Virome Analysis of Small Mammals from the Brazilian Amazon
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction and cDNA Synthesis
2.3. Library Preparation and Sequencing
2.4. Bioinformatic Analysis
2.4.1. Data Filtering and Quality Control
2.4.2. Assembly and Alignment
2.4.3. Viral Diversity Analysis
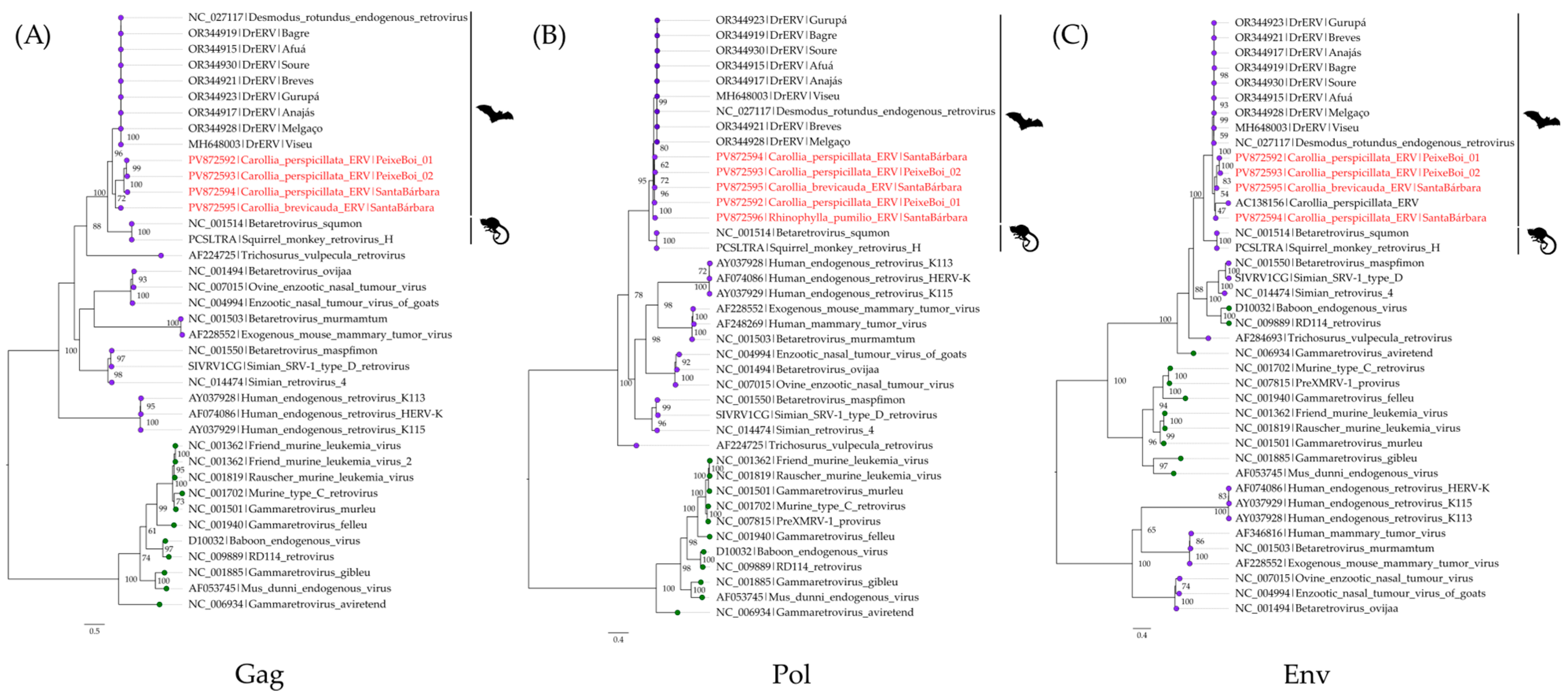
2.4.4. Inspection and Phylogenetic Analysis
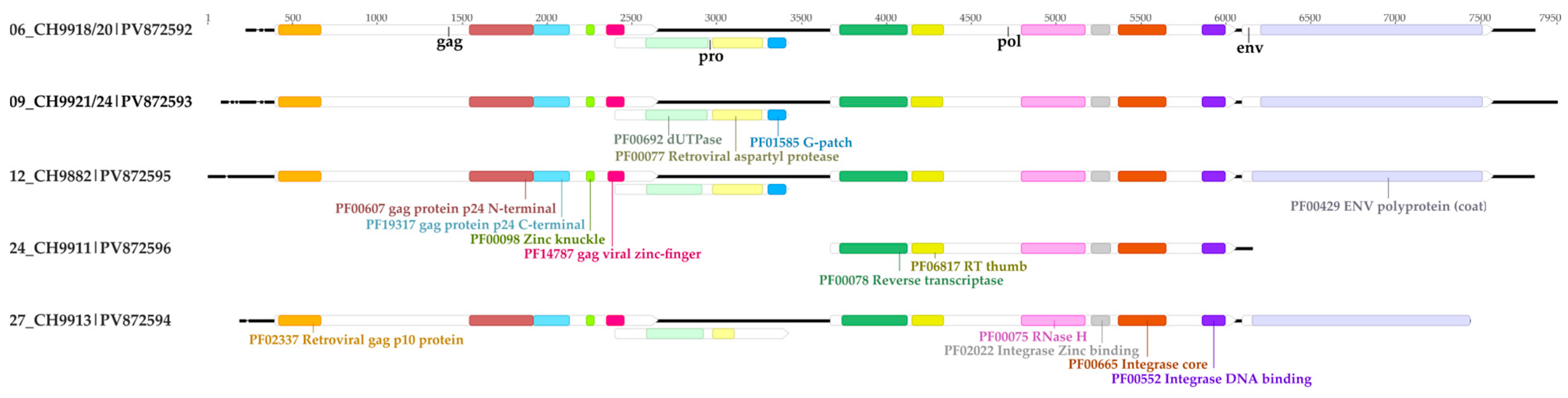
2.4.5. Functional Domains Annotation
3. Results
