RNA Polymerase III Regulates HIV Replication and Latency
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Reagents and Cell Culture
2.3. RNA Extraction and Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
2.4. Preparation of Pseudotyped HIV DuoFluo Virus
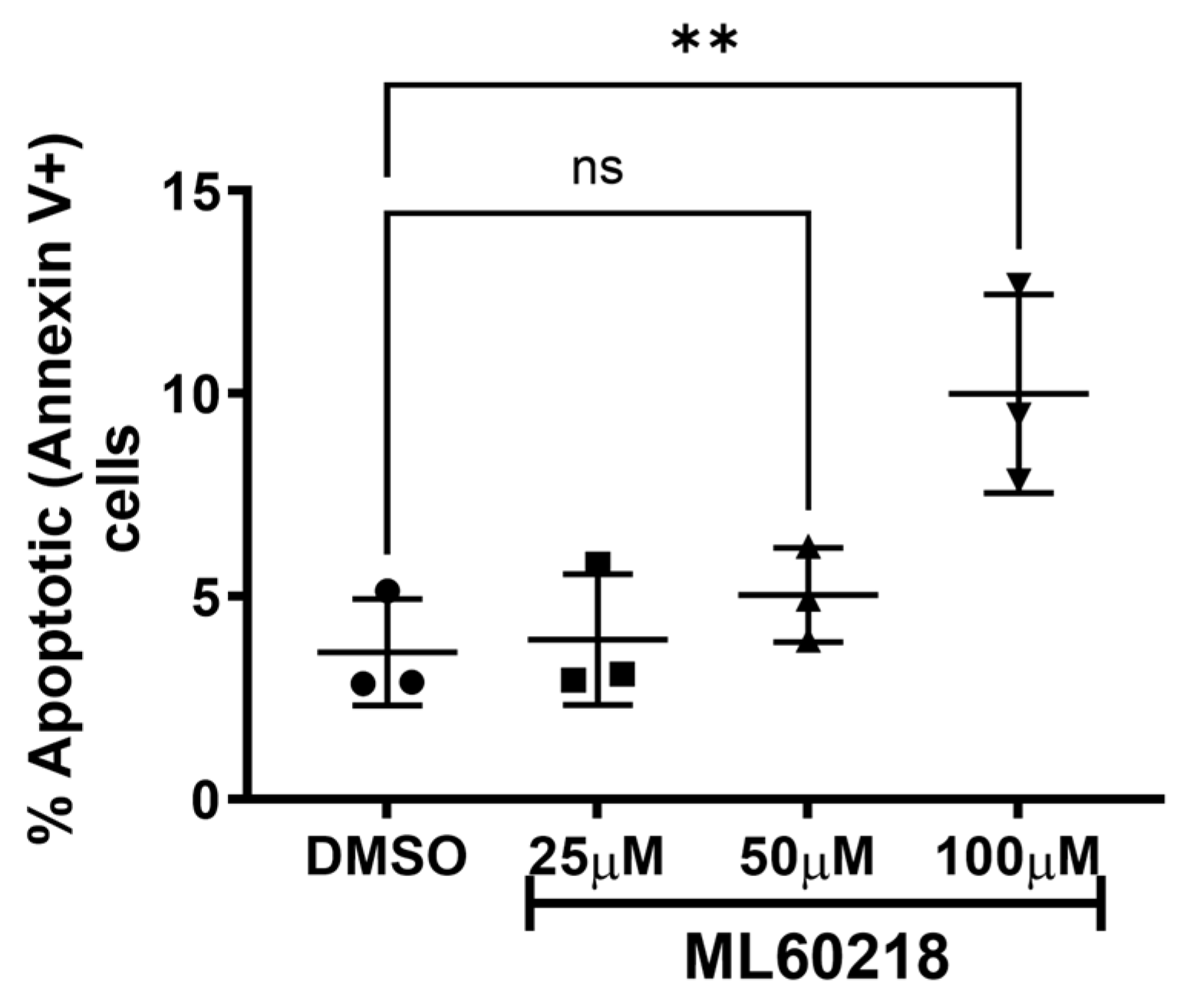
2.5. Cellular Death Assays
2.6. Statistical Analysis
3. Results
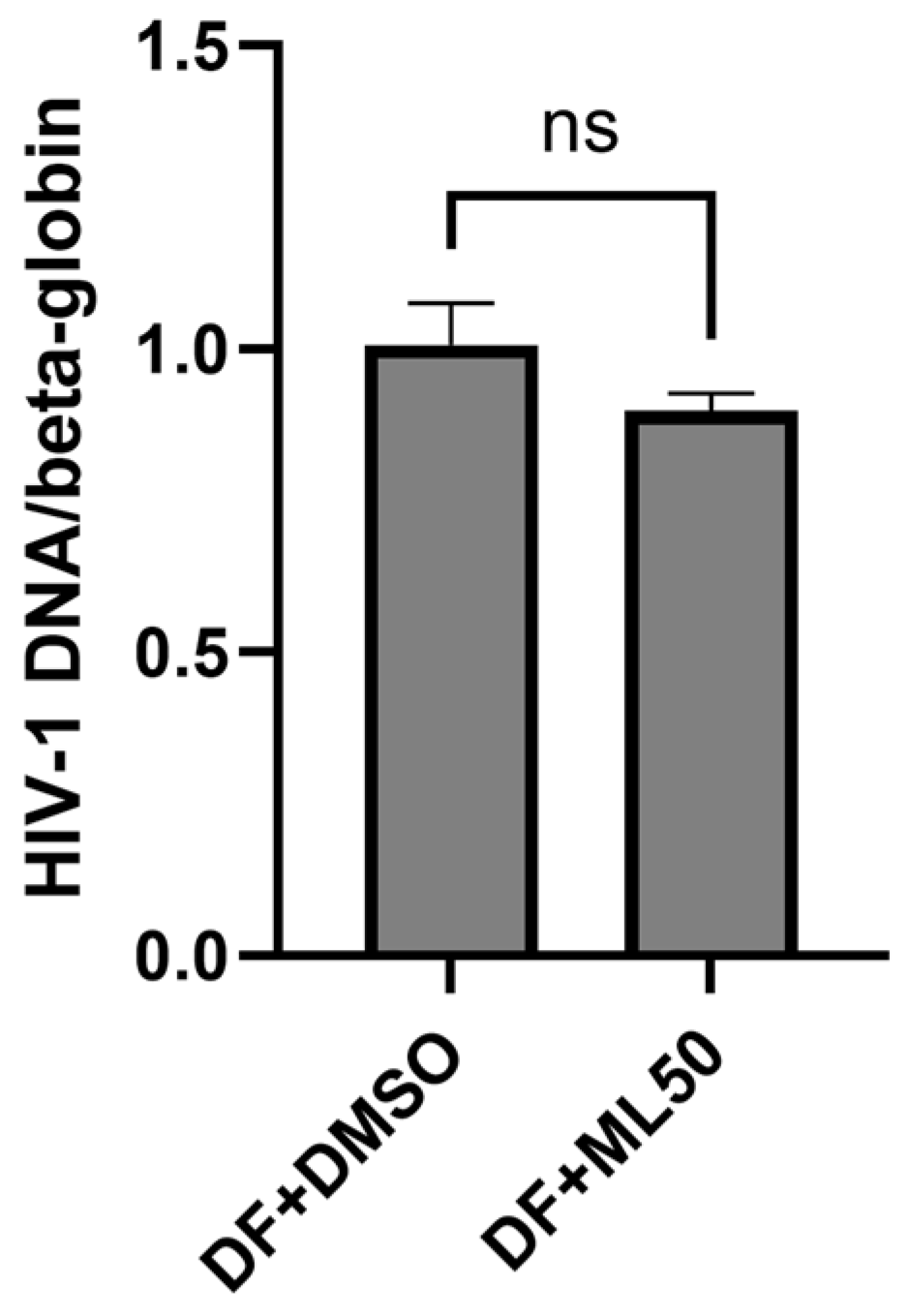
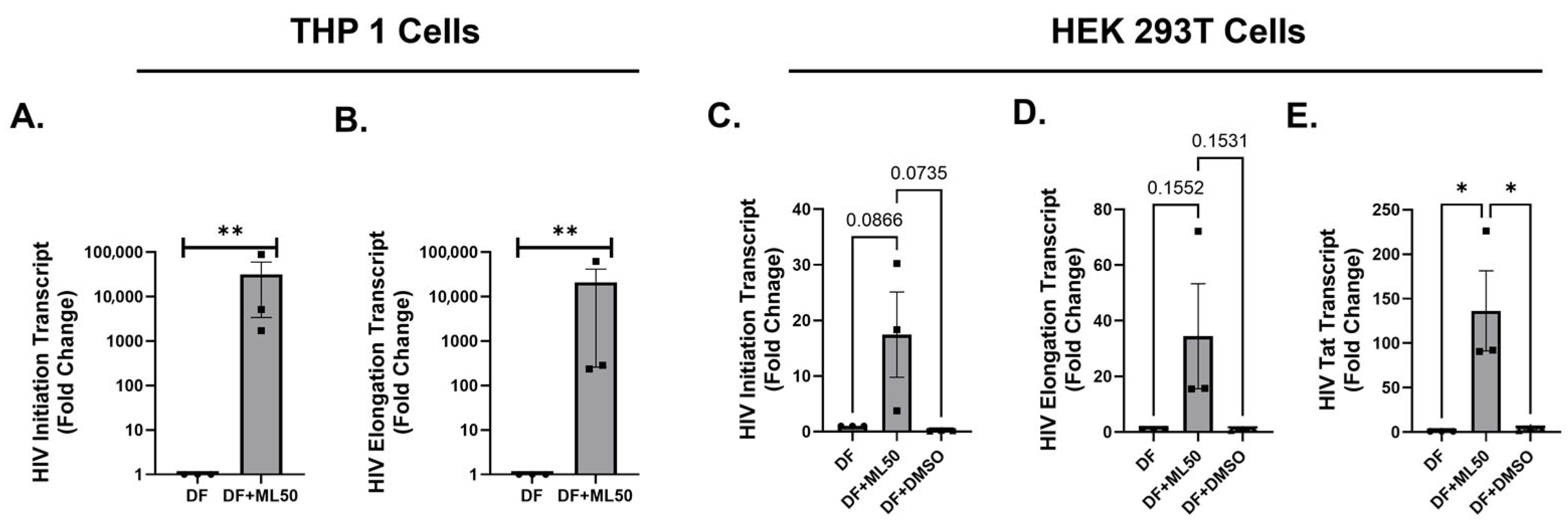
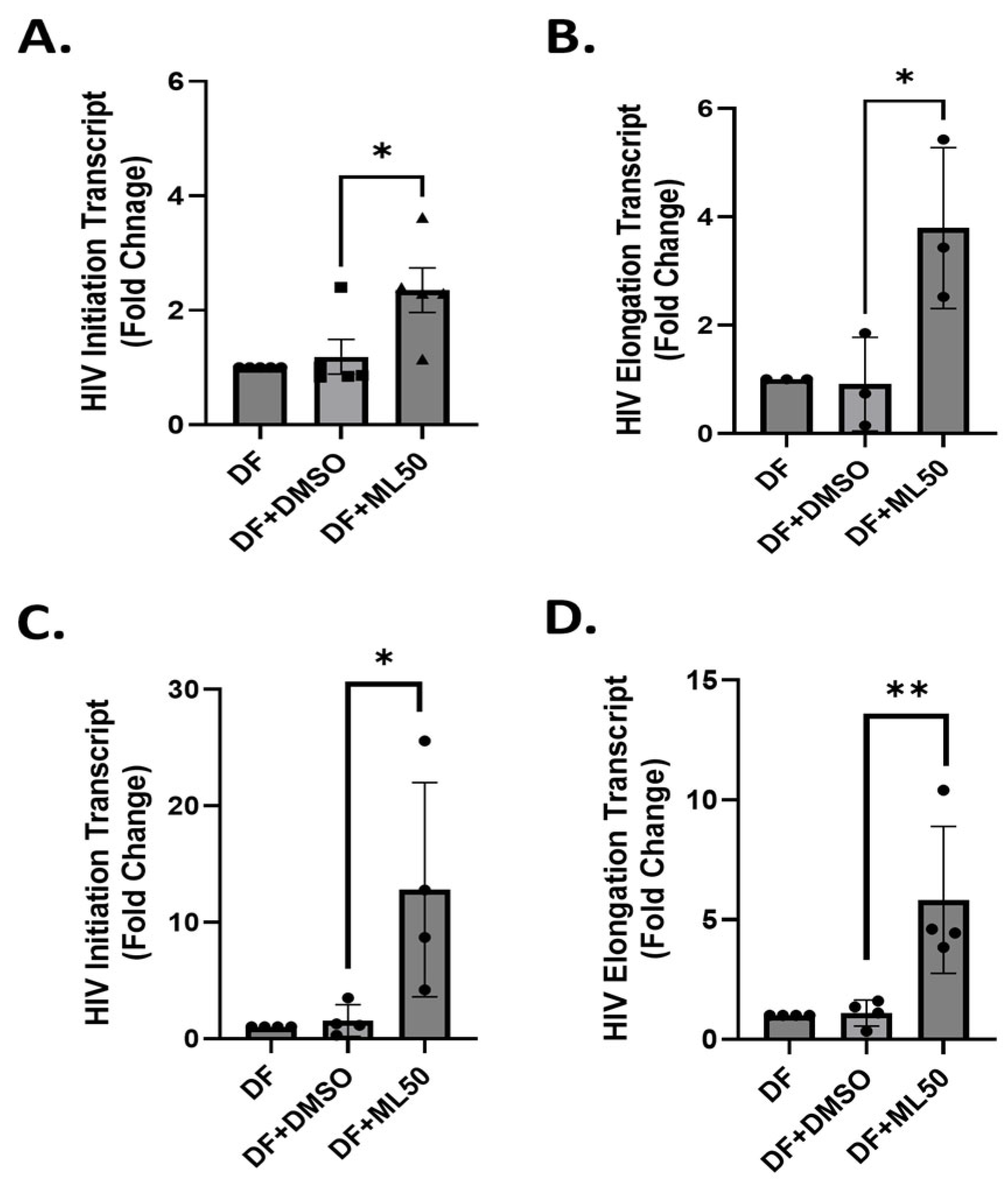
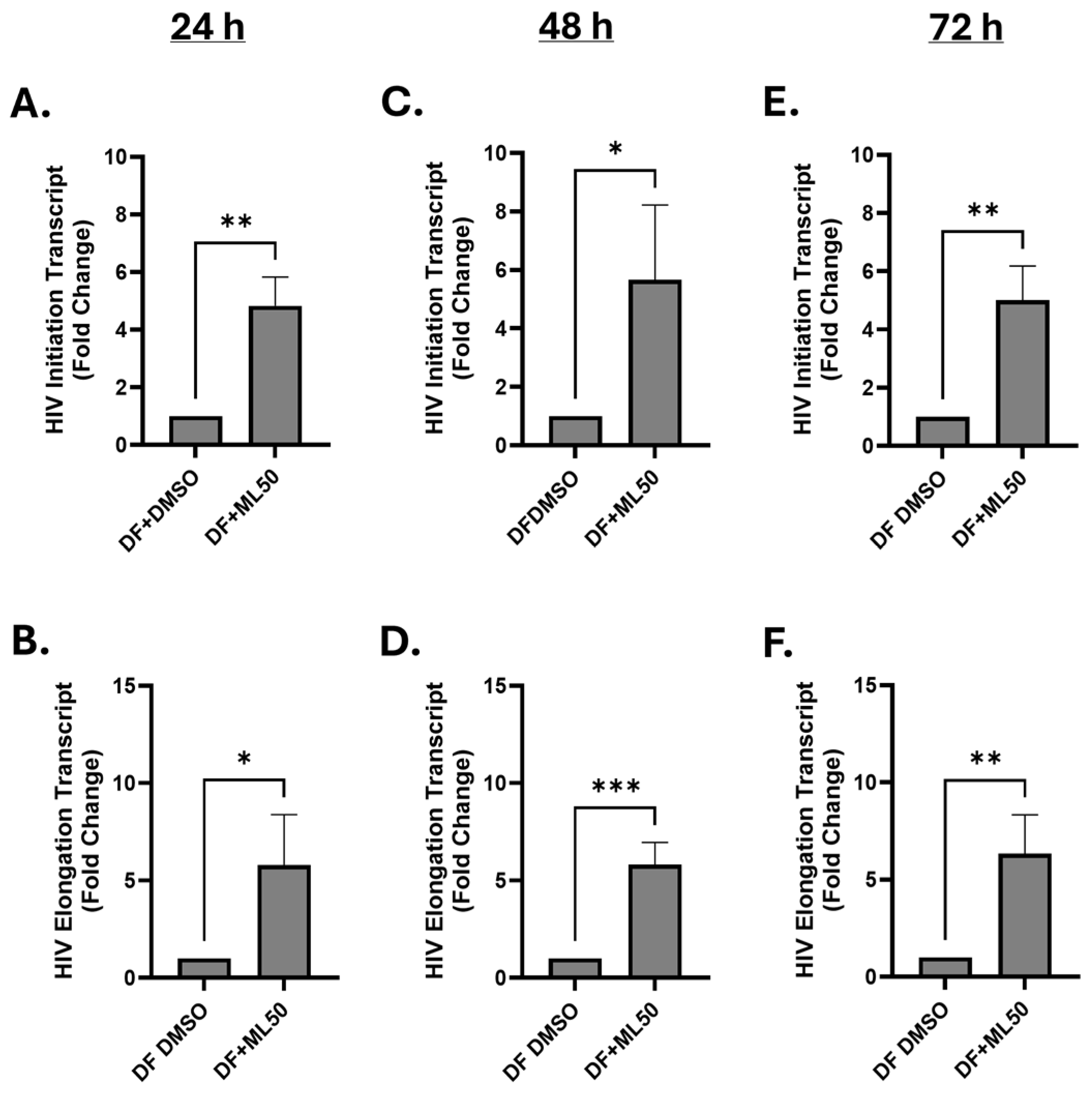
3.1. RNAP III Inhibition Promotes Latency Reactivation
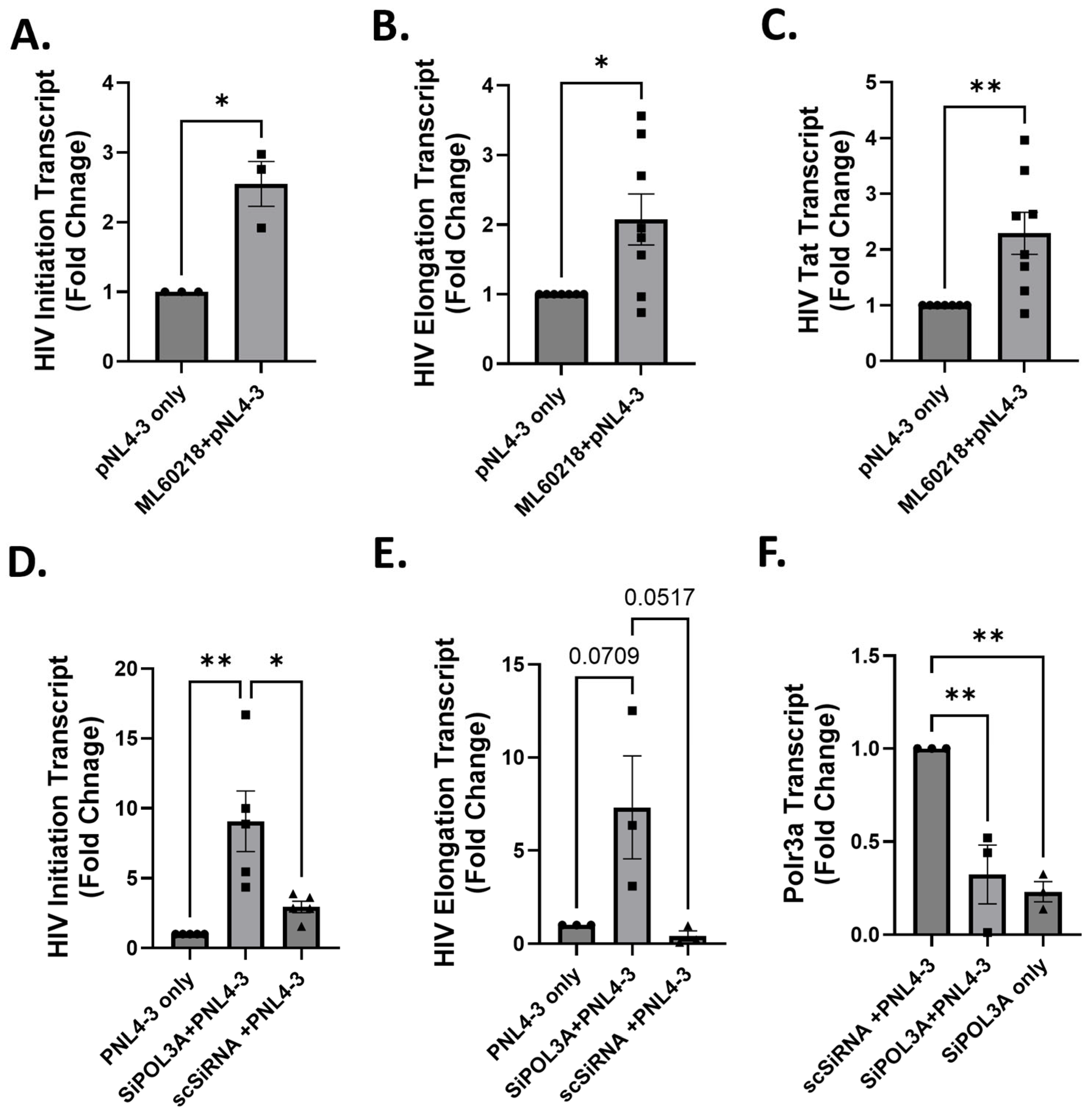
3.2. RNA Pol III Regulates HIV Transcription
3.3. RNA Pol III Regulates HIV Transcription in T Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
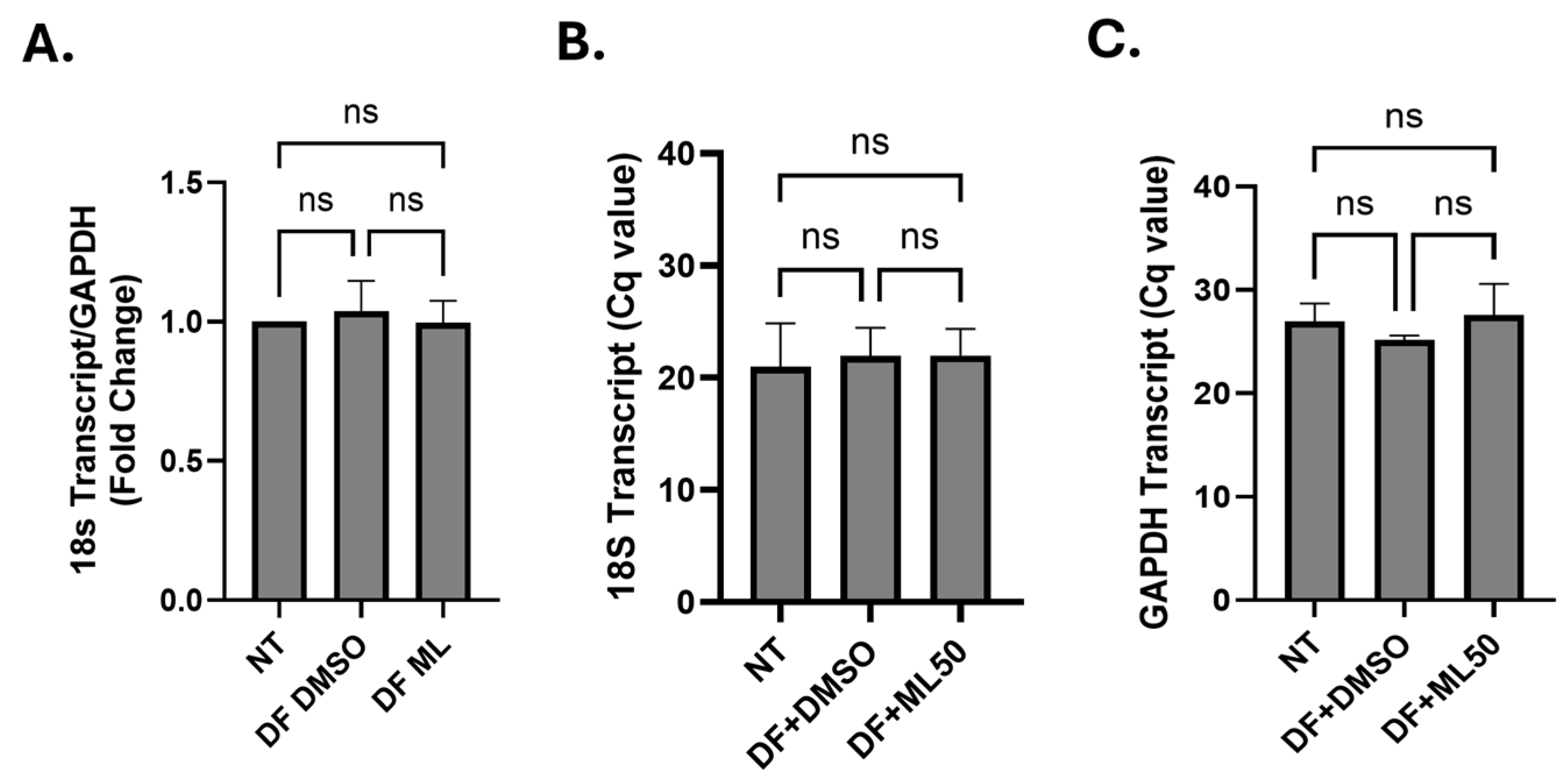
Appendix A

References
- Dufour, C.; Gantner, P.; Fromentin, R.; Chomont, N. The multifaceted nature of HIV latency. J. Clin. Investig. 2020, 130, 3381–3390. [Google Scholar] [CrossRef] [PubMed]
- Davey, R.T., Jr.; Bhat, N.; Yoder, C.; Chun, T.W.; Metcalf, J.A.; Dewar, R.; Natarajan, V.; Lempicki, R.A.; Adelsberger, J.W.; Miller, K.D.; et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc. Natl. Acad. Sci. USA 1999, 96, 15109–15114. [Google Scholar] [CrossRef]
- Blankson, J.N.; Persaud, D.; Siliciano, R.F. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 2002, 53, 557–593. [Google Scholar] [CrossRef]
- Crooks, A.M.; Bateson, R.; Cope, A.B.; Dahl, N.P.; Griggs, M.K.; Kuruc, J.D.; Gay, C.L.; Eron, J.J.; Margolis, D.M.; Bosch, R.J.; et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J. Infect. Dis. 2015, 212, 1361–1365. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.J.; Kwon, K.J.; Farber, D.L.; Siliciano, R.F. The Latent Reservoir for HIV-1: How Immunologic Memory and Clonal Expansion Contribute to HIV-1 Persistence. J. Immunol. 2016, 197, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Banga, R.; Procopio, F.A.; Noto, A.; Pollakis, G.; Cavassini, M.; Ohmiti, K.; Corpataux, J.M.; de Leval, L.; Pantaleo, G.; Perreau, M. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat. Med. 2016, 22, 754–761. [Google Scholar] [CrossRef]
