observed (Figure 2G). Infectious mature virions consisted of an envelope containing a 116.5 ± 2.68 nm in diameter icosahedral capsid with capsomeres of approximately 11.66 ± 1.87 nm. The CIDMTR strain of GPCMV presented similar cytopathic and pathogenic effects as those demonstrated by ATCC strain 22122 [33,34,35]. Nevertheless, some differences were noted between these two strains. The ATCC strain in contrast to the CIDMTR strain, demonstrated more efficient infection of fibroblasts, even at the same multiplicity of infection (MOI; MOI of 1), with over 80% of fibroblastic cells exhibiting cytopathic effect. The nuclear and cytoplasmic factories were less pronounced in the CIDMTR-infected cells, with generation of reduced quantities of infectious and non-infectious enveloped viral particles and dense bodies (data not shown).
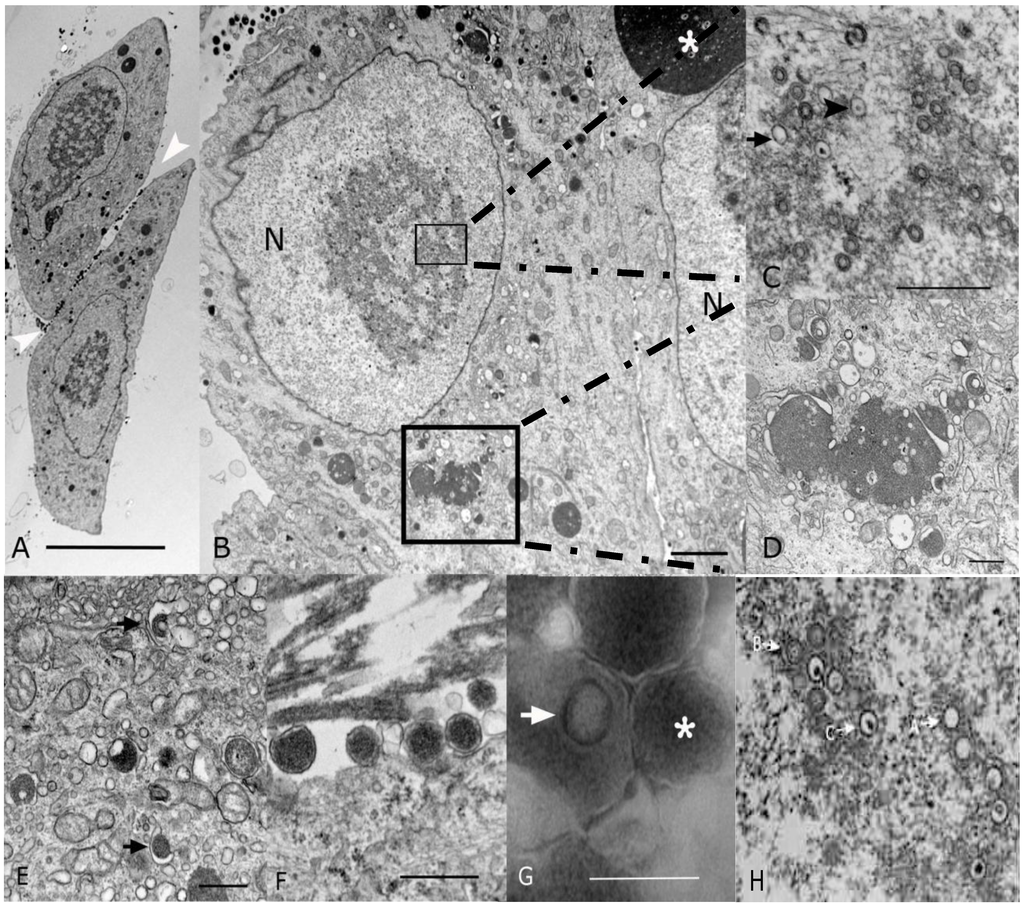
Figure 2.
Transmission electron microscopy (EM) microphotographs of fibroblasts infected with the CIDMTR Strain of GPCMV. (A) Plastic embedded preparation contrasted with uranyl acetate/lead citrate of two enlarged fibroblasts showing cell intranuclear and cytoplasmic viral inclusions and numerous dense bodies (white arrowheads) in the intercellular space (bar = 10 µm). (B) Magnification of another CIDMTR-infected cell revealed nuclear (N) viral replication and nucleocapsid assembly sites (small square), as well as a large maturation site (asterisk) and dense body formation in the cytoplasm (large square, bar = 2 µm). (C) Additional magnification in which replication and capsid assembly in the nucleus (small square) is appreciated; note empty capsids (arrow) and DNA containing nucleocapsids (arrowhead; bar = 0.5 µm). (D) Magnification of virus maturation sites within the cytoplasm (large square in panel (B)) reveals electron dense material formed by aggregation of nucleocapsids and cell organelles such as Golgi systems, endoplasmic reticulum and vesicles (bar = 0.5 µm). (E) Capsid is demonstrated becoming coated with tegument proteins and then acquiring its final envelope by budding into vesicles (arrows; bar = 0.5 µm). (F) Several dense bodies are present in the intercellular space (bar = 0.5 µm). (G) Negative contrast preparation of enveloped B-capsid (arrow) and dense body (asterisk, bar = 0.5 µm) is demonstrated. (H) EM of strain 22122-infected cells demonstrating virtually identical morphology; A, B and C capsids are identified as described in text.
2.3. Sequencing and Sequence Analysis
Viral DNA (P2) was purified from the CIDMTR strain after two passages in GLFs and subjected to sequence analysis using the MiSeq and PacBio platforms as described in Section 3. The CIDMTR genome was 232,778 nt in length. Overall sequence homology to strain 22122 (accession #KC503762.1) was 98%, consistent with the hypothesis suggested by immunoblotting that CIDMTR is a GPCMV and not a novel and distinct betaherpesvirus of guinea pigs. As discussed further in Section 4, a possibility existed that genetic diversity of GPCMV isolates could be restricted due to bottlenecks in the breeding history of Hartley guinea pigs, from which both 22122 and CIDMTR are derived. Historically, HCMV strains were differentiated on the basis of restriction pattern polymorphisms. Epidemiologically unrelated isolates, considered unique HCMV strains, exhibit predominantly identical restriction patterns with occasional unique fragments. To compare CIDMTR to 22122 in these terms, genomic sequences for each virus were used to generate in silico-predicted restriction patterns for three enzymes (EcoR I, Hind III, and Xba I). As shown in Figure 3a, the majority of fragments matched between the two virus genomes in all three predicted restriction enzyme patterns, while for each restriction pattern a few unique (unmatched) fragments were evident. These results are consistent with CIDMTR and 22122 being distinct GPCMV strains in a manner similar to that of HCMV strains.
Sequence comparison revealed three regions of striking discrepancies between the two genome sequences (Figure 3b). The first was located between nucleotides 199227 and 203071 of the 22122 sequence. Within this 3,845-bp region, 1,437 bps were entirely missing from the CIDMTR genome, while the remainder corresponded with a poorly conserved (50% average nucleotide conservation) 2,366-bp region in CIDMTR. As a result, one of the three gp138 family ORFs previously annotated in 22122 was missing from CIDMTR (Figure 3c). Curiously, this region corresponds to a hot-spot for spontaneous deletions that occur in response to over length genomes in GPCMV that also roughly corresponds to regions of instability/rearrangements in HCMV and rhesus cytomegalovirus [36].
The second region of discrepancy was located between nucleotides 221608 and 224743 of the CIDMTR sequence. Within this 3,136-bp region, 1,424 bps were entirely missing from the 22122 genome, and the remainder correspond with a poorly conserved (68% average nucleotide conservation) 1,684-bp region in 22122. Surprisingly, the additional sequences in CIDMTR encoded a fourth putative MHC class I homolog, annotated gp147.1, that lies adjacent to three MHC class I homologs (gp147, gp148, and gp149) that were previously identified in 22122 (Figure 3c).
The third region of discrepancy was located between nucleotides 230176 and 231697 of the 22122 sequence. Within this 1,522-bp region, 107 bps were missing from the CIDMTR genome, and the remainder correspond with a poorly conserved (51% average nucleotide conservation) 1,383-bp region in CIDMTR. This discrepant region partially overlaps gp149 and results in CIDMTR- and 22129-encoded gp149 proteins that lack amino acid homology in residues 1-134 (22122) or 1-164 (CIDMTR) but are highly conserved within the remaining C-terminal 494 residues.
Finally, like 22122, the CIDMTR genome was found to have direct terminal repeat sequences, although the CIDMTR repeats were slightly shorter (841 nt) than those in 22122 (953 nt).
Figure 3b highlights the regions of DNA sequence dissimilarity when the CIDMTR strain is compared to strain 22122. Figure 3c illustrates in map format the regions of genomic discontinuity. To confirm that these large-scale rearrangements did not arise during passage of virus in cell culture, PCR and sequence analyses were performed directly on both DNA purified from the original salivary gland homogenate (no tissue culture passage), and on DNA from the tissue culture-derived isolate (P0), with identical results (see Section 2.5 and Figure 4). The sequences of the PCR-generated fragments were identical to the Illumina/PacBio genome sequence, confirming that discontinuities with strain 22122 did not arise as an artifact of tissue culture passage. It therefore appears that, as it appears in its natural host, the CIDMTR strain lacks the gp138.1 gene found in 22122. Similarly, 22122 lacks the fourth MHC class I homolog gene (gp147.1) found in CIDMTR. As the original SG extract from which 22122 was cultured is no longer available, it is not possible to conduct similar studies to determine whether gp147.1 was absent from 22122 prior to its initial isolation in cell culture or was lost during subsequent in vitro/in vivo passage. Nevertheless, these results suggest that naturally occurring strains of GPCMV may be polymorphic with respect to the presence or absence of entire genes. Similar findings have been observed when comparing the Smith strain of murine cytomegalovirus to strains isolated from wild mice [37]. Further analyses of other primary virus isolates from “naturally infected” guinea pigs will be necessary to confirm this hypothesis and to determine if such polymorphisms are limited to specific genes, such as gp138.1 and perhaps gp147.1, or include others.
Table 1 summarizes the predicted ORFs identified in the CIDMTR strain. ORFs that are highly conserved in cytomegaloviruses are noted in the table in upper case/bold font (e.g., GP55). In contrast, ORFs appearing unique to GPCMV are noted in lower case (e.g., gp138). The “C” designation in Table 1 refers to the complimentary strand. Splicing sites are predicted based on previously published reports and these need empiric confirmation in future studies unless otherwise indicated (Figure 4 and Figure 5); exons are described as coding exons.
