Discovery of Landscape Phage Probes Against Cellular Communication Network Factor 1 (CCN1/Cyr61)
Abstract
1. Introduction
2. Materials and Methods
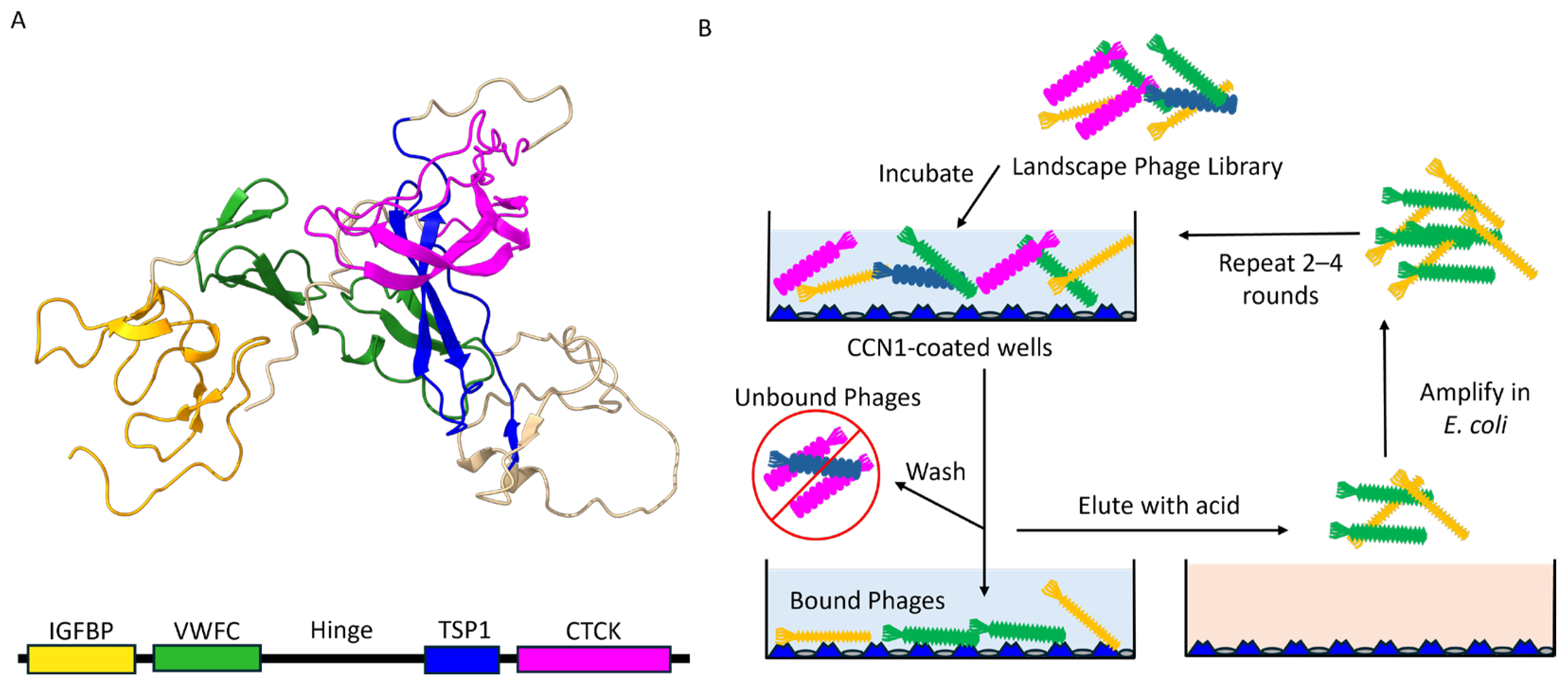
2.1. Landscape Phage Display Library
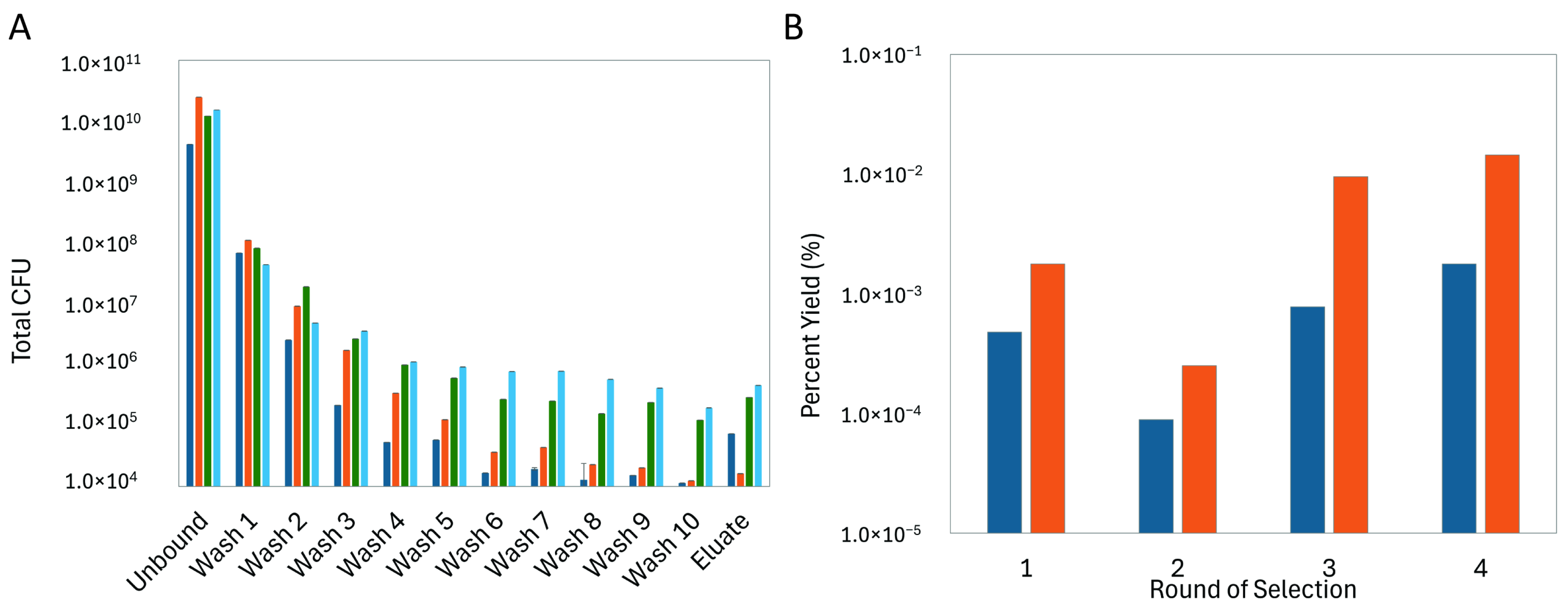
2.2. Selection of CCN1 Binding Phages
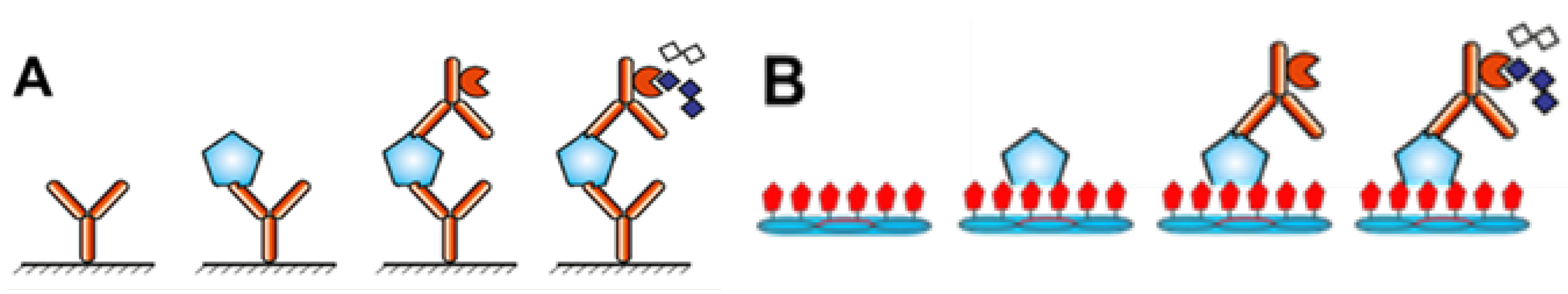
2.3. Screening Candidate CCN1-Binding Phages by Phage ELISA
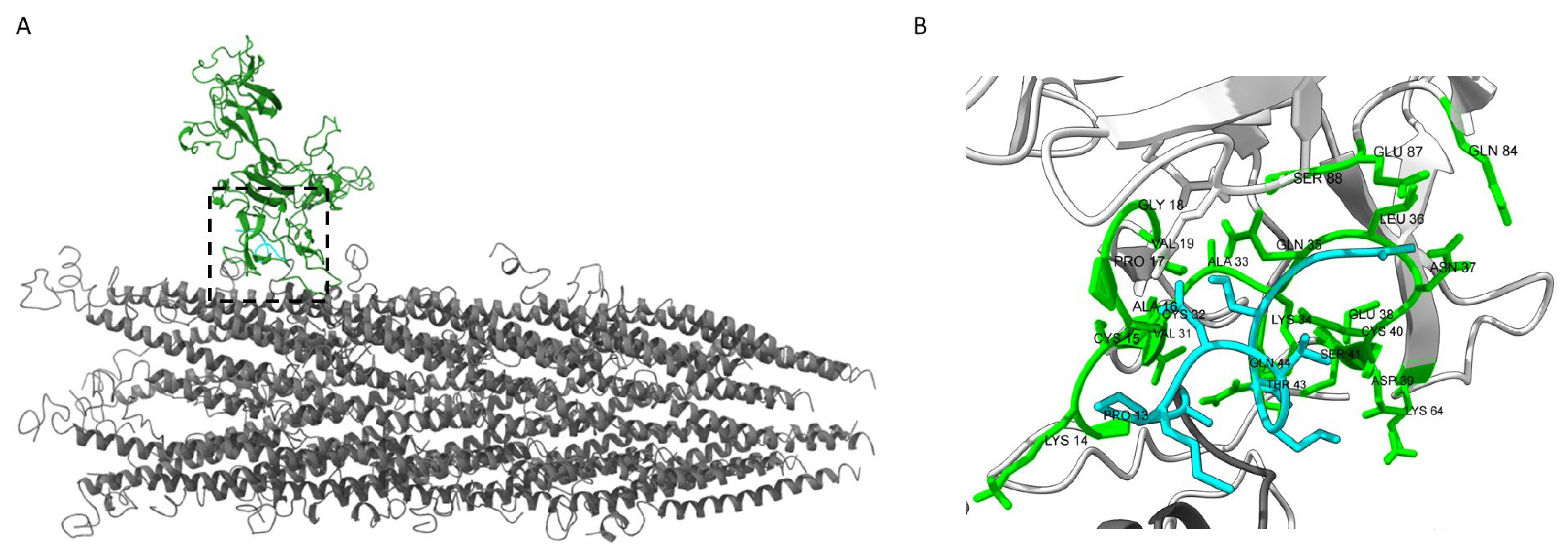
2.4. Molecular Modeling
3. Results
3.1. Isolation of CCN1-Binding Landscape Phages
3.2. Discovery and Analysis of CCN1-Binding Phages
3.3. Specificity of CCN1-Binding Phage Clones
3.4. Identification of Putative CCN1-Binding Sites by Molecular Docking
4. Discussion and Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jia, Q.; Xu, B.; Zhang, Y.; Ali, A.; Liao, X. CCN Family Proteins in Cancer: Insight Into Their Structures and Coordination Role in Tumor Microenvironment. Front. Genet. 2021, 12, 649387. [Google Scholar] [CrossRef] [PubMed]
- Kerek, R.; Sawma Awad, J.; Bassam, M.; Hajjar, C.; Ghantous, F.; Rizk, K.; Rima, M. The multifunctional protein CCN1/CYR61: Bridging physiology and disease. Exp. Mol. Pathol. 2025, 142, 104969. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, K.A.; Hasib, T.A.; Paul, S.K.; Saddam, M.; Mimi, A.; Saikat, A.S.M.; Faruque, H.A.; Rahman, M.A.; Uddin, M.J.; Kim, B. Potential Role of CCN Proteins in Breast Cancer: Therapeutic Advances and Perspectives. Curr. Oncol. 2021, 28, 4972–4985. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Chu, P.Y. Current and Developing Liquid Biopsy Techniques for Breast Cancer. Cancers 2022, 14, 2052. [Google Scholar] [CrossRef]
- Welch, H.G.; Bergmark, R. Cancer Screening, Incidental Detection, and Overdiagnosis. Clin. Chem. 2024, 70, 179–189. [Google Scholar] [CrossRef]
- Franzen, C.A.; Chen, C.C.; Todorovic, V.; Juric, V.; Monzon, R.I.; Lau, L.F. Matrix protein CCN1 is critical for prostate carcinoma cell proliferation and TRAIL-induced apoptosis. Mol. Cancer Res. 2009, 7, 1045–1055. [Google Scholar] [CrossRef]
- Balijepalli, P.; Yue, G.; Prasad, B.; Meier, K.E. Global Proteomics Analysis of Lysophosphatidic Acid Signaling in PC-3 Human Prostate Cancer Cells: Role of CCN1. Int. J. Mol. Sci. 2024, 25, 2067. [Google Scholar] [CrossRef]