- Chun, T.W.; Nickle, D.C.; Justement, J.S.; Meyers, J.H.; Roby, G.; Hallahan, C.W.; Kottilil, S.; Moir, S.; Mican, J.M.; Mullins, J.I.; et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J. Infect. Dis. 2008, 197, 714–720. [Google Scholar] [CrossRef]
- Churchill, M.J.; Gorry, P.R.; Cowley, D.; Lal, L.; Sonza, S.; Purcell, D.F.; Thompson, K.A.; Gabuzda, D.; McArthur, J.C.; Pardo, C.A.; et al. Use of laser capture microdissection to detect integrated HIV-1 DNA in macrophages and astrocytes from autopsy brain tissues. J. Neurovirol. 2006, 12, 146–152. [Google Scholar] [CrossRef]
- Jenabian, M.A.; Costiniuk, C.T.; Mehraj, V.; Ghazawi, F.M.; Fromentin, R.; Brousseau, J.; Brassard, P.; Belanger, M.; Ancuta, P.; Bendayan, R.; et al. Immune tolerance properties of the testicular tissue as a viral sanctuary site in ART-treated HIV-infected adults. AIDS 2016, 30, 2777–2786. [Google Scholar] [CrossRef]
- Durand, C.M.; Ghiaur, G.; Siliciano, J.D.; Rabi, S.A.; Eisele, E.E.; Salgado, M.; Shan, L.; Lai, J.F.; Zhang, H.; Margolick, J.; et al. HIV-1 DNA is detected in bone marrow populations containing CD4+ T cells but is not found in purified CD34+ hematopoietic progenitor cells in most patients on antiretroviral therapy. J. Infect. Dis. 2012, 205, 1014–1018. [Google Scholar] [CrossRef] [PubMed]
- Hansen, E.C.; Ransom, M.; Hesselberth, J.R.; Hosmane, N.N.; Capoferri, A.A.; Bruner, K.M.; Pollack, R.A.; Zhang, H.; Drummond, M.B.; Siliciano, J.M.; et al. Diverse fates of uracilated HIV-1 DNA during infection of myeloid lineage cells. Elife 2016, 5, e18447. [Google Scholar] [CrossRef]