3.1. Data Processing
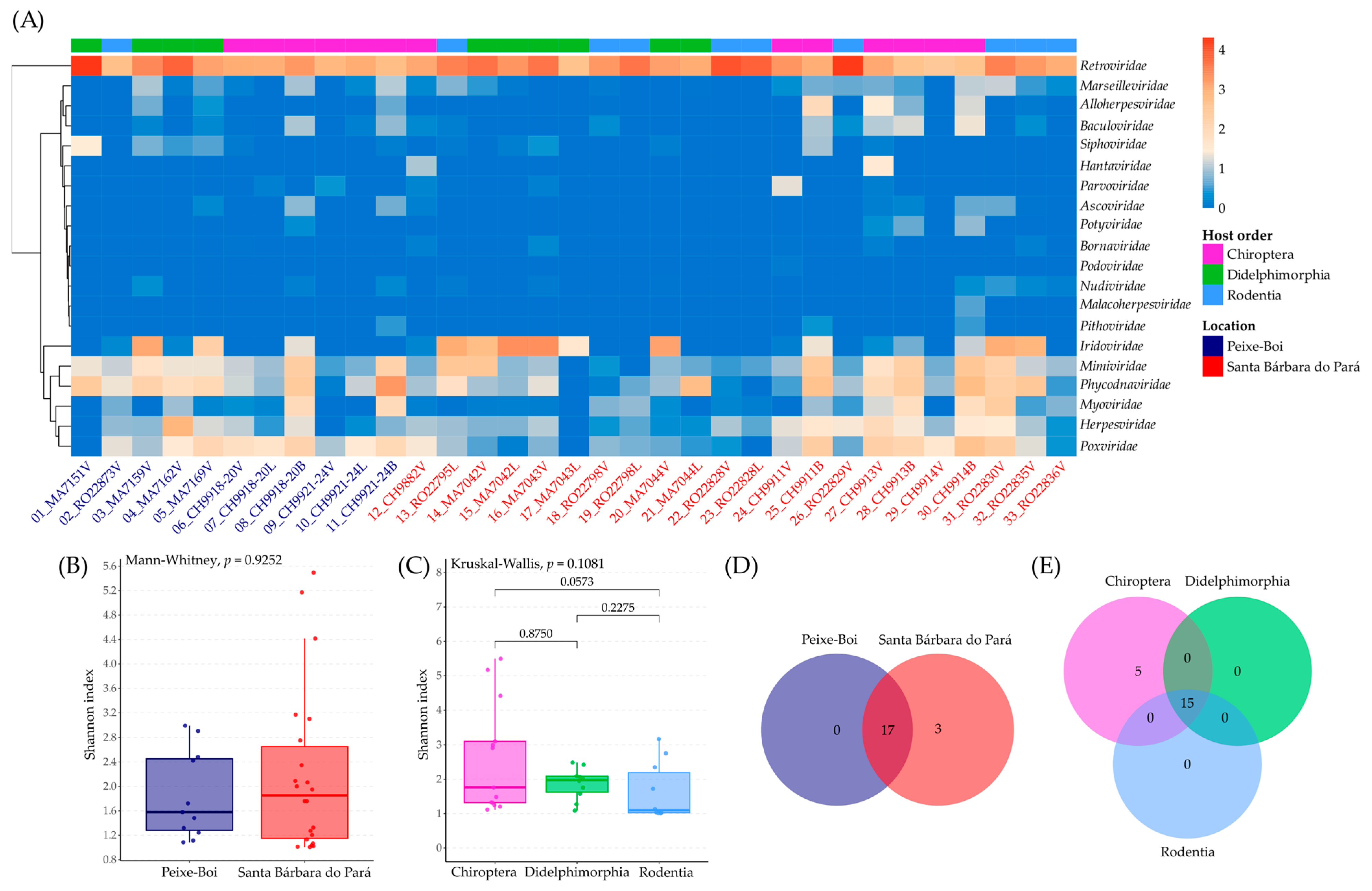
3.2. Viral Diversity
3.3. Bat Endogenous Retroviruses (Retroviridae)
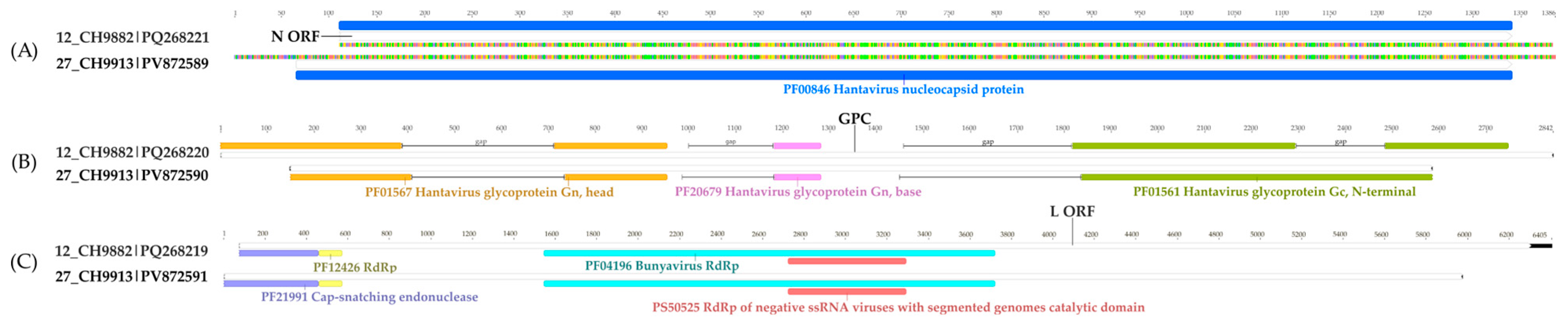
3.4. Mobatvirus (Hantaviridae)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Horefti, E. The Importance of the One Health Concept in Combating Zoonoses. Pathogens 2023, 12, 977. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global Trends in Emerging Infectious Diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Lapola, D.M.; Pinho, P.; Barlow, J.; Aragão, L.E.O.C.; Berenguer, E.; Carmenta, R.; Liddy, H.M.; Seixas, H.; Silva, C.V.J.; Silva-Junior, C.H.L.; et al. The drivers and impacts of Amazon forest degradation. Science 2023, 379, eabp8622. [Google Scholar] [CrossRef] [PubMed]
- Brazilian National Institute for Space Research. Legal Amazon Deforestation Rates. Available online: https://terrabrasilis.dpi.inpe.br/app/dashboard/deforestation/biomes/legal_amazon/rates (accessed on 21 July 2025).