- Alorda-Clara, M.; Torrens-Mas, M.; Morla-Barcelo, P.M.; Martinez-Bernabe, T.; Sastre-Serra, J.; Roca, P.; Pons, D.G.; Oliver, J.; Reyes, J. Use of Omics Technologies for the Detection of Colorectal Cancer Biomarkers. Cancers 2022, 14, 817. [Google Scholar] [CrossRef]
- Kok, S.H.; Chang, H.H.; Tsai, J.Y.; Hung, H.C.; Lin, C.Y.; Chiang, C.P.; Liu, C.M.; Kuo, M.Y. Expression of Cyr61 (CCN1) in human oral squamous cell carcinoma: An independent marker for poor prognosis. Head Neck 2010, 32, 1665–1673. [Google Scholar] [CrossRef]
- Malapelle, U.; Pisapia, P.; Pepe, F.; Russo, G.; Buono, M.; Russo, A.; Gomez, J.; Khorshid, O.; Mack, P.C.; Rolfo, C.; et al. The evolving role of liquid biopsy in lung cancer. Lung Cancer 2022, 172, 53–64. [Google Scholar] [CrossRef]
- Tsai, Y.T.; Schlom, J.; Donahue, R.N. Blood-based biomarkers in patients with non-small cell lung cancer treated with immune checkpoint blockade. J. Exp. Clin. Cancer Res. 2024, 43, 82. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yang, Y.; Zhu, Y.; Chen, H.; Yang, Y.; Zhang, L.; Li, W. Blood protein biomarkers in lung cancer. Cancer Lett. 2022, 551, 215886. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wei, W.; Tu, S.; Liang, B.; Li, C.; Li, Y.; Luo, W.; Wu, Y.; Dai, X.; Wang, Y.; et al. Upregulation of CYR61 by TGF-beta and YAP signaling exerts a counter-suppression of hepatocellular carcinoma. J. Biol. Chem. 2024, 300, 107208. [Google Scholar]
- Espinoza, I.; Kurapaty, C.; Park, C.H.; Vander Steen, T.; Kleer, C.G.; Wiley, E.; Rademaker, A.; Cuyas, E.; Verdura, S.; Buxo, M.; et al. Depletion of CCN1/CYR61 reduces triple-negative/basal-like breast cancer aggressiveness. Am. J. Cancer Res. 2022, 12, 839–851. [Google Scholar]
- Bartkowiak, K.; Heidrich, I.; Kwiatkowski, M.; Banys-Paluchowski, M.; Andreas, A.; Wurlitzer, M.; Geffken, M.; Voss, H.; Zeller, T.; Blankenberg, S.; et al. Circulating Cellular Communication Network Factor 1 Protein as a Sensitive Liquid Biopsy Marker for Early Detection of Breast Cancer. Clin. Chem. 2022, 68, 344–353. [Google Scholar] [CrossRef]
- Trier, N.H.; Houen, G. Antibodies as Diagnostic Targets and as Reagents for Diagnostics. Antibodies 2020, 9, 15. [Google Scholar] [CrossRef]