Amino acid sequences predicted from CIDMTR genes encoding conserved envelope glycoproteins were compared to those from strain 22122. There was striking sequence conservation of the gB homolog; although 13 SNPs were noted, only 2 coding changes were observed (overall identity of 99%). In contrast, sequence analyses of GP74 (gO) demonstrated the greatest degree of sequence divergence, with 303/374 (81% identity) noted, followed by GP75 (gH), with 613/726 (84%) identity compared to the 22122 strain. GP73, the homolog of HCMV glycoprotein N, a protein known to exhibit substantial sequence divergence across clinical isolates [19,38,39], demonstrated 92% identity between the CIDMTR and 22122 strains. These results confirm that CIDMTR represents a GPCMV strain distinct from 22122 as the amino acid sequences of gH, gO, and gN clearly diverged through gradual evolutionary processes and cannot be attributed to abrupt, limited events such as insertions, deletions, or duplications. This further implies that despite potential bottlenecks imposed by selective breeding, GPCMV, as it exists within commercial breeding colonies, exhibits significant strain diversity of a nature similar to that of HCMV. These findings suggest that GPCMV may provide a useful model to study the impact of naturally occurring strain variation on viral pathogenesis, particularly in the area of nonprimary infection and disease.
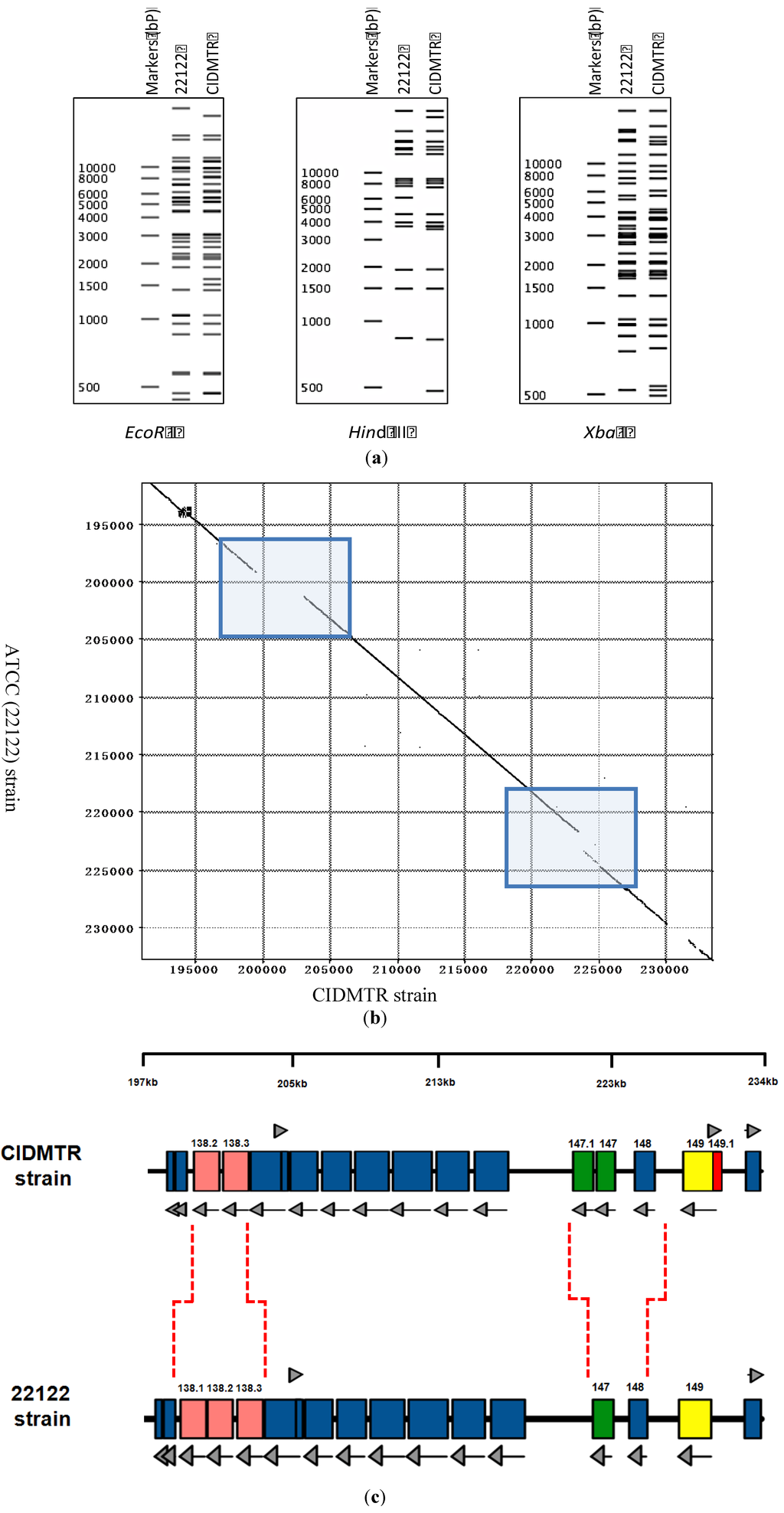
Figure 3.
Strain-specific genome structure differences between CIDMTR and 22122. (a) In silico analysis of predicted restriction endonuclease profiles for CIDMTR strain and 22122 (ATCC) strain with enzymes EcoRI, HindIII, and XbaI. Predictions and profile generated using program CLC Main Workbench 6 [40]. (b) Dot-matrix comparison of CIDMTR vs. 22122 genomic sequences. Two genome regions demonstrating sequence variability resulting in ORFs unique to each strain are indicated by shaded boxes. (c) Linear map comparison of the major areas of genome discontinuity. Linear maps prepared using XPlasMap [41]. Colored boxes in blue represent predicted conserved ORFs. Light red and green colored ORFs demonstrate discordance between the two strains. In the CIDMTR strain, an additional ORF not annotated in 22122, 149.1, is also noted (dark red). Dashed lines indicate the positions where the sequences differ. See Table 1 for a full list of sequenced gene annotations.

Table 1.
GPCMV Strain CIDMTR Predicted ORFs.
| GPCMV Open Reading Frames (ORFs): CIDMTR Strain | |||||
|---|---|---|---|---|---|
| ORF | Strand | Begin | End | Codons | Notes |
| gp1 | C | 12,464 | 12,769 | 101 | GPCMV MIP 1α; CC chemokine homolog |
| gp2 | 14,840 | 15,685 | 281 | Homology to MCMV M69a | |
| gp3 | C | 17,197 | 19,563 | 788 | Homology to THV T5b; US22 superfamily |
| gp4 | C | 20,829 | 21,152 | 107 | Homology to RCMV r136d |
| gp5 | C | 26,721 | 27,818 | 365 | Homology to MCMV m32a |
| GP23 | C | 33,299 | 34,501 | 400 | UL23 homolog; US22 gene superfamily |
| GP24 | C | 34,739 | 35,956 | 405 | UL24 homolog; US22 superfamily |
| GP25 | 36,542 | 38,194 | 550 | UL25 homolog; tegument protein | |
| GP26 | C | 38,360 | 39,043 | 227 | UL26 homolog |
| GP27 | C | 39,166 | 41,211 | 681 | UL27 homolog |
| GP28 | C | 41,311 | 42,378 | 355 | UL28 homolog; US22 superfamily |
| GP28.1 | C | 42,866 | 44,287 | 473 | UL28 homolog; US22 superfamily |
| GP29 | C | 44,653 | 46,623 | 656 | UL29 homolog; US22 superfamily |
| gp29.1 | C | 47,247 | 47,861 | 204 | US22 superfamily |
| GP30 | C | 49,082 | 50,800 | 572 | UL30 homolog |
| GP31 | 51,094 | 52,572 | 492 | UL31 homolog | |
| GP32 | C | 52,665 | 54,365 | 566 | UL32 homolog |
| GP33 | 54,585 | 55,868 | 427 | UL33 homolog; 7-TMR GPCR superfamily | |
| GP34 | 56,221 | 57,804 | 527 | UL34 homolog | |
| GP35 | 58,008 | 59,666 | 552 | UL35 homolog | |
| GP37 | C | 59,788 | 60,711 | 307 | UL37 homolog |
| GP38 | C | 61,068 | 61,988 | 306 | UL38 homolog |
| gp38.1 | C | 62,705 | 63,262 | 185 | Positional homolog of HCMV UL40 |
| gp38.2 | C | 63,620 | 64,933 | 437 | Positional homolog of HCMV UL41a |
| gp38.3 | C | 65,624 | 66,478 | 284 | Positional homolog of HCMV UL42 |
| gp38.4 | C | 66,997 | 67,362 | 121 | Homology to RCMV r42d |
| GP43 | C | 67,951 | 68,964 | 337 | UL43 homolog |
| GP44 | C | 68,952 | 70,175 | 407 | UL44 homolog |
| GP45 | C | 70,887 | 73,673 | 928 | UL45 homolog |
| GP46 | C | 73,779 | 74,867 | 362 | UL46 homolog |
| GP47 | 74,674 | 77,787 | 1037 | UL47 homolog | |
| GP48 | 77,784 | 84,155 | 2123 | UL48 homolog | |
| GP48.2 | C | 84,238 | 84,468 | 76 | UL48a homolog |
| GP49 | C | 84,479 | 86,119 | 546 | UL49 homolog |
| GP50 | C | 86,088 | 87,158 | 356 | UL50 homolog |
| GP51 | C | 87,281 | 87,580 | 99 | UL51 homolog; terminase subunit |
| GP52 | 87,900 | 89,480 | 526 | UL52 homolog | |
| GP53 | 89,473 | 90,459 | 328 | UL53 homolog | |
| gp53.1 | C | 90,196 | 90,537 | 113 | Homology to RhCMV rh86; YP_068179.1 |
| GP54 | C | 90,551 | 93,904 | 1117 | UL54 homolog; DNA polymerase |
| GP55 | C | 93,947 | 96,652 | 901 | UL55 homolog; glycoprotein B |
| GP56 | C | 96,549 | 98,816 | 755 | UL56 homolog; terminase subunit |
| GP57 | C | 98,967 | 102,650 | 1227 | UL57 homolog |
| gp57.1 | C | 104,411 | 104,926 | 171 | Homolog to RCMV r23.1d |
| GP69 | C | 108,260 | 111,430 | 1056 | UL69 homolog |
| GP70 | C | 112,136 | 115,339 | 1067 | UL70 homolog; helicase-primase |
| GP71 | 115,338 | 116,114 | 258 | UL71 homolog | |
| GP72 | C | 116,277 | 117,350 | 357 | UL72 homolog; dUTPase |
| GP73 | 117,432 | 117,833 | 133 | UL73 homolog; glycoprotein N | |
| GP73.5ex1 | 117,860 | 117,869 | 3 | ||
| GP73.5ex2 | 119,005 | 119,198 | 64 | ||
| GP74 | C | 117,780 | 118,904 | 374 | UL74 homolog; glycoprotein O |
| GP75 | C | 119,349 | 121,529 | 726 | UL75 homolog; glycoprotein H |
| GP76 | 121,694 | 122,533 | 279 | UL76 homolog | |
| GP77 | 122,247 | 124,091 | 614 | UL77 homolog | |
| GP78 | 124,473 | 125,717 | 414 | UL78 homolog; 7-TMR GPCR superfamily | |
| GP79 | C | 125,914 | 126,861 | 315 | UL79 homolog |
| GP80 | 126,722 | 129,031 | 769 | UL80 homolog; CMV protease | |
| GP80.5 | 127,610 | 129,031 | 473 | UL80.5 | |
| GP82 | C | 129,324 | 130,889 | 521 | UL82 homolog; pp71 |
| GP83 | C | 131,109 | 132,809 | 566 | UL83 homolog; pp65 |
| GP84 | C | 133,051 | 134,487 | 478 | UL84 homolog |
| GP85 | C | 134,785 | 135,696 | 303 | UL85 homolog |
| GP86 | C | 135,977 | 140,026 | 1349 | UL86 homolog |
| GP87 | 140,407 | 143,328 | 973 | UL87 homolog | |
| GP88 | 143,231 | 144,499 | 422 | UL88 homolog | |
| GP89ex2 | C | 144,545 | 145,675 | 376 | UL89 homolog; terminase subunit, exon 2 |
| GP91 | 146,102 | 146,365 | 87 | UL91 homolog | |
| GP92 | 146,362 | 146,991 | 209 | UL92 homolog | |
| GP93 | 146,957 | 148,732 | 591 | UL93 homolog | |
| GP94 | 148,644 | 149,681 | 345 | UL94 homolog | |
| GP89ex1 | C | 150,032 | 150,913 | 291 | UL89 homolog; terminase subunit, exon 1 |