- Ellwanger, J.H.; Fearnside, P.M.; Ziliotto, M.; Valverde-Villegas, J.M.; Veiga, A.B.G.D.; Vieira, G.F.; Bach, E.; Cardoso, J.C.; Müller, N.F.D.; Lopes, G.; et al. Synthesizing the connections between environmental disturbances and zoonotic spillover. An. Acad. Bras. Cienc. 2022, 94, e20211530. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Crameri, G. Emerging zoonotic viral diseases. Rev. Sci. Tech. 2014, 33, 569–581. [Google Scholar] [CrossRef]
- Van Brussel, K.; Holmes, E.C. Zoonotic disease and virome diversity in bats. Curr. Opin. Virol. 2022, 52, 192–202. [Google Scholar] [CrossRef]
- Tian, J.; Sun, J.; Li, D.; Wang, N.; Wang, L.; Zhang, C.; Meng, X.; Ji, X.; Suchard, M.A.; Zhang, X.; et al. Emerging viruses: Cross-species transmission of coronaviruses, filoviruses, henipaviruses, and rotaviruses from bats. Cell Rep. 2022, 39, 110969. [Google Scholar] [CrossRef]
- Du, H.; Zhang, L.; Zhang, X.; Yun, F.; Chang, Y.; Tuersun, A.; Aisaiti, K.; Ma, Z. Metagenome-Assembled Viral Genomes Analysis Reveals Diversity and Infectivity of the RNA Virome of Gerbillinae Species. Viruses 2022, 14, 356. [Google Scholar] [CrossRef]
- Wu, Z.; Lu, L.; Du, J.; Yang, L.; Ren, X.; Liu, B.; Jiang, J.; Yang, J.; Dong, J.; Sun, L.; et al. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 2018, 6, 178. [Google Scholar] [CrossRef]
- Ko, K.K.K.; Chng, K.R.; Nagarajan, N. Metagenomics-enabled microbial surveillance. Nat. Microbiol. 2022, 7, 486–496. [Google Scholar] [CrossRef]
- Purushothaman, S.; Meola, M.; Egli, A. Combination of Whole Genome Sequencing and Metagenomics for Microbiological Diagnostics. Int. J. Mol. Sci. 2022, 23, 9834. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 25 July 2025).
- Kolde, R. Package ‘pheatmap’. R. Package 2015, 1, 790. [Google Scholar]
- Oksanen, J.; Simpson, G.L.; Guillaume Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Package ‘vegan’. R. Package 2013, 2, 1–125. [Google Scholar]
- Wickham, H. ggplot2. WIREs Comp. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega for Making Accurate Alignments of Many Protein Sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Wong, T.K.F.; Ly-Trong, N.; Ren, H.; Baños, H.; Roger, A.J.; Susko, E.; Bielow, C.; De Maio, N.; Goldman, N.; Hahn, M.W.; et al. IQ-TREE 3: Phylogenomic Inference Software using Complex Evolutionary Models. Ecoevorxiv 2025, X2P62N. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 14 June 2025).
- Inkscape. Available online: https://inkscape.org/release/inkscape-1.1/ (accessed on 14 June 2025).