- Wang, X.; Ding, Q.; Groleau, R.R.; Wu, L.; Mao, Y.; Che, F.; Kotova, O.; Scanlan, E.M.; Lewis, S.E.; Li, P.; et al. Fluorescent Probes for Disease Diagnosis. Chem. Rev. 2024, 124, 7106–7164. [Google Scholar] [CrossRef]
- Yuan, M.; Wu, Y.; Zhao, C.; Chen, Z.; Su, L.; Yang, H.; Song, J. Activated molecular probes for enzyme recognition and detection. Theranostics 2022, 12, 1459–1485. [Google Scholar] [CrossRef]
- Kuhn, P.; Fuhner, V.; Unkauf, T.; Moreira, G.M.; Frenzel, A.; Miethe, S.; Hust, M. Recombinant antibodies for diagnostics and therapy against pathogens and toxins generated by phage display. Proteom. Clin. Appl. 2016, 10, 922–948. [Google Scholar] [CrossRef]
- Kothari, M.; Wanjari, A.; Acharya, S.; Karwa, V.; Chavhan, R.; Kumar, S.; Kadu, A.; Patil, R. A Comprehensive Review of Monoclonal Antibodies in Modern Medicine: Tracing the Evolution of a Revolutionary Therapeutic Approach. Cureus 2024, 16, e61983. [Google Scholar] [CrossRef]
- Alfaleh, M.A.; Alsaab, H.O.; Mahmoud, A.B.; Alkayyal, A.A.; Jones, M.L.; Mahler, S.M.; Hashem, A.M. Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Front. Immunol. 2020, 11, 1986. [Google Scholar] [CrossRef] [PubMed]
- Song, B.P.C.; Ch’ng, A.C.W.; Lim, T.S. Review of phage display: A jack-of-all-trades and master of most biomolecule display. Int. J. Biol. Macromol. 2024, 256 Pt 2, 128455. [Google Scholar] [CrossRef]
- Petrenko, V.A. Landscape Phage: Evolution from Phage Display to Nanobiotechnology. Viruses 2018, 10, 311. [Google Scholar] [CrossRef]
- Petrenko, V.A.; Smith, G.P. Phages from landscape libraries as substitute antibodies. Protein Eng. 2000, 13, 589–592. [Google Scholar] [CrossRef]
- Petrenko, V.A. Phage Display’s Prospects for Early Diagnosis of Prostate Cancer. Viruses 2024, 16, 277. [Google Scholar] [CrossRef]
- Han, L.; Xia, H.; Yin, L.; Petrenko, V.A.; Liu, A. Selected landscape phage probe as selective recognition interface for sensitive total prostate-specific antigen immunosensor. Biosens. Bioelectron. 2018, 106, 1–6. [Google Scholar] [CrossRef]
- Lang, Q.; Wang, F.; Yin, L.; Liu, M.; Petrenko, V.A.; Liu, A. Specific probe selection from landscape phage display library and its application in enzyme-linked immunosorbent assay of free prostate-specific antigen. Anal. Chem. 2014, 86, 2767–2774. [Google Scholar] [CrossRef]