| GP95 | 150,967 | 152,235 | 422 | UL95 homolog | |
| GP96 | 152,468 | 152,830 | 120 | UL96 homolog | |
| GP97 | 152,910 | 154,727 | 605 | UL97 homolog; protein kinase | |
| GP98 | 154,747 | 156,531 | 594 | UL98 homolog; alkaline nuclease | |
| GP99 | 156,444 | 156,965 | 173 | UL99 homolog; pp28 | |
| gp99.1 | 157,155 | 157,769 | 204 | Homology to RCMV r4d | |
| GP100 | C | 157,278 | 158,327 | 349 | UL100 homolog; glycoprotein M |
| GP102 | 158,657 | 160,942 | 761 | UL102 homolog | |
| GP103 | C | 161,127 | 161,852 | 241 | UL103 homolog |
| GP104 | C | 161,815 | 163,908 | 697 | UL104 homolog; portal |
| GP105 | 163,748 | 166,531 | 927 | UL105 homolog; helicase-primase | |
| GP112ex1 | 176,745 | 177,498 | 315 | UL112 homolog; replication accessory, ex 1 | |
| GP112ex2 | 177,606 | 177,782 | UL112 homolog; replication accessory, ex 2 | ||
| GP112ex3 | 178,115 | 178,131 | UL122 homolog; replication accessory, ex 3 | ||
| GP114 | C | 179,126 | 179,920 | 264 | UL114 homolog; uracil glycosylase |
| GP115 | C | 179,986 | 180,762 | 258 | UL115 homolog; glycoprotein L |
| GP116 | C | 180,755 | 181,654 | 299 | Homology to THV t116b; Fc receptor//Ig |
| GP117 | C | 181,877 | 183,262 | 461 | UL117 homolog |
| gp119.1 | C | 184,418 | 185,167 | 249 | Similar to MCMV in ACE95619.1 |
| GP121.2 | C | 185,160 | 185,834 | 224 | Betaherpesvirus B7D8, accession AFK83957 |
| GP121.4 | C | 186,299 | 187,174 | 291 | UL121 homolog; Tupaia t121.4, NP_116476 |
| GP122ex3 | C | 187,993 | 189,677 | 677 | UL122 homolog; HCMV IE2 |
| GP122ex2 | C | 191,079 | 191,311 | ||
| §GP122ex1 | C | 191,403 | 191,518 | ||
| GP123ex3 | C | 189,907 | 190,985 | 475 | UL123 homolog; HCMV IE1 |
| GP123ex2 | C | 191,079 | 191,311 | ||
| §GP123ex1 | C | 191,403 | 191,518 | ||
| GP128 | 195,400 | 196,455 | 351 | Similar to Bat HSV B126; US22 Family | |
| gp130 | 196,655 | 196,999 | 114 | ||
| GP129ex3 | C | 196,432 | 196,690 | 178 | Homolog of HCMV UL128 |
| GP129ex2 | 196,768 | 196,890 | |||
| GP129ex1 | C | 196,974 | 197,128 | ||
| GP131ex2 | C | 197,133 | 197,469 | 191 | Homolog of HCMV UL130 |
| GP131ex1 | C | 197,550 | 197,788 | ||
| GP133 | C | 197,788 | 198,174 | 128 | Homolog of HCMV UL131 |
| GP134 | C | 198,268 | 198,951 | 227 | |
| gp138.2 | C | 199,367 | 200,875 | 502 | |
| gp138.3 | C | 201,090 | 202,607 | 505 | |
| gp139 | C | 202,706 | 204,793 | 695 | THV T5; US22 superfamily |
| gp140 | 204,522 | 204,929 | 135 | Homology to CCMV UL132 | |
| gp141 | C | 205,053 | 206,660 | 535 | HCMV US23; US22 superfamily |
| gp142 | C | 206,928 | 208,622 | 564 | HCMV US24; US22 superfamily |
| gp143 | C | 208,875 | 210,866 | 663 | THV T5; US22 superfamily |
| gp144 | C | 211,109 | 213,403 | 764 | US26; US22 gene superfamily |
| gp145 | C | 213,677 | 215,575 | 632 | HCMV IRS1/TRS1; US22 superfamily |
| gp146 | C | 215,932 | 217,914 | 660 | HCMV IRS1/TRS1; US22 superfamily |
| gp147.1 | C | 221,739 | 222,935 | 398 | MHC class I homolog |
| gp147 | C | 223,124 | 224,218 | 364 | MHC class I homolog |
| gp148 | C | 225,379 | 226,560 | 393 | MHC class I homolog |
| gp149 | C | 228,236 | 230,209 | 657 | MHC class I homolog |
| gp149.1 | 230,163 | 230,465 | 100 | Unique ORF sequence in CIDMTR | |
§ An observation of note is the codon, ATA, spanning nucleotides 191516–191518 (minus strand). This codon is ATG in strain 22122, which was annotated as the start codon for both IE2 (GP122; exons 3, 4 and 5) and IE1 (GP123; exons 3, 4 and 6) [42,43,44]. Whether alternative splicing is occurring in the CIDMTR strain, or the start codon for the IE1/IE2 proteins is different between the two strains of GPCMV (i.e., in exon 4, not exon 3), requires further evaluation; this is described in greater detail in Section 2.4.
2.4. Sequence Characterization of DNA from Original SG Homogenate in Region of IE1/2 Start Codon
A surprising finding from the DNA sequence analysis was the finding of an ATA codon as the putative start codon of the IE1/IE2 protein product [42]. To examine this issue, sequence analyses of PCR-amplified DNA from the original salivary gland homogenates, as well as salivary gland homogenates from further in vivo passages, were undertaken. DNA from the original salivary gland homogenate that was the source of the CIDMTR isolate was compared to DNA from GPL cells infected with CIDMTR following cell culture passage. The PCR was done with the primer pair designated IE splice 3' P1 (5'-TGCGAAGCGATCTCTCTCAAC-3') and IE splice 5' P1 (5'-GTGGTTGTACGTGTCGTCGTCA-3'), which was predicted to produce an 864-bp product from CIDMTR DNA. The purified DNA was cloned and three clones from each reaction were sequenced. This analysis indicated that the original salivary gland source of the CIDMTR strain contained viral DNA that encoded an ATG codon (as does 22122), the putative start codon for the IE1/IE2 protein product, but that the tissue cultured-derived CIDMTR strain, following two passages in fibroblasts, had a DNA sequence corresponding to an ATA codon.
To further examine splicing of the IE region from the CIDMTR strain (tissue culture-derived; CIDMTR-TC) virus, reverse-transcriptase PCR was performed on RNA harvested at immediate early times post-infection. RNA was extracted from GPL cells infected with 22122 or CIDTMR at 4 hours post-inoculation cDNA was synthesized from 1 μg of total RNA. Conventional PCR was carried out using cDNA as template. PCR used primer pair IE exon4 F1 (5'-CCGCATTTTCTGAGGGTGTT-3') and IE exon1 R1 (5'-CATGCCAGTTCCCTGTGCTG-3'), and primer pair IE exon5 F1 (5'- GGAAGATGTCCACTTGGAAG-3') and IE exon2 R1 (5'- ACGTAGCCGAGAAGTAAAGT-3'). The expected product sizes for primer pair IE exon4 F1/ IE exon1 R1 and IE exon5 F1/IE exon2 R1 were 473 bp and 508 bp, respectively.
A product of the expected size was purified, cloned into pCR2.1, and multiple clones with inserts from each reaction were sequenced. Notably, all CIDMTR-derived clones contained ATA at the putative (collinear) IE1/IE2 gene product start codon, confirming the DNA sequencing analysis. In contrast, all the 22122 clones contained an ATG at this position. The observed exon junctions were as described in the 22122 strain. The ATG codon in exon 3 was originally annotated as the putative IE1/2 start codon in 22122 [41] and the DNA sequence analysis of DNA from the CIDMTR strain purified directly from the salivary gland homogenate also demonstrated an ATG codon in exon 3. Thus, the finding of this ATA by DNA and RT-PCR sequencing in tissue culture-passaged CIDMTR virus was surprising. Since IE1/2 is abundantly expressed and ATA is known as an inefficient initiator of protein translation, we looked for ATGs that might serve as alternative start sites. Tissue culture‑passaged CIDMTR strain may employ a start codon in exon four for the IE1 and IE2 gene products (Figure 4), although the Kozak consensus sequence of this putative start codon (T at −3, A at +4) is relatively weak [45].