- Massanella, M.; Bakeman, W.; Sithinamsuwan, P.; Fletcher, J.L.K.; Chomchey, N.; Tipsuk, S.; Chalermchai, T.; Routy, J.P.; Ananworanich, J.; Valcour, V.G.; et al. Infrequent HIV Infection of Circulating Monocytes during Antiretroviral Therapy. J. Virol. 2019, 94, e01174-19. [Google Scholar] [CrossRef]
- Wong, M.E.; Jaworowski, A.; Hearps, A.C. Corrigendum: The HIV Reservoir in Monocytes and Macrophages. Front. Immunol. 2019, 10, 2517. [Google Scholar] [CrossRef]
- Zhu, T.; Muthui, D.; Holte, S.; Nickle, D.; Feng, F.; Brodie, S.; Hwangbo, Y.; Mullins, J.I.; Corey, L. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 2002, 76, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Maldarelli, F.; Wu, X.; Su, L.; Simonetti, F.R.; Shao, W.; Hill, S.; Spindler, J.; Ferris, A.L.; Mellors, J.W.; Kearney, M.F.; et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014, 345, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Einkauf, K.B.; Lee, G.Q.; Gao, C.; Sharaf, R.; Sun, X.; Hua, S.; Chen, S.M.; Jiang, C.; Lian, X.; Chowdhury, F.Z.; et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Invest. 2019, 129, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Stevens, S.W.; Griffith, J.D. Human immunodeficiency virus type 1 may preferentially integrate into chromatin occupied by L1Hs repetitive elements. Proc. Natl. Acad. Sci. USA 1994, 91, 5557–5561. [Google Scholar] [CrossRef]
- Ruelas, D.S.; Greene, W.C. An integrated overview of HIV-1 latency. Cell 2013, 155, 519–529. [Google Scholar]
- Romani, B.; Allahbakhshi, E. Underlying mechanisms of HIV-1 latency. Virus Genes 2017, 53, 329–339. [Google Scholar] [CrossRef]
- Choudhary, S.K.; Margolis, D.M. Curing HIV: Pharmacologic approaches to target HIV-1 latency. Annu. Rev. Pharmacol. Toxicol. 2011, 51, 397–418. [Google Scholar] [CrossRef][Green Version]
- Hakre, S.; Chavez, L.; Shirakawa, K.; Verdin, E. HIV latency: Experimental systems and molecular models. FEMS Microbiol. Rev. 2012, 36, 706–716. [Google Scholar] [CrossRef]