- Brigati, J.; Williams, D.D.; Sorokulova, I.B.; Nanduri, V.; Chen, I.H.; Turnbough, C.L., Jr.; Petrenko, V.A. Diagnostic probes for Bacillus anthracis spores selected from a landscape phage library. Clin. Chem. 2004, 50, 1899–1906. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Liu, P.; Petrenko, V.A.; Liu, A. A Label-Free Electrochemical Impedance Cytosensor Based on Specific Peptide-Fused Phage Selected from Landscape Phage Library. Sci. Rep. 2016, 6, 22199. [Google Scholar] [CrossRef]
- Petrenko, V.A. Landscape Phage as a Molecular Recognition Interface for Detection Devices. Microelectron. J. 2008, 39, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lakshmanan, R.S.; Petrenko, V.A.; Chin, B.A. Phage-Based Pathogen Biosensors. In Phage Nanobiotechnology; Petrenko, V.A., Smith, G.P., Eds.; RSC Publishing: Cambridge, UK, 2011; p. 273. [Google Scholar]
- Brigati, J.R.; Petrenko, V.A. Thermostability of landscape phage probes. Anal. Bioanal. Chem. 2005, 382, 1346–1350. [Google Scholar] [CrossRef]
- Kuzmicheva, G.A.; Jayanna, P.K.; Eroshkin, A.M.; Grishina, M.A.; Pereyaslavskaya, E.S.; Potemkin, V.A.; Petrenko, V.A. Mutations in fd phage major coat protein modulate affinity of the displayed peptide. Protein Eng. Des. Sel. 2009, 22, 631–639. [Google Scholar] [CrossRef]
- Kuzmicheva, G.A.; Jayanna, P.K.; Sorokulova, I.B.; Petrenko, V.A. Diversity and censoring of landscape phage libraries. Protein Eng. Des. Sel. 2009, 22, 9–18. [Google Scholar] [CrossRef]
- Brigati, J.R.; Samoylova, T.I.; Jayanna, P.K.; Petrenko, V.A. Phage display for generating peptide reagents. Curr. Protoc. Protein Sci. 2008, 51, 18.9.1–18.9.27. [Google Scholar] [CrossRef] [PubMed]
- Petrenko, V.A.; Gillespie, J.W.; Xu, H.; O’Dell, T.; De Plano, L.M. Combinatorial Avidity Selection of Mosaic Landscape Phages Targeted at Breast Cancer Cells-An Alternative Mechanism of Directed Molecular Evolution. Viruses 2019, 11, 785. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Bohning, J.; Graham, M.; Letham, S.C.; Davis, L.K.; Schulze, U.; Stansfeld, P.J.; Corey, R.A.; Pearce, P.; Tarafder, A.K.; Bharat, T.A.M. Biophysical basis of filamentous phage tactoid-mediated antibiotic tolerance in P. aeruginosa. Nat. Commun. 