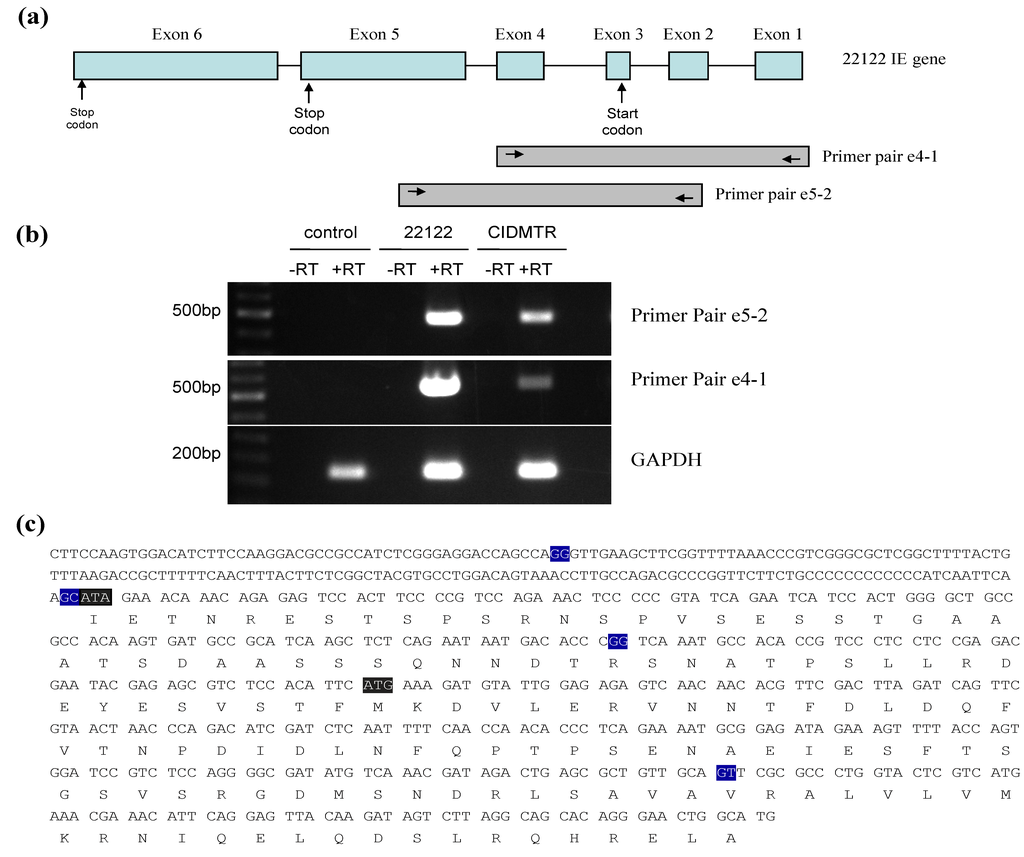
Figure 4.
RT-PCR mapping of CIDMTR strain and 22122 strain splice sites. (a) Cartoon representation of the IE1/2 gene locus illustrating positions of primer pairs used for RT‑PCR. Introns are indicated by straight lines and exons are blue boxes, although exon 1 and 2 are non-coding. (b) Results of RT-PCR reactions e4-1, e5-2, and GAPDH (control) using RNA from uninfected cells or cells infected with 22122 or CIDMTR. (c) RT-PCR consensus sequence of CIDMTR strain. Exon junctions are highlighted in blue. The first gray highlighted sequence is an ATA codon; this is an ATG codon and the putative start codon for the 22122 IE1/2 proteins, but is not conserved in the CIDMTR sequence in tissue culture passaged virus. The second gray highlighted sequence may therefore represent the putative start codon for CIDMTR TC IE1/2.
To further define IE transcription from the CIDMTR-TC virus, 5' RACE was performed. RNA was extracted and cDNA was synthesized with a specific sequence attached to the 5' UTR for capped mRNAs. After ligation of a specific RNA oligo to the 5'UTR of mRNAs, cDNA was synthesized using oligo-dT and random hexamers. The cDNA was used as template for a first round of PCR using GeneRacer 5' Primer and primer IE2 P5 (5'-GGCGTCAATGGGCTCGGGTTTGAT-3'). Two nested PCR reactions were then performed with the GeneRacer 5' Nested Primer, per the manufacturer’s specifications, and IE2 P5 or IE exon3 P1 (5'-GGCAGCCCCAGTGGATGATTCTGATA-3') (Figure 5b). Purified DNA was cloned and sequenced. Results matched the previously described splice sites for strain 22122 [42]. Using this primer combination, the RT-PCR product was 734 nt in size (5 nt longer than the homologous ATCC/22122 cDNA). The substitution of ATA for the putative ATCC/22122 IE2/3 start codon was once again noted. We did not observe other in-frame start codons for the CIDMTR strain IE1/2 other than the one identified in exon 4. Further experiments will be required to identify the start codon for IE1/2; possibly, both 22122 and CIDMTR strains use the start codon in exon 4, in spite of its suboptimal Kozak consensus. Alternatively, growth of the CIDMTR strain in fibroblasts may select for mutation of the exon 3 start codon, even upon minimal passage. Development of guinea pig epithelial/endothelial cell for isolation and passage of CIDMTR virus is an important future priority, to determine whether in vivo and in vitro passage result in different selective pressure on viral sequences.
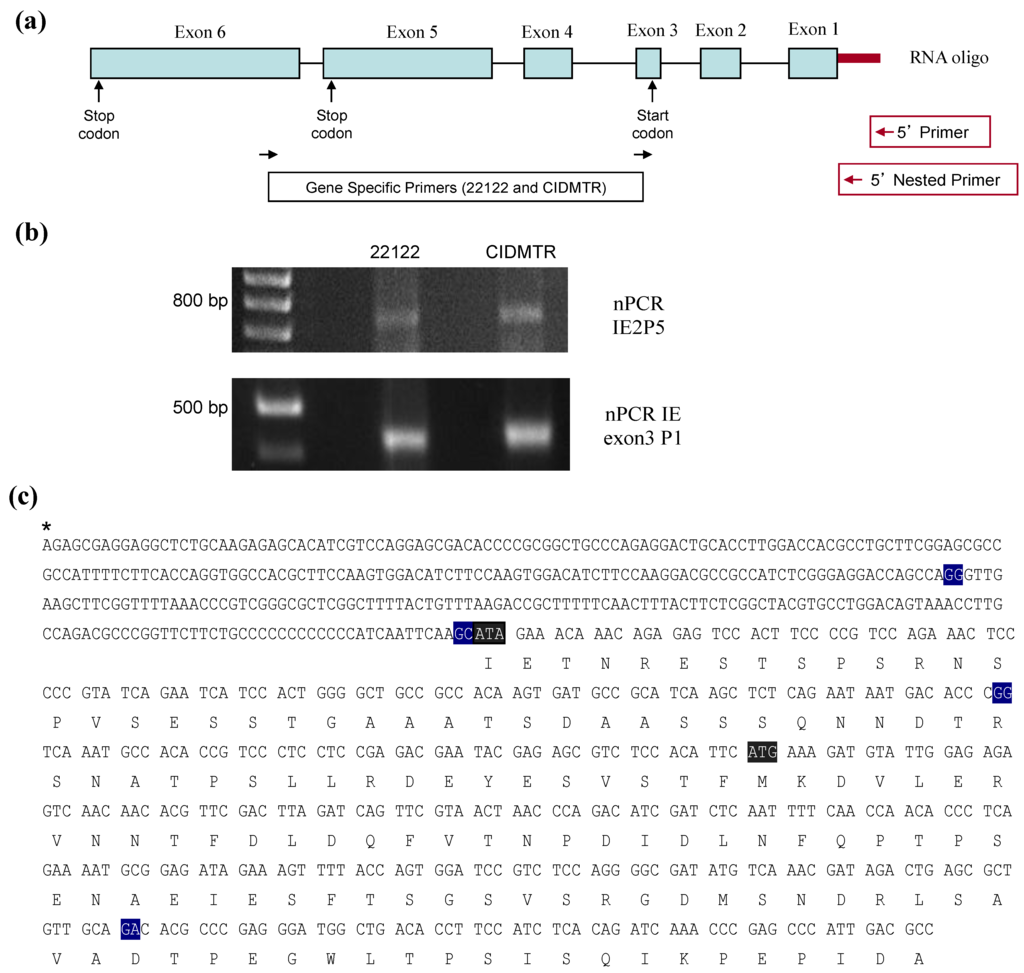
Figure 5.
5' RACE analysis of CIDMTR IE region. (a) Schematic of IE gene indicating primers pair used in RACE. (b) nPCR gels for 22122 and CIDMTR using RNA purified under IE conditions. The CIDMTR RACE product is 734 nt compared to 729 nt for the 22122 strain, explaining its slightly higher migration by gel electrophoresis. (c) RACE consensus sequence of CIDMTR IE. Exon junctions highlighted in blue. First gray highlighted sequence is co-linear to the start codon for 22122; second highlighted sequence represents putative start codon for CIDMTR IE1/2.
2.5. PCR Confirmation of GPCMV-CIDMTR Genome Structure
To confirm the structure of the CIDMTR strain compared to the 22122 strain, PCR was performed on viral DNA from both strains, using primers spanning the mismatched regions observed in the sequence analysis comparisons (Figure 3b, boxed/shaded regions). Two primers pairs were used for each region of mismatch (mismatch region 1 and mismatch region 2). PCR was performed using primer pairs mismatch-region 1 F1/R1 and mismatch-region 1 F2/R2, that amplify a ~4-kb region for the 22122 strain, but a ~2.5-kb region for the CIDMTR strain. The amplification region using primer pairs mismatch‑region 2 F1/R1 or mismatch-region 2 F2/R2 was predicted to be ~2.2 kb for the 22122 strain and ~3.7 kb for the CIDMTR strain. Primer sequences are indicated in Table 2. The results of these experiments confirmed that the genome configuration was precisely as predicted from the deep sequence analysis. Moreover, the PCR was also performed on DNA purified directly from the original salivary gland homogenate from which the CIDMTR isolate was obtained (data not shown). These results confirmed that the insertions and deletions identified by sequencing (Figure 3c) did not arise as an artifact of limited passage of virus in cell culture. The PCR results are shown in Figure 6.

Table 2.