- Jaafoura, S.; de Goer de Herve, M.G.; Hernandez-Vargas, E.A.; Hendel-Chavez, H.; Abdoh, M.; Mateo, M.C.; Krzysiek, R.; Merad, M.; Seng, R.; Tardieu, M.; et al. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4(+) memory T Cells. Nat. Commun. 2014, 5, 5407. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.A.; Tolstrup, M.; Brinkmann, C.R.; Olesen, R.; Erikstrup, C.; Solomon, A.; Winckelmann, A.; Palmer, S.; Dinarello, C.; Buzon, M.; et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: A phase 1/2, single group, clinical trial. Lancet HIV 2014, 1, e13–e21. [Google Scholar] [CrossRef] [PubMed]
- Golumbeanu, M.; Cristinelli, S.; Rato, S.; Munoz, M.; Cavassini, M.; Beerenwinkel, N.; Ciuffi, A. Single-Cell RNA-Seq Reveals Transcriptional Heterogeneity in Latent and Reactivated HIV-Infected Cells. Cell Rep. 2018, 23, 942–950. [Google Scholar] [CrossRef]
- Matsuda, Y.; Kobayashi-Ishihara, M.; Fujikawa, D.; Ishida, T.; Watanabe, T.; Yamagishi, M. Epigenetic heterogeneity in HIV-1 latency establishment. Sci. Rep. 2015, 5, 7701. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.; Das, B.; Dobrowolski, C.; Karn, J. Multiple Histone Lysine Methyltransferases Are Required for the Establishment and Maintenance of HIV-1 Latency. mBio 2017, 8, e00133-17. [Google Scholar] [CrossRef]
- Park, J.L.; Lee, Y.S.; Kunkeaw, N.; Kim, S.Y.; Kim, I.H.; Lee, Y.S. Epigenetic regulation of noncoding RNAtranscription by mammalian RNA polymerase III. Epigenomics 2017, 9, 171–187. [Google Scholar] [CrossRef]
- Schramm, L.; Hernandez, N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002, 16, 2593–2620. [Google Scholar] [CrossRef]
- Dieci, G.; Conti, A.; Pagano, A.; Carnevali, D. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim. Biophys. Acta 2013, 1829, 296–305. [Google Scholar] [CrossRef]