2023, 14, 8429. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Honorato, R.V.; Trellet, M.E.; Jimenez-Garcia, B.; Schaarschmidt, J.J.; Giulini, M.; Reys, V.; Koukos, P.I.; Rodrigues, J.; Karaca, E.; van Zundert, G.C.P.; et al. The HADDOCK2.4 web server for integrative modeling of biomolecular complexes. Nat. Protoc. 2024, 19, 3219–3241. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef]
- Honorato, R.V.; Koukos, P.I.; Jimenez-Garcia, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef] [PubMed]
- Fagbohun, O.A.; Bedi, D.; Grabchenko, N.I.; Deinnocentes, P.A.; Bird, R.C.; Petrenko, V.A. Landscape phages and their fusion proteins targeted to breast cancer cells. Protein Eng. Des. Sel. 2012, 25, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Samoylova, T.I.; Petrenko, V.A.; Morrison, N.E.; Globa, L.P.; Baker, H.J.; Cox, N.R. Phage probes for malignant glial cells. Mol. Cancer Ther. 2003, 2, 1129–1137. [Google Scholar] [PubMed]
- Fagbohun, O.A.; Kazmierczak, R.A.; Petrenko, V.A.; Eisenstark, A. Metastatic prostate cancer cell-specific phage-like particles as a targeted gene-delivery system. J. Nanobiotechnology 2013, 11, 31. [Google Scholar] [CrossRef]
- Romanov, V.I.; Durand, D.B.; Petrenko, V.A. Phage display selection of peptides that affect prostate carcinoma cells attachment and invasion. Prostate 2001, 47, 239–251. [Google Scholar] [CrossRef]
- Gillespie, J.W.; Wei, L.; Petrenko, V.A. Selection of Lung Cancer-Specific Landscape Phage for Targeted Drug Delivery. Comb. Chem. High. Throughput Screen. 2016, 19, 412–422. [Google Scholar] [CrossRef]
- Gillespie, J.W.; Yang, L.; De Plano, L.M.; Stackhouse, M.A.; Petrenko, V.A. Evolution of a Landscape Phage Library in a Mouse Xenograft Model of Human Breast Cancer. Viruses 2019, 11, 988. [Google Scholar] [CrossRef]
- Bakhshinejad, B.; Sadeghizadeh, M. A polystyrene binding target-unrelated peptide isolated in the screening of phage display library. Anal. Biochem. 2016, 512, 120–128. [Google Scholar] [CrossRef]
- Adey, N.B.; Mataragnon, A.H.; Rider, J.E.; Carter, J.M.; Kay, B.K. Characterization of phage that bind plastic from phage-displayed random peptide libraries. Gene 1995, 156, 27–31. [Google Scholar] [CrossRef]