Primer Sequences.
| Primer Name | Primer Sequence |
|---|---|
| Mismatch Region 1 F1 | 5'-GTGAGACGTAAGAATAGCTTGC-3' |
| Mismatch Region 1 F2 | 5'-GATCCTTAGACTCTATCACGG-3' |
| Mismatch Region 1 R1 | 5'-GTGTTGTCACAATTGGCACATG-3' |
| Mismatch Region 1 R2 | 5'-ACATGGTCACGACAGAATC-3' |
| Mismatch Region 2 F1 | 5'-GTGGACAGGATCCCCAAATT-3' |
| Mismatch Region 2 F2 | 5'-CCAAATTTCTGTCGTCGGCG-3' |
| Mismatch Region 2 R1 | 5'-TGTTTCCGTGTCTGTCTCCGT-3' |
| Mismatch Region 2 R2 | 5'-GTCTTAGCCCGAGACCTTC-3' |
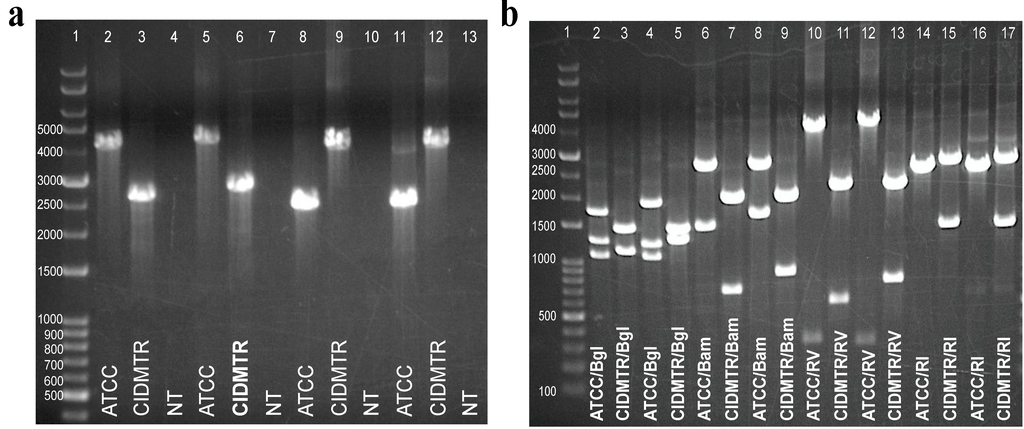
Figure 6.
Strain-specific PCR assays differentiate ATCC 22122 strain of GPCMV from CIDMTR strain. (a) Primers were designed to amplify across discontinuous/unique regions as demonstrated in Figure 3b. Lane 1, kb ladder; lane 2, mismatch region 1 F1/R1, ATCC; lane 3, mismatch region 1, F1/R1, CIDMTR; lane 4, mismatch region 1, no template control; lane 5, mismatch region 1 F2/R2, ATCC; lane 6, mismatch region 1 F2/R2, CIDMTR; lane 7, mismatch region 1 F2/R2, no template control; lane 8, mismatch region 2 F1/R1, ATCC; lane 9, mismatch region 2 F1/R2, CIDMTR; lane 10, mismatch region 2 F1/R1, no template control; lane 11, mismatch region 2 F2/R2, ATCC; lane 12, mismatch region 2 F2/R2, CIDMTR; lane 13, mismatch region 2 F2/R2, no template control. (b) Restriction polymorphisms were as predicted by DNA sequence. PCR amplification products were digested with enzymes as indicated. Lane 1, kb ladder; lane 2, mismatch region 1 F1/R1 ATCC, Bgl 2; lane 3, mismatch region 1 F1/R1 CIDMTR, Bgl 2; lane 4, mismatch region 1 F2/R2 ATCC, Bgl 2; lane 5, mismatch region 1 F2/R2 CIDMTR, Bgl 2; lane 6, mismatch region 1 F1/R1 ATCC, BamH I; lane 7, mismatch region 1 F1/R1 CIDMTR, BamH I; lane 8, mismatch region 1 F2/R2 ATCC, BamH I; lane 9, mismatch region 1 F2/R2, BamH I; lane 10, mismatch region 1 F1R1 ATCC, EcoR V; lane 11, mismatch region 1 F1/R1 CIDMTR, EcoR V; lane 12, mismatch region 1 F2/R2 ATCC, EcoR V; lane 13, mismatch region 1 F2/R2, EcoR V; lane 14, mismatch region 2 F1R1 ATCC, EcoR I; lane 15, mismatch region 2 F1R1 CIDMTR, EcoR I; lane 16, mismatch region 2 F2/R2 ATCC, EcoR I; lane 17, mismatch region 2 F2/R2, EcoR I.
2.6. Infection In Vivo and Development of a Real-Time PCR Assay for Detection of CIDMTR DNA
To evaluate DNAemia and end-organ infection in the course of in vivo studies, a strain-specific real-time PCR assay was developed in order to differentiate the GPCMV-CIDMTR strain from the 22122 strain. This assay focused on the amplification of sequences corresponding to the CIDMTR strain GP147.1 ORF (Table 1), since this sequence is absent in the 22122 strain. A GPCMV 147.1 specific PCR primer pair, consisting of CIDMTR147.1_464F (5'-ATGCAACATAGCGTGCTGAC-3') and CIDMTR147.1_583R (5'-GGGACAAAAGCACGATGAAC-3') was designed and utilized for the real-time PCR assay (described in detail in 3.5). These primers amplified a 120 bp region of the 147.1 gene specific for the CIDMTR strain. DNA was extracted from either 100 μL citrated blood or from fresh frozen tissues samples, as described in the Methods section. For quantitative PCR, both previously validated primers for the GP83 gene [sequences shared by both strains] and novel primers for the GPCMV 147.1 gene [sequences only found in the CIDMTR strain] were used for real‑time PCR assay. There was strong concordance between the viral load estimates identified by the gp147.1 and GP83 real-time primers (data not shown). As a negative control, GPCMV 147.1 primers were used in several PCR assays of 22122 DNA, with consistently negative results (data not shown).
Next, twelve young, GPCMV-seronegative outbred Hartley guinea pigs were divided into two groups of six/group. Each group was inoculated with CIDMTR strain virus (p1) at a dose of 1 × 105 pfu, administered subcutaneously, as described in Section 3. Group 1 (n = 6) was treated with 200 mg/kg cyclophosphamide on day −1 and 50 mg/kg on day +6 following viral inoculation as described previously [32]; group 2 (n = 6) was sham-treated (PBS only). Whole blood and sera samples were collected on day 0, 3, 7, and 21. Animals were sacrificed on day 21 and tissue, including lung, liver, spleen, and brain, were collected for PCR analysis. In group 1, 5/6 animals were DNAemic, peaking at day 7 (mean, 3.2 +/− 0.3 log10 genomes/mL) while in group 2, the prevalence of DNAemia was lower (3/6 animals; 2.5 +/− 0.35 log10 genomes/mL, p = 0.06 compared to group 1). Viral DNA was most readily recovered from spleen upon dissection at day 21 post-infection. All 6 animals from each group had recoverable CIDMTR strain DNA in the spleen. Total spleen viral load in group 1 was 2.4 +/− 0.07 log10 copies/mg and was 2.4 +/− 0.1 log10 copies/mg in group 2 (p = NS compared to group 1).
The ability of the CIDMTR strain to infect pregnant animals previously inoculated with the 22122 strain was next assessed. A total of 6 female seronegative Hartley guinea pigs received a primary infection with SG homogenate GPCMV (22122) at a dose of 1 × 104 pfu, administered subcutaneously. A parallel group of animals were sham-treated (PBS only). Then, 5 weeks after inoculation, the animals in both groups were mated with GPCMV-seronegative breeders. Dams originally inoculated with the SG virus (22122 strain) were documented to undergo seroconversion; in addition, all were demonstrated to have low-level DNAemia after inoculation with 22122 virus (data not shown). Approximately 5 weeks after the initiation of mating, group 1 was challenged subcutaneously with CIDMTR strain at a titer of 6 × 105 pfu. A total of 19 pups were born in the experimental group; all pups were liveborn. When pup tissues were analyzed, 4/19 (21%) of these animals had recoverable CIDMTR strain-specific DNA isolated from the salivary gland. One of these animals also had detectable DNA in the brain.
3. Experimental Section
3.1. Cells, Virus and DNA Preparation
GPCMV (strain 22122, ATCC VR682), and CPCMV/CIDMTR were propagated on guinea pig lung fibroblast cells (GPL; ATCC CCL 158) in F-12 medium supplemented with 10% fetal calf serum (FCS; Gibco-BRl), 10,000 IU of penicillin/liter, 10 mg of streptomycin/liter (Gibco-BRL), and 7.5% NaHCO3 (Gibco-BRL). When the P1 passaged CIDMTR virus exhibited extensive CPE in GPL cell culture, cells and virions were pelleted. Aliquots were fixed in phosphotungstic acid for EM studies (see below) and the remainder washed in 10 mM Tris (pH 8.0)/1 mM EDTA (TE), and lysis buffer (200 mM NaCl, 2% SDS, 200 μg/mL proteinase K, in TE), was added. Cells and virions were gently suspended, inverted, and incubated at 68 °C overnight. Following transfer to a 37 °C water bath, three successive phenol-chloroform extractions were performed, followed by ethanol precipitation.
3.2. Transmission Electron Microscopy
Cells were fixed in 1 mL of 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer and post fixed with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (all reagents from Electron Microscopy Sciences, Hatfield, PA, USA). After three washes in distilled water, samples were dehydrated using a 25%–100% ethyl alcohol gradient. Samples were then infiltrated with 2:1 ethanol: Embed 812 resin (Electron Microscopy Sciences, Hatfield, PA, USA) for 1 hour and subsequently transferred to a 1:2 ethanol: Embed 812 resin mixture for 1 hour. Cells were further infiltrated with 100% resin and were embedded and incubated at 58 °C for 24 hours to polymerize the resin. Embedded samples were trimmed and sectioned on a Leica UC6 Ultramicrotome (Leica Microsystems, Vienna, Austria). Thin sections (60–70 nm) were obtained and collected on a 200 mesh copper grid (Electron Microscopy Sciences, Hatfield, PA, USA) using a perfect loop. Grids were contrasted with 5% uranyl acetate for 20 minutes and Santos’ lead citrate for 6 minutes. For negative contrast, virions and dense bodies were collected from P1 supernatants of infected fibroblasts, transferred to airfuge tubes (Beckman-Coulter, Brea, CA, USA), and centrifuged at 30 PSI using an airfuge (Beckman-Coulter, Brea, CA, USA) for 20 minutes on parafilm and formvar coated copper grids (Electron Microscopy Sciences, Hatfield, PA, USA). Excess liquid was wicked and the grids were stained with 1% phosphotungstic acid for one minute. All sections were observed under JEOL 1200 EX II transmission electron microscope (JEOL LTD, Tokyo, Japan). Images were obtained using a Veleta 2K × 2K camera with iTEM software (Olympus SIS, Munster, Germany) [46].