- Alla, R.K.; Cairns, B.R. RNA polymerase III transcriptomes in human embryonic stem cells and induced pluripotent stem cells, and relationships with pluripotency transcription factors. PLoS ONE 2014, 9, e85648. [Google Scholar] [CrossRef]
- Carriere, L.; Graziani, S.; Alibert, O.; Ghavi-Helm, Y.; Boussouar, F.; Humbertclaude, H.; Jounier, S.; Aude, J.C.; Keime, C.; Murvai, J.; et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res. 2012, 40, 270–283. [Google Scholar] [CrossRef]
- Moqtaderi, Z.; Wang, J.; Raha, D.; White, R.J.; Snyder, M.; Weng, Z.; Struhl, K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat. Struct. Mol. Biol. 2010, 17, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Chepelev, I.; Liko, D.; Cuddapah, S.; Fleming, A.B.; Birch, J.; Cui, K.; White, R.J.; Zhao, K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat. Struct. Mol. Biol. 2010, 17, 629–634. [Google Scholar] [CrossRef]
- Canella, D.; Praz, V.; Reina, J.H.; Cousin, P.; Hernandez, N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010, 20, 710–721. [Google Scholar] [CrossRef] [PubMed]
- White, R.J. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008, 24, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhong, Q.; Evans, A.G.; Levy, D.; Zhong, S. Phosphorylation of histone H3 serine 28 modulates RNA polymerase III-dependent transcription. Oncogene 2011, 30, 3943–3952. [Google Scholar] [CrossRef]
- Schwartz, L.B.; Sklar, V.E.; Jaehning, J.A.; Weinmann, R.; Roeder, R.G. Isolation and partial characterization of the multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in the mouse myeloma, MOPC 315. J. Biol. Chem. 1974, 249, 5889–5897. [Google Scholar] [CrossRef]
- Dunker, W.; Zhao, Y.; Song, Y.; Karijolich, J. Recognizing the SINEs of Infection: Regulation of Retrotransposon Expression and Modulation of Host Cell Processes. Viruses 2017, 9, 386. [Google Scholar] [CrossRef]