- Qiang, X.; Sun, K.; Xing, L.; Xu, Y.; Wang, H.; Zhou, Z.; Zhang, J.; Zhang, F.; Caliskan, B.; Wang, M.; et al. Discovery of a polystyrene binding peptide isolated from phage display library and its application in peptide immobilization. Sci. Rep. 2017, 7, 2673. [Google Scholar] [CrossRef]
- Vodnik, M.; Zager, U.; Strukelj, B.; Lunder, M. Phage display: Selecting straws instead of a needle from a haystack. Molecules 2011, 16, 790–817. [Google Scholar] [CrossRef]
- Kapp, T.G.; Rechenmacher, F.; Neubauer, S.; Maltsev, O.V.; Cavalcanti-Adam, E.A.; Zarka, R.; Reuning, U.; Notni, J.; Wester, H.J.; Mas-Moruno, C.; et al. A Comprehensive Evaluation of the Activity and Selectivity Profile of Ligands for RGD-binding Integrins. Sci. Rep. 2017, 7, 39805. [Google Scholar] [CrossRef]
- Takagi, J. Structural basis for ligand recognition by RGD (Arg-Gly-Asp)-dependent integrins. Biochem. Soc. Trans. 2004, 32 Pt 3, 403–406. [Google Scholar] [CrossRef]
- Won, J.H.; Choi, J.S.; Jun, J.I. CCN1 interacts with integrins to regulate intestinal stem cell proliferation and differentiation. Nat. Commun. 2022, 13, 3117. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Leu, S.J.; Todorovic, V.; Lam, S.C.; Lau, L.F. Identification of a novel integrin alphavbeta3 binding site in CCN1 (CYR61) critical for pro-angiogenic activities in vascular endothelial cells. J. Biol. Chem. 2004, 279, 44166–44176. [Google Scholar] [CrossRef]
- Leu, S.J.; Liu, Y.; Chen, N.; Chen, C.C.; Lam, S.C.; Lau, L.F. Identification of a novel integrin alpha 6 beta 1 binding site in the angiogenic inducer CCN1 (CYR61). J. Biol. Chem. 2003, 278, 33801–33808. [Google Scholar] [CrossRef]
- Ackar, L.; Casjens, S.; Andreas, A.; Raiko, I.; Bruning, T.; Geffken, M.; Peine, S.; Kollmeier, J.; Johnen, G.; Bartkowiak, K.; et al. Blood-based detection of lung cancer using cysteine-rich angiogenic inducer 61 (CYR61) as a circulating protein biomarker: A pilot study. Mol. Oncol. 2021, 15, 2877–2890. [Google Scholar] [CrossRef]
- Zeri, A.C.; Mesleh, M.F.; Nevzorov, A.A.; Opella, S.J. Structure of the coat protein in fd filamentous bacteriophage particles determined by solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. USA 2003, 100, 6458–6463. [Google Scholar] [CrossRef] [PubMed]