3.3. Deep Sequencing and Sequence Analyses
For sequence analysis of CIDMTR viral DNA, virions were purified as described previously [44], and lysis buffer (200 mM NaCl, 2% SDS, and 200 µg/mL proteinase K in Tris-EDTA [TE]) was added. Following incubation at 68 °C overnight, three phenol-chloroform extractions were performed, followed by ethanol precipitation of viral DNA. Genomic sequencing was performed using Illumina MiSeq and Pacific Biosciences PacBio RS platforms. Approximately 5.2 million 151-bp paired-end MiSeq reads were generated at the University of Minnesota’s Biomedical Genomics Center with a nominal insert size of 400 bp. Removal of low quality reads and PhiX sequence resulted in a set of 4.0 million cleaned reads, approximately 11,000× coverage. Initial scaffolds were generated from the cleaned Illumina reads using the ABySS assembler (version 1.3.4) [47]. Scaffold quality was assessed manually by comparison with the reference strain, 22122 [44,48,49], and by remapping the reads using Bowtie 2 and scrutinizing local coverage and consensus using Tablet [50] and SAMtools [51]. Special attention was paid to correct alignment and orientation of the paired ends. Regions of weak coverage and scaffold gaps were identified and closed either by manual local assembly [52] or by Sanger sequencing. Independent validation of the pseudomolecule was also performed using the longer PacBio RS reads, validating the Sanger sequencing and manual assembly. SMRT Analysis software [53] produced 998 high quality (“corrected”) reads ranging between 509–15,898 bp, median 6,257 bp (approximately 27× coverage), as well as another set of scaffolds. These data were used to evaluate the Illumina assembly, specifically its structural correctness, and to correct misassembled repeat regions. The resulting complete genome was deposited with the EMBL Nucleotide Sequence Database (accession number HG531783).
3.4. RT-PCR and RACE Analyses
Reverse-transcriptase PCR was performed on RNA harvested at immediate early times post‑infection. RNA was extracted from GPL cells infected with 22122 or CIDTMR, at 4 hours post‑inoculation, using the RNeasy mini kit (Qiagen, Hilden, North Rhine-Westphalia, Germany) according to the manufacturer’s instructions. RNA was treated with RNase-free DNase Set (Qiagen) while in the column according to manufacturer’s instructions. cDNA was synthesized from 1 μg of total RNA using Quantitect Reverse Transcription kit (Qiagen). Conventional PCR was carried out using cDNA as template and AmpliTaq Gold Fast PCR Master Mix (Invitrogen, Carlsbad, CA, USA). Primers are as described in Section 2.4. The PCR products were run in a 0.7% agarose gel. A band of the expected size was cut form the gel and purified using QIAquick Gel Extraction Kit (Qiagen). The purified DNA was cloned into pCR2.1 (Invitrogen) using the TA Cloning Kit (Invitrogen). Three of clones with inserts from each reaction were sequenced with T3 promoter, M13 Reverse and the PCR primers.
For 5' RACE analysis, RNA was used as template for cDNA synthesis using the GeneRacer kit (Invitrogen) in order to generate a cDNA with a specific sequence attached to the 5' UTR for only mRNAs that were capped. After ligation of a specific RNA oligo to the 5'UTR of mRNAs, cDNA was synthesized using a combination of oligo-dT and random hexamers. The cDNA was used as template for a first round of PCR using GeneRacer 5' Primer and primer IE2 P5 (5'-GGCGTCAATGGGCTCGGGTTTGAT-3'). In order to increase the specificity and quantity of the PCR product, nested PCR (nPCR) was performed in a dilution (1:20) of the PCR product. Two nPCR reactions were performed with the GeneRacer 5' Nested Primer, per the manufacturer’s specifications, and IE2 P5 or IE exon3 P1 (5'- GGCAGCCCCAGTGGATGATTCTGATA-3'). A band of the expected size was cut form the gel and purified using QIAquick Gel Extraction Kit (Qiagen). The purified DNA was cloned into pCR4-TOPO (Invitrogen) using the TOPO TA Cloning. Clones were analyzed for the present of the insert by digestion with EcoR I. Three of the clones with insert from each reaction were sequenced with T3 promoter, M13 forward (−20) and the PCR primers.
3.5. GPCMV-CIDMTR Strain PCR Assay
For confirmation of viral genome structural differences noted by deep sequencing, PCR was performed for each virus using primers indicated in Table 2. The PCR reaction was performed in a 50 μL of total volume using GoTaq long PCR Master Mix from Promega and 1.0 μM primers. The DNA template was total genomic and viral DNA extracted from GPL cells infected with 22122 or CIDMTR tissue culture strain. The conditions for the PCR were: initial denaturation at 95 °C for 2 min, followed by 95 °C for 30 s, 53 °C for 30 s, 72 °C for 4 min for a total of 35 cycles, and elongation at 72 °C for 10 min. The PCR product (4 μL) was subjected to electrophoresis in a 0.7% agarose gel. The PCR product from mismatch 1 F2/R2 and mismatch 2 F2/R2 were purified from the gel using the Geneclean® II kit (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s instructions. The cleaned PCR product was then sequenced by Sanger sequencing (Functional Biosciences, Madison, WI, USA). Purified PCR products were also subjected to restriction endonuclease comparisons as indicated in Figure 5b.
The real-time PCR assay focused on the amplification of sequences corresponding to the CIDMTR strain gp147.1 ORF (Table 1), since this sequence is absent in the 22122 strain sequence. A GPCMV gp147.1 specific real-time PCR primer pair, consisting of CIDMTR147.1_464F (5'-ATGCAACATAGCGTGCTGAC-3') and CIDMTR147.1_583R (5'-GGGACAAAAGCACGATGAAC-3') was designed and utilized for the real-time PCR assay. These primers amplified a 120 bp region of the gp147.1 gene specific for the CIDMTR strain. The specific hydrolysis probe used for detection was CIDMTR147.1_494P (FAM-GTGTTCGTGTCCTTGATCGTACGCA-BHQ1). A second GPCMV 147.1 specific primer pair, CIDMTR147.1_225F (5'-AATGGTTCGCTACGGACATC-3') and CIDMTR147.1_368R (5'-CGGACAACGGAACATACTTG-3') was also utilized in real-time PCR assays. These primers amplified a 144 bp region of 147.1 specific for the CIDMTR strain. The specific hydrolysis probe used for detection with this primer pair was CIDMTR147.1_262P (FAM‑TTCCTCGACGAAGCTCGCGGTATAAT-BHQ1). In each instance, the PCR reactions were performed in a 25 μL reaction, using LightCycler 480 Probes Master from Roche (Penzberg, Bavaria, Germany); as well as 0.4 μM primers, 0.1 μM probe and 0.4 u/μL of UNG. PCR was performed using the LightCycler 480 Real-Time PCR System (Roche) under the following conditions: initial denaturation at 95 °C for 10 min, followed by 95 °C for 10 s, 56 °C for 15 s, 72 °C for 10 s for a total of 45 cycles, then a final hold step at 40 °C. The first primer pair (464F and 583R) was chosen for detection of viral genome for in vivo studies. Data were analyzed with the LightCycler Data Analysis Software (version 1.5; Roche) [54] using standard curves generated using serial dilutions of plasmid pCR2.1 with gp147.1 at known concentrations. Negative results were arbitrarily assigned a level of 50 for the purpose of statistical comparisons, based upon limit-of-detection analyses observed in other real-time PCR experiments [55].
3.6. Animal Challenge Studies
All animal studies were performed with the approval of the University of Minnesota Institutional Animal Care and Use Committee (IACUC). Some animals were immune suppressed at day −1 (200 mg/kg) and day +6 (50 mg/kg) with cyclophosphamide delivered by intraperitoneal injection. For experiments described in Section 2.6, both cyclophosphamide-treated and untreated animals (n = 6/group) were challenged with CIDMTR strain virus (P1 workpool) at a dose of 1 × 105 pfu by subcutaneous injection. Blood samples were collected at day 0, 3, 7 and 21 post-inoculation and animals humanely sacrificed at day 21 for collection of tissue samples for PCR analyses. Pregnancy/challenge studies were conducted as described in Section 2.6. Liveborn pups were sacrificed within 72 hours of delivery for DNA extraction and subsequent PCR.
4. Conclusions
The 22122 strain was originally isolated by Hartley in 1957 [27]. As the only characterized isolate of GPCMV it was used in virtually all subsequent GPCMV research. However, GPCMV infection is common among animals in commercial breeding colonies. In a longitudinal study conducted by Hsiung and colleagues from 1974 to 1979, GPCMV-neutralizing antibody was observed in 25% of Hartley strain guinea pigs obtained from commercial sources while virus was isolated from only 6 of 204 animals [56]. The percentages of antibody-positive animals obtained from different sources varied from shipment to shipment, ranging from 8%–50%. It does not appear that any of these viral isolates were retained. Thus, given only a single characterized isolate, the extent to which diverse GPCMV strains have been maintained within these domesticated populations was not previously known. Given that Hartley guinea pigs are descended from animals imported from South America to Europe in the 16th century, and that these animals subsequently underwent centuries of selective breeding, first as pets and later as research animals, the possibility existed that bottlenecks in the breeding history of Hartley guinea pigs could have limited the genetic diversity of GPCMV strains currently endemic in commercial breeding facilities.
In the current study, a second virus was independently isolated and characterized, again from a Hartley strain guinea pig obtained from a commercial supplier in the United States. Based on a high degree of genetic relatedness to 22122, predominantly similar restriction endonuclease patterns, and an conserved overall genomic structure, we conclude that CIDMTR is a GPCMV and not a novel and distinct betaherpesvirus. However, based on restriction pattern polymorphisms and significant divergence of amino acid sequences for several envelope glycoproteins (particularly gH and gO), we conclude that CIDMTR and 22122 represent two distinct strains of GPCMV and are not minor variants of the same strain. This further suggests that GPCMV strain diversity has been sustained within commercial breeding colonies and that other strains exhibiting similar levels of divergence may exist and could potentially be exploited to further extend this important animal model.