- Karijolich, J.; Zhao, Y.; Alla, R.; Glaunsinger, B. Genome-wide mapping of infection-induced SINE RNAs reveals a role in selective mRNA export. Nucleic Acids Res. 2017, 45, 6194–6208. [Google Scholar] [CrossRef]
- Kutsch, O.; Benveniste, E.N.; Shaw, G.M.; Levy, D.N. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 2002, 76, 8776–8786. [Google Scholar] [CrossRef]
- Huang, C.B.; Alimova, Y.V.; Strange, S.; Ebersole, J.L. Polybacterial challenge enhances HIV reactivation in latently infected macrophages and dendritic cells. Immunology 2011, 132, 401–409. [Google Scholar] [CrossRef]
- Martinez-Bonet, M.; Clemente, M.I.; Serramia, M.J.; Moreno, S.; Munoz, E.; Munoz-Fernandez, M.A. Immunological and pharmacological strategies to reactivate HIV-1 from latently infected cells: A possibility for HIV-1 paediatric patients? J. Virus Erad. 2015, 1, 148–152. [Google Scholar] [CrossRef]
- Shishido, T.; Wolschendorf, F.; Duverger, A.; Wagner, F.; Kappes, J.; Jones, J.; Kutsch, O. Selected drugs with reported secondary cell-differentiating capacity prime latent HIV-1 infection for reactivation. J. Virol. 2012, 86, 9055–9069. [Google Scholar] [CrossRef] [PubMed]
- Darcis, G.; Kula, A.; Bouchat, S.; Fujinaga, K.; Corazza, F.; Ait-Ammar, A.; Delacourt, N.; Melard, A.; Kabeya, K.; Vanhulle, C.; et al. An In-Depth Comparison of Latency-Reversing Agent Combinations in Various In Vitro and Ex Vivo HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression. PLoS Pathog. 2015, 11, e1005063. [Google Scholar] [CrossRef]
- Dickey, L.L.; Martins, L.J.; Planelles, V.; Hanley, T.M. HIV-1-induced type I IFNs promote viral latency in macrophages. J. Leukoc. Biol. 2022, 112, 1343–1356. [Google Scholar] [CrossRef]
- Calvanese, V.; Chavez, L.; Laurent, T.; Ding, S.; Verdin, E. Dual-color HIV reporters trace a population of latently infected cells and enable their purification. Virology 2013, 446, 283–292. [Google Scholar] [CrossRef]