- Marvin, D.A.; Welsh, L.C.; Symmons, M.F.; Scott, W.R.; Straus, S.K. Molecular structure of fd (f1, M13) filamentous bacteriophage refined with respect to X-ray fibre diffraction and solid-state NMR data supports specific models of phage assembly at the bacterial membrane. J. Mol. Biol. 2006, 355, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Rakonjac, J.; Russel, M.; Khanum, S.; Brooke, S.J.; Rajic, M. Filamentous Phage: Structure and Biology. Adv. Exp. Med. Biol. 2017, 1053, 1–20. [Google Scholar]
- Conners, R.; Leon-Quezada, R.I.; McLaren, M.; Bennett, N.J.; Daum, B.; Rakonjac, J.; Gold, V.A.M. Cryo-electron microscopy of the f1 filamentous phage reveals insights into viral infection and assembly. Nat. Commun. 2023, 14, 2724. [Google Scholar] [CrossRef]
- Barozet, A.; Chacon, P.; Cortes, J. Current approaches to flexible loop modeling. Curr. Res. Struct. Biol. 2021, 3, 187–191. [Google Scholar] [CrossRef]
- Petrenko, V.A.; Gillespie, J.W.; De Plano, L.M.; Shokhen, M.A. Phage-Displayed Mimotopes of SARS-CoV-2 Spike Protein Targeted to Authentic and Alternative Cellular Receptors. Viruses 2022, 14, 384. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Huo, R.; Wang, L.; Zhou, Z.; Sun, Y.; Shen, B.; Wang, R.; Li, N. A novel anti-Cyr61 antibody inhibits breast cancer growth and metastasis in vivo. Cancer Immunol. Immunother. 2012, 61, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Tiessen, A.; Perez-Rodriguez, P.; Delaye-Arredondo, L.J. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res. Notes 2012, 5, 85. [Google Scholar] [CrossRef] [PubMed]
- Niwa, Y.; Suzuki, T.; Dohmae, N.; Simizu, S. O-Fucosylation of CCN1 is required for its secretion. FEBS Lett. 2015, 589, 3287–3293. [Google Scholar] [CrossRef]
- Ishizawa, Y.; Niwa, Y.; Suzuki, T.; Kawahara, R.; Dohmae, N.; Simizu, S. Identification and characterization of collagen-like glycosylation and hydroxylation of CCN1. Glycobiology 2019, 29, 696–704. [Google Scholar] [CrossRef]
- Zhu, Y.; Almuntashiri, S.; Han, Y.; Wang, X.; Somanath, P.R.; Zhang, D. The Roles of CCN1/CYR61 in Pulmonary Diseases. Int. J. Mol. Sci. 2020, 21, 7810. [Google Scholar] [CrossRef]

| Sequence * | Round 3 (Plastic and BSA) | Round 3 (BSA only) | Round 4 (10 µg CCN1) | Round 4 (1 µg CCN1) |