The passage history of 22122 is uncertain. In the original report by Hartley, two parallel isolations of GPCMV were reported: one, from salivary gland homogenate obtained from guinea pigs purchased by a commercial supplier in Yonkers, NY, and a second isolate obtained from a second supplier in New York state. Reactivation of virus may have been driven by allogeneic responses engendered from injecting SG extract into an animal with a different allotype, as has been suggested may drive HCMV and MCMV reactivation during transplantation [57,58]. Allogeneic responses may similarly have played a role, along with cyclophosphamide immunosuppression, in reactivation of the CIDMTR strain, since repeated tissue culture of salivary gland explants directly obtained from seropositive animals failed to result in growth of virus (data not shown). With respect to the original isolation of 22122, it is worth noting that serial in vivo passages may have occurred in the context, for some animals, of “mixed infection”, since Hartley noted that 7.7% (3/39) of “control NIH” strain guinea pigs aged 5 months or older were naturally infected, as evidenced by infrequent salivary gland inclusions [27]. This strain underwent 22 additional passages in cell culture of fibroblasts, and appears to be the strain that eventually was deposited with ATCC. The precise date of deposit is unclear, but reports of GPCMV research from the 1960s [59] and early 1970s describe obtaining this strain directly from the NIH, while studies since the late 1970s describe obtaining the virus from ATCC [60]. Between its original isolation by Hartley and its submission to ATCC, it appears to have undergone 54 additional passages in guinea pigs, and six additional passages in cell culture (3 passages in guinea pig embryo fibroblasts, and 3 passages in CCL 158 cells [61]. The 22122 strain derives from multiple rounds of both in vivo passage, some possibly occurring in the context of mixed infection, and 25–30 passages in cell culture. Thus, it is possible that 22122 underwent changes after isolation from the initial animal, either in cell culture or during in vivo passage.
In contrast, CIDMTR was subjected to minimal passage (one passage in vivo and two passages in cell culture) prior to genomic characterization. Thus, the genomic structure and sequence of the CIDMTR strain may more likely represent a bona fide “wild-type” GPCMV sequence than does the 22122 strain.
Sequence comparison with the 22122 strain revealed generally good conservation of protein coding sequences, although three areas of substantial discontinuity were noted. Thus, each strain contains unique sequences that can be used as markers to distinguish the strains during in vivo coinfection experiments. One of the regions unique to CIDMTR contains an ORF encoding a fourth putative MHC class I homolog not found in the 22122 strain. Since the three putative MHC class I homologs found in 22122 appear to be important for the in vivo pathogenesis of infection [55], further functional comparisons of the two strains will be of interest. In spite of minimal cell culture passage, at least one mutation, in the IE1/IE2 start codon, was observed in the tissue culture-adapted CIDMTR virus, compared to sequences amplified from salivary gland homogenate. This observation is similar to reported nucleotide sequence comparisons between several open reading frames in the DNA of different laboratory-adapted strains and clinical isolates of HCMV that has revealed amino-terminal sequence extensions of ORFs with alternate start codon usage [62]. Sequence differences with respect to start codon usage have also been noted upon comparison of laboratory-passaged and “wild” isolates of MCMV [37]. Future sequence analysis of CIDMTR DNA propagated solely in vivo is therefore warranted. Studies in immune competent and immune compromised guinea pigs confirm the ability of the CIDMTR virus to disseminate and produce viremia (DNAemia). Variation in glycoprotein protein coding sequences were noted, particularly for the gH and gO proteins, suggesting that this virus may be useful for the study of re-infection of immune guinea pigs in the guinea pig model of congenital cytomegalovirus infection. Cross-neutralization studies examining strain-specific antibody responses to envelope glycoproteins would enhance the usefulness of this new strain for the modeling of vaccine-mediated protection against re-infection and congenital transmission in this uniquely valuable model.
Acknowledgments
Support from NIH HD038416, HD044864 and HD068322 is acknowledged.
Author Contributions
M.R.S. conceived of the experiments and approach. A.A. performed electron microscopy studies. S.M., N.H.-A., C.F.-A., J.C.Z., and M.R.S. conducted experiments, and performed data analysis. T.A.R. and J.C. performed sequence analysis. M.M. analyzed data and assisted in writing the manuscript. All other authors also assisted in writing the manuscript.
Conflict of Interest
The authors declare no conflict of interest relevant to this work.
EMBL Accession Number
The genome sequence for this strain has been deposited with the EMBL Nucleotide Sequence Database under the accession number HG531783. The CIDMTR strain has been deposited with ATCC (Rockville, MD, USA; accession number PTA-120714), or is available upon request from the corresponding author.
References and Notes
- Sung, H.; Schleiss, M.R. Update on the current status of cytomegalovirus vaccines. Expert Rev. Vaccines 2010, 9, 1303–1314. [Google Scholar] [CrossRef]
- Nigro, G.; Adler, S.P.; La Torre, R.; Best, A.M. Passive immunization during pregnancy for congenital cytomegalovirus infection. N. Engl. J. Med. 2005, 353, 1350–1362. [Google Scholar] [CrossRef]
- Griffiths, P.; Plotkin, S.; Mocarski, E.; Pass, R.; Schleiss, M.; Krause, P.; Bialek, S. Desirability and feasibility of a vaccine against cytomegalovirus. Vaccine 2013, 31, B197–B203. [Google Scholar] [CrossRef]
- Krause, P.R.; Bialek, S.R.; Boppana, S.B.; Griffiths, P.D.; Laughlin, C.A.; Ljungman, P.; Mocarski, E.S.; Pass, R.F.; Read, J.S.; Schleiss, M.R.; et al. Priorities for cmv vaccine development. Vaccine 2013, 32, 4–10. [Google Scholar] [CrossRef]
- Pass, R.F.; Zhang, C.; Evans, A.; Simpson, T.; Andrews, W.; Huang, M.L.; Corey, L.; Hill, J.; Davis, E.; Flanigan, C.; et al. Vaccine prevention of maternal cytomegalovirus infection. N. Engl. J. Med. 2009, 360, 1191–1199. [Google Scholar] [CrossRef]
- Griffiths, P.D.; Stanton, A.; McCarrell, E.; Smith, C.; Osman, M.; Harber, M.; Davenport, A.; Jones, G.; Wheeler, D.C.; O'Beirne, J.; et al. Cytomegalovirus glycoprotein-b vaccine with mf59 adjuvant in transplant recipients: A phase 2 randomised placebo-controlled trial. Lancet 2011, 377, 1256–1263. [Google Scholar] [CrossRef]
- Ahlfors, K.; Ivarsson, S.A.; Harris, S. Report on a long-term study of maternal and congenital cytomegalovirus infection in sweden. Review of prospective studies available in the literature. Scand. J. Infect. Dis. 1999, 31, 443–457. [Google Scholar] [CrossRef]
- Ahlfors, K.; Harris, S.; Ivarsson, S.; Svanberg, L. Secondary maternal cytomegalovirus infection causing symptomatic congenital infection. N. Engl. J. Med. 1981, 305, 284. [Google Scholar]
- Boppana, S.B.; Rivera, L.B.; Fowler, K.B.; Mach, M.; Britt, W.J. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 2001, 344, 1366–1371. [Google Scholar] [CrossRef]
- Ross, S.A.; Arora, N.; Novak, Z.; Fowler, K.B.; Britt, W.J.; Boppana, S.B. Cytomegalovirus reinfections in healthy seroimmune women. J. Infect. Dis. 2010, 201, 386–389. [Google Scholar] [CrossRef]
- Yamamoto, A.Y.; Mussi-Pinhata, M.M.; Boppana, S.B.; Novak, Z.; Wagatsuma, V.M.; Oliveira Pde, F.; Duarte, G.; Britt, W.J. Human cytomegalovirus reinfection is associated with intrauterine transmission in a highly cytomegalovirus-immune maternal population. Am. J. Obstet. Gynecol. 2010, 202, e291–298, 297 e291–298. [Google Scholar]
- Mussi-Pinhata, M.M.; Yamamoto, A.Y.; Moura Brito, R.M.; de Lima Isaac, M.; de Carvalho e Oliveira, P.F.; Boppana, S.; Britt, W.J. Birth prevalence and natural history of congenital cytomegalovirus infection in a highly seroimmune population. Clin. Infect. Dis. 2009, 49, 522–528. [Google Scholar] [CrossRef]
- Ross, S.A.; Fowler, K.B.; Ashrith, G.; Stagno, S.; Britt, W.J.; Pass, R.F.; Boppana, S.B. Hearing loss in children with congenital cytomegalovirus infection born to mothers with preexisting immunity. J. Pediatr. 2006, 148, 332–336. [Google Scholar] [CrossRef]
- Yamamoto, A.Y.; Mussi-Pinhata, M.M.; Isaac, M.D.; Amaral, F.R.; Carvalheiro, C.G.; Aragon, D.C.; Manfredi, A.K.; Boppana, S.B.; Britt, W.J. Congenital cytomegalovirus infection as a cause of sensorineural hearing loss in a highly immune population. J. Pediatr. Infect. Dis. 2011, 30, 1043–1046. [Google Scholar] [CrossRef]
- Novak, Z.; Ross, S.A.; Patro, R.K.; Pati, S.K.; Reddy, M.K.; Purser, M.; Britt, W.J.; Boppana, S.B. Enzyme-linked immunosorbent assay method for detection of cytomegalovirus strain-specific antibody responses. Clin. Vaccine Immunol. 2009, 16, 288–290. [Google Scholar] [CrossRef]
- Yamamoto, A.Y.; Mussi-Pinhata, M.M.; de Deus Wagatsuma, V.M.; Marin, L.J.; Duarte, G.; Figueiredo, L.T. Human cytomegalovirus glycoprotein b genotypes in brazilian mothers and their congenitally infected infants. J. Med. Virol. 2007, 79, 1164–1168. [Google Scholar] [CrossRef]