- Linkner, T.R.; Ambrus, V.; Kunkli, B.; Szojka, Z.I.; Kallo, G.; Csosz, E.; Kumar, A.; Emri, M.; Tozser, J.; Mahdi, M. Cellular Proteo-Transcriptomic Changes in the Immediate Early-Phase of Lentiviral Transduction. Microorganisms 2021, 9, 2207. [Google Scholar] [CrossRef] [PubMed]
- Krizova, I.; Dostalkova, A.; Castro, E.; Prchal, J.; Hadravova, R.; Kaufman, F.; Hrabal, R.; Ruml, T.; Llano, M.; Echegoyen, L.; et al. Fullerene Derivatives Prevent Packaging of Viral Genomic RNA into HIV-1 Particles by Binding Nucleocapsid Protein. Viruses 2021, 13, 2451. [Google Scholar] [CrossRef] [PubMed]
- Boehm, D.; Ott, M. A flow cytometry-based assay to investigate HIV-1 expression in SMYD5 shRNA containing primary CD4(+) T cells. STAR Protoc. 2023, 4, 102694. [Google Scholar] [CrossRef]
- Yu, D.; Wang, W.; Yoder, A.; Spear, M.; Wu, Y. The HIV envelope but not VSV glycoprotein is capable of mediating HIV latent infection of resting CD4 T cells. PLoS Pathog. 2009, 5, e1000633. [Google Scholar] [CrossRef]
- Roman, A.C.; Gonzalez-Rico, F.J.; Fernandez-Salguero, P.M. B1-SINE retrotransposons: Establishing genomic insulatory networks. Mob. Genet. Elem. 2011, 1, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Huang, J.; Tian, K.; Yi, X.; Zheng, H.; Zhu, Y.; Guo, T.; Ji, X. Cross-regulome profiling of RNA polymerases highlights the regulatory role of polymerase III on mRNA transcription by maintaining local chromatin architecture. Genome Biol. 2022, 23, 246. [Google Scholar] [CrossRef] [PubMed]
- Rajendra, K.C.; Cheng, R.; Zhou, S.; Lizarazo, S.; Smith, D.; Van Bortle, K. Evidence of RNA polymerase III recruitment and transcription at protein-coding gene promoters. bioRxiv 2024. [Google Scholar] [CrossRef]
- Wu, L.; Pan, J.; Thoroddsen, V.; Wysong, D.R.; Blackman, R.K.; Bulawa, C.E.; Gould, A.E.; Ocain, T.D.; Dick, L.R.; Errada, P.; et al. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot. Cell 2003, 2, 256–264. [Google Scholar] [CrossRef]
- Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef]
- Petrie, J.L.; Swan, C.; Ingram, R.M.; Frame, F.M.; Collins, A.T.; Dumay-Odelot, H.; Teichmann, M.; Maitland, N.J.; White, R.J. Effects on prostate cancer cells of targeting RNA polymerase III. Nucleic Acids Res. 2019, 47, 3937–3956. [Google Scholar] [CrossRef]
- Vabret, N.; Najburg, V.; Solovyov, A.; Gopal, R.; McClain, C.; Sulc, P.; Balan, S.; Rahou, Y.; Beauclair, G.; Chazal, M.; et al. Y RNAs are conserved endogenous RIG-I ligands across RNA virus infection and are targeted by HIV-1. iScience 2022, 25, 104599. [Google Scholar] [CrossRef]
- Rosa, M.D.; Gottlieb, E.; Lerner, M.R.; Steitz, J.A. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol. Cell. Biol. 1981, 1, 785–796. [Google Scholar]
- Weinmann, R.; Brendler, T.G.; Raskas, H.J.; Roeder, R.G. Low molecular weight viral RNAs transcribed by RNA polymerase III during adenovirus 2 infection. Cell 1976, 7, 557–566. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thompson, L.; Jamal, I.; Das, J.; Dang, C.; Hong, Z.; Katz, D.; Bosque, A.; Singh, V.B. RNA Polymerase III Regulates HIV Replication and Latency. Viruses 2025, 17, 1278. https://doi.org/10.3390/v17091278
Thompson L, Jamal I, Das J, Dang C, Hong Z, Katz D, Bosque A, Singh VB. RNA Polymerase III Regulates HIV Replication and Latency. Viruses. 2025; 17(9):1278. https://doi.org/10.3390/v17091278
Chicago/Turabian StyleThompson, Landon, Imran Jamal, Juthika Das, Casey Dang, Zhenzi Hong, Doran Katz, Alberto Bosque, and Vir B. Singh. 2025. "RNA Polymerase III Regulates HIV Replication and Latency" Viruses 17, no. 9: 1278. https://doi.org/10.3390/v17091278
APA StyleThompson, L., Jamal, I., Das, J., Dang, C., Hong, Z., Katz, D., Bosque, A., & Singh, V. B. (2025). RNA Polymerase III Regulates HIV Replication and Latency. Viruses, 17(9), 1278. https://doi.org/10.3390/v17091278