|---|---|---|---|---|
| ADMSSDFPD | - | 1 | - | 1 |
| AFVYDDAAD | - | - | 9 | 2 |
| ASDSDAFSG | - | - | 1 | 3 |
| AWSGYEGSD | 1 | - | - | - |
| DIGVMAENE | - | - | 1 | - |
| DIVILIIRN | - | - | 1 | - |
| DIVYFDNSD | - | - | 7 | - |
| DTTGSGVDG | - | 1 | - | - |
| EDDDSSFPD | - | 1 | - | - |
| ENRFVGDTD | - | - | - | 1 |
| EQNSVDLPD | 1 | - | - | - |
| ESPYSEFPD | 2 | - | - | 1 |
| ETYGDPSLG | - | - | 1 | - |
| EYYSLNGSD | 1 | - | - | - |
| GPGEALNTD | 1 | - | - | - |
| GQYEQSVAE | - | - | - | 1 |
| GSDSVDMPA | - | - | - | 1 |
| GSDSVEMPA | - | - | - | 1 |
| GSESVDMPA | 10 | 7 | 2 | 7 |
| VDNPQSGSD | 1 | - | - | - |
| VGRGDGNED | - | 1 | - | - |
| VQSSASSEG | - | - | 1 | - |
| VVDRNESMD | - | 3 | 1 | - |
| VVQDRSADD | 1 | - | - | - |
| VYGDSSFSD | - | 1 | - | - |
| VYTGSGSED | 1 | - | - | - |
| Round 3 (Plastic & BSA Depletion) | ||
|---|---|---|
| GS[D/E] | SVD | SGS[D/E] |
| AWSGYEGSD | EQNSVDLPD | VDNPQSGSD |
| EYYSLNGSD | GSESVDMPA | VYTGSGSED |
| GSESVDMPA | ||
| VDNPQSGSD | ||
| VYTGSGSED | ||
| Round 3 (BSA Depletion) | ||
| DXXSXFPD | RGD | |
| ADMSSDFPD | VGRGDGNED | |
| EDDDSSFPD | ||
| Round 4 (10 µg CCN1) | ||
| DIV | S[D/E]S | |
| DIVILIIRN | ASDSDAFSG | |
| DIVYFDNSD | GSESVDMPA | |
| Round 4 (1 µg CCN1) | ||
| S[D/E]FPD | GS[D/E]SV[D/E]MPA | |
| ADMSSDFPD | GSDSVDMPA | |
| ESPYSEFPD | GSDSVEMPA | |
| GSESVDMPA | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gillespie, J.W.; Petrenko, V.A. Discovery of Landscape Phage Probes Against Cellular Communication Network Factor 1 (CCN1/Cyr61). Viruses 2025, 17, 1273. https://doi.org/10.3390/v17091273
Gillespie JW, Petrenko VA. Discovery of Landscape Phage Probes Against Cellular Communication Network Factor 1 (CCN1/Cyr61). Viruses. 2025; 17(9):1273. https://doi.org/10.3390/v17091273
Chicago/Turabian StyleGillespie, James W., and Valery A. Petrenko. 2025. "Discovery of Landscape Phage Probes Against Cellular Communication Network Factor 1 (CCN1/Cyr61)" Viruses 17, no. 9: 1273. https://doi.org/10.3390/v17091273
APA StyleGillespie, J. W., & Petrenko, V. A. (2025). Discovery of Landscape Phage Probes Against Cellular Communication Network Factor 1 (CCN1/Cyr61). Viruses, 17(9), 1273. https://doi.org/10.3390/v17091273