- Murthy, S.; Hayward, G.S.; Wheelan, S.; Forman, M.S.; Ahn, J.H.; Pass, R.F.; Arav-Boger, R. Detection of a single identical cytomegalovirus (cmv) strain in recently seroconverted young women. PLoS One 2011, 6, e15949. [Google Scholar]
- Kropff, B.; Burkhardt, C.; Schott, J.; Nentwich, J.; Fisch, T.; Britt, W.; Mach, M. Glycoprotein n of human cytomegalovirus protects the virus from neutralizing antibodies. PLoS Pathog. 2012, 8, e1002999. [Google Scholar] [CrossRef]
- Pati, S.K.; Novak, Z.; Purser, M.; Arora, N.; Mach, M.; Britt, W.J.; Boppana, S.B. Strain-specific neutralizing antibody responses against human cytomegalovirus envelope glycoprotein n. Clin. Vaccine Immunol. 2012, 19, 909–913. [Google Scholar] [CrossRef]
- Burkhardt, C.; Himmelein, S.; Britt, W.; Winkler, T.; Mach, M. Glycoprotein n subtypes of human cytomegalovirus induce a strain-specific antibody response during natural infection. J. Gen. Virol. 2009, 90, 1951–1961. [Google Scholar] [CrossRef]
- Sabbaj, S.; Pass, R.F.; Goepfert, P.A.; Pichon, S. Glycoprotein b vaccine is capable of boosting both antibody and cd4 t-cell responses to cytomegalovirus in chronically infected women. J. Infect. Dis. 2011, 203, 1534–1541. [Google Scholar] [CrossRef]
- Schleiss, M.R. Could therapeutic vaccination of cytomegalovirus-seropositive persons prevent reinfection and congenital virus transmission? J. Infect. Dis. 2011, 203, 1513–1516. [Google Scholar] [CrossRef]
- Kern, E.R. Pivotal role of animal models in the development of new therapies for cytomegalovirus infections. Antivir. Res. 2006, 71, 164–171. [Google Scholar] [CrossRef]
- Schleiss, M.R. Nonprimate models of congenital cytomegalovirus (cmv) infection: Gaining insight into pathogenesis and prevention of disease in newborns. ILAR J. 2006, 47, 65–72. [Google Scholar] [CrossRef]
- Yue, Y.; Wang, Z.; Abel, K.; Li, J.; Strelow, L.; Mandarino, A.; Eberhardt, M.K.; Schmidt, K.A.; Diamond, D.J.; Barry, P.A. Evaluation of recombinant modified vaccinia ankara virus-based rhesus cytomegalovirus vaccines in rhesus macaques. Med. Microbiol. Immunol. 2008, 197, 117–123. [Google Scholar] [CrossRef]
- Bia, F.J.; Miller, S.A.; Davidson, K.H. The guinea pig cytomegalovirus model of congenital human cytomegalovirus infection. Birth. Defects Orig. Artic. Ser. 1984, 20, 233–241. [Google Scholar]
- Hartley, J.W.; Rowe, W.P.; Huebner, R.J. Serial propagation of the guinea pig salivary gland virus in tissue culture. Proc. Soc. Exp. Biol. Med. 1957, 96, 281–285. [Google Scholar] [CrossRef]
- Johnson, K.P.; Connor, W.S. Guinea pig cytomegalovirus: Transplacental transmission. Brief report. Arch. Virol. 1979, 59, 263–267. [Google Scholar] [CrossRef]
- Schleiss, M.R.; Bourne, N.; Stroup, G.; Bravo, F.J.; Jensen, N.J.; Bernstein, D.I. Protection against congenital cytomegalovirus infection and disease in guinea pigs, conferred by a purified recombinant glycoprotein b vaccine. J. Infect. Dis. 2004, 189, 1374–1381. [Google Scholar] [CrossRef]
- Bratcher, D.F.; Bourne, N.; Bravo, F.J.; Schleiss, M.R.; Slaoui, M.; Myers, M.G.; Bernstein, D.I. Effect of passive antibody on congenital cytomegalovirus infection in guinea pigs. J. Infect. Dis. 1995, 172, 944–950. [Google Scholar] [CrossRef]
- Britt, W.J.; Harrison, C. Identification of an abundant disulfide-linked complex of glycoproteins in the envelope of guinea pig cytomegalovirus. Virology 1994, 201, 294–302. [Google Scholar] [CrossRef]
- Schleiss, M.R.; Bernstein, D.I.; McVoy, M.A.; Stroup, G.; Bravo, F.; Creasy, B.; McGregor, A.; Henninger, K.; Hallenberger, S. The non-nucleoside antiviral, bay 38–4766, protects against cytomegalovirus (cmv) disease and mortality in immunocompromised guinea pigs. Antivir. Res. 2005, 65, 35–43. [Google Scholar] [CrossRef]
- Fong, C.K.; Bia, F.; Hsiung, G.D. Ultrastructural development and persistence of guinea pig cytomegalovirus in duet cells of guinea pig submaxillary gland. Arch. Virol. 1980, 64, 97–108. [Google Scholar] [CrossRef]
- Fong, C.K.; Bia, F.; Hsiung, G.D.; Madore, P.; Chang, P.W. Ultrastructural development of guinea pig cytomegalovirus in cultured guinea pig embryo cells. J. Gen. Virol. 1979, 42, 127–140. [Google Scholar] [CrossRef]
- Fong, C.K.; Brigati, D. Ultrastructural localization of viral antigen in nuclear inclusions of cytomegalovirus infected guinea pig cells. Arch. Virol. 1982, 74, 125–133. [Google Scholar] [CrossRef]
- Cui, X.; McGregor, A.; Schleiss, M.R.; McVoy, M.A. The impact of genome length on replication and genome stability of the herpesvirus guinea pig cytomegalovirus. Virology 2009, 386, 132–138. [Google Scholar] [CrossRef]
- Smith, L.M.; McWhorter, A.R.; Masters, L.L.; Shellam, G.R.; Redwood, A.J. Laboratory strains of murine cytomegalovirus are genetically similar to but phenotypically distinct from wild strains of virus. J. Virol. 2008, 82, 6689–6696. [Google Scholar] [CrossRef]
- Pignatelli, S.; Dal Monte, P.; Rossini, G.; Chou, S.; Gojobori, T.; Hanada, K.; Guo, J.J.; Rawlinson, W.; Britt, W.; Mach, M.; et al. Human cytomegalovirus glycoprotein n (gpul73-gn) genomic variants: Identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J. Gen. Virol. 2003, 84, 647–655. [Google Scholar] [CrossRef]
- Pignatelli, S.; Dal Monte, P.; Rossini, G.; Lazzarotto, T.; Gatto, M.R.; Landini, M.P. Intrauterine cytomegalovirus infection and glycoprotein n (gn) genotypes. J. Clin. Virol. 2003, 28, 38–43. [Google Scholar] [CrossRef]
- CLC Workbench software, version 6.0; CLC: Cambridge, MA, USA. Available online: http://www.clcbio.com/products/clc-main-workbench/ (accessed on 23 January 2014).
- XPlasMap, version 0.99; Available online: http://www.iayork.com/XPlasMap/ (accessed on 16 January 2014).
- Yamada, S.; Nozawa, N.; Katano, H.; Fukui, Y.; Tsuda, M.; Tsutsui, Y.; Kurane, I.; Inoue, N. Characterization of the guinea pig cytomegalovirus genome locus that encodes homologs of human cytomegalovirus major immediate-early genes, ul128, and ul130. Virology 2009, 391, 99–106. [Google Scholar] [CrossRef]
- Nozawa, N.; Yamamoto, Y.; Fukui, Y.; Katano, H.; Tsutsui, Y.; Sato, Y.; Yamada, S.; Inami, Y.; Nakamura, K.; Yokoi, M.; et al. Identification of a 1.6 kb genome locus of guinea pig cytomegalovirus required for efficient viral growth in animals but not in cell culture. Virology 2008, 379, 45–54. [Google Scholar] [CrossRef]
- Yang, D.; Tamburro, K.; Dittmer, D.; Cui, X.; McVoy, M.A.; Hernandez-Alvarado, N.; Schleiss, M.R. Complete genome sequence of pathogenic guinea pig cytomegalovirus from salivary gland homogenates of infected animals. Genome Announc. 2013, 1, e0005413. [Google Scholar]
- Kozak, M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger rnas. Nucleic Acids Res. 1987, 15, 8125–8148. [Google Scholar] [CrossRef]
- iTEM software, Version 9.0; Olympus SIS: Münster, Germany, 2014.
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. Abyss: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef]
- Kanai, K.; Yamada, S.; Yamamoto, Y.; Fukui, Y.; Kurane, I.; Inoue, N. Re-evaluation of the genome sequence of guinea pig cytomegalovirus. J. Gen. Virol. 2011, 92, 1005–1020. [Google Scholar] [CrossRef]
- Schleiss, M.R.; McGregor, A.; Choi, K.Y.; Date, S.V.; Cui, X.; McVoy, M.A. Analysis of the nucleotide sequence of the guinea pig cytomegalovirus (gpcmv) genome. Virol. J. 2008, 5, 139. [Google Scholar] [CrossRef]
- Milne, I.; Bayer, M.; Cardle, L.; Shaw, P.; Stephen, G.; Wright, F.; Marshall, D. Tablet--next generation sequence assembly visualization. Bioinformatics 2010, 26, 401–402. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- SMRT Analysis software, Version 1.4.0; Pacific Biosciences: Menlo Park, CA, USA, 2014. Available online: http://www.pacificbiosciences.com/products/software/secondary-analysis/ (accessed on 16 January 2014).
- LightCycler Data Analysis Software, version 1.5; Roche: Indianapolis, IN, USA, 2014.
- Crumpler, M.M.; Choi, K.Y.; McVoy, M.A.; Schleiss, M.R. A live guinea pig cytomegalovirus vaccine deleted of three putative immune evasion genes is highly attenuated but remains immunogenic in a vaccine/challenge model of congenital cytomegalovirus infection. Vaccine 2009, 27, 4209–4218. [Google Scholar] [CrossRef]
- Hsiung, G.D.; Bia, F.J.; Fong, C.K. Viruses of guinea pigs: Considerations for biomedical research. Microbiol. Rev. 1980, 44, 468–490. [Google Scholar]
- Hummel, M.; Zhang, Z.; Yan, S.; DePlaen, I.; Golia, P.; Varghese, T.; Thomas, G.; Abecassis, M.I. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: A model for reactivation from latency. J. Virol. 2001, 75, 4814–4822. [Google Scholar] [CrossRef]
- Soderberg-Naucler, C.; Fish, K.N.; Nelson, J.A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 1997, 91, 119–126. [Google Scholar] [CrossRef]
- Middelkamp, J.N.; Patrizi, G.; Reed, C.A. Light and electron microscopic studies of the guinea pig cytomegalovirus. J. Ultrastruct. Res. 1967, 18, 85–101. [Google Scholar] [CrossRef]
- Choi, Y.C.; Hsiung, G.D. Cytomegalovirus infection in guinea pigs. II. Transplacental and horizontal transmission. J. Infect. Dis. 1978, 138, 197–202. [Google Scholar] [CrossRef]
- ATCC History of GPCMV. Available online: http://www.atcc.org/products/all/VR-682.aspx#history/ (accessed on 23 January 2014).
- Brondke, H.; Schmitz, B.; Doerfler, W. Nucleotide sequence comparisons between several strains and isolates of human cytomegalovirus reveal alternate start codon usage. Arch. Virol. 2007, 152, 2035–2046. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).