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Hernández, L.H.A.; da Paz, T.Y.B.; da Silva, S.P.; de Barros, B.C.V.; Coelho, T.F.S.B.; Cruz, A.C.R. First description of a mobatvirus (Hantaviridae) in the Amazon region. Virus Res. 2024, 350, 199494. [Google Scholar] [CrossRef]
- Ellwanger, J.H.; Kulmann-Leal, B.; Kaminski, V.L.; Valverde-Villegas, J.M.; Veiga, A.B.G.D.; Spilki, F.R.; Fearnside, P.M.; Caesar, L.; Giatti, L.L.; Wallau, G.L.; et al. Beyond diversity loss and climate change: Impacts of Amazon deforestation on infectious diseases and public health. An. Acad. Bras. Cienc. 2020, 92, e20191375. [Google Scholar] [CrossRef]
- Ellwanger, J.H.; Veiga, A.B.G.; Kaminski, V.L.; Valverde-Villegas, J.M.; Freitas, A.W.Q.; Chies, J.A.B. Control and prevention of infectious diseases from a One Health perspective. Genet. Mol. Biol. 2021, 44, e20200256. [Google Scholar] [CrossRef]
- Guo, Y.; Su, C.; Liang, H.; Jiang, X.; Yang, R.; Ye, J.; Gillespie, T.R.; Gao, Z.; Xu, L. Virome diversity and potential sharing of wild mammals in a biodiversity hotspot, Yunnan, China. Virol. J. 2025, 22, 79. [Google Scholar] [CrossRef]
- Carlson, C.J.; Zipfel, C.M.; Garnier, R.; Bansal, S. Global estimates of mammalian viral diversity accounting for host sharing. Nat. Eco Evol. 2019, 3, 1070–1075. [Google Scholar] [CrossRef] [PubMed]
- Olival, K.J.; Hosseini, P.R.; Zambrana-Torrelio, C.; Ross, N.; Bogich, T.L.; Daszak, P. Host and viral traits predict zoonotic spillover from mammals. Nature 2017, 548, 612. [Google Scholar] [CrossRef] [PubMed]
- Prestes, N.G.O.; Hernández, L.H.A.; da Silva, F.S.; da Paz, T.Y.B.; Aragão, A.O.; de Barros, B.C.V.; Guimarães, R.J.P.S.; Ramos, R.T.J.; Casseb, L.M.N.; da Silva, S.P.; et al. Discovery of a Rodent Hepacivirus in the Brazilian Amazon. Viruses 2025, 17, 830. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.F.M.; da Silva, F.S.; Cruz, A.C.R.; da Silva, S.P.; Queiroz, A.L.N.; Casseb, L.M.N.; Martins, L.C.; Medeiros, D.B.A. Viral diversity in wild rodents in the regions of Canaã de Carajás and Curionopólis, State of Pará, Brazil. Front. Microbiol. 2025, 15, 1502462. [Google Scholar] [CrossRef]
- Tirera, S.; de Thoisy, B.; Donato, D.; Bouchier, C.; Lacoste, V.; Franc, A.; Lavergne, A. The Influence of Habitat on Viral Diversity in Neotropical Rodent Hosts. Viruses 2021, 13, 1690. [Google Scholar] [CrossRef]
- de Moraes Pires, W.M.; Cruz, A.C.R.; de Souza, A.J.S.; Silva, S.P.; Souza Barbosa Coelho, T.F.; Dias, D.D.; Rosa Júnior, J.W.; Mendes, S.B.; da Costa Fraga, E.; Barros, M.C.; et al. Genomic characterization of a novel Hepatovirus identified in Maranhão state, Brazil. Sci. Rep. 2024, 14, 7981. [Google Scholar] [CrossRef]
- Short, S.M. The ecology of viruses that infect eukaryotic algae. Environ. Microbiol. 2012, 14, 2253–2271. [Google Scholar] [CrossRef]
- Chinchar, V.G.; Hick, P.; Ince, I.A.; Jancovich, J.K.; Marschang, R.; Qin, Q.; Subramaniam, K.; Waltzek, T.B.; Whittington, R.; Williams, T.; et al. ICTV Virus Taxonomy Profile: Iridoviridae. J. Gen. Virol. 2017, 98, 890–891. [Google Scholar] [CrossRef]
- Temmam, S.; Davoust, B.; Berenger, J.M.; Raoult, D.; Desnues, C. Viral metagenomics on animals as a tool for the detection of zoonoses prior to human infection? Int. J. Mol. Sci. 2014, 15, 10377–10397. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Shi, M.; Holmes, E.C. Using Metagenomics to Characterize an Expanding Virosphere. Cell 2018, 172, 1168–1172. [Google Scholar] [CrossRef]
- Luis, A.D.; Hayman, D.T.; O’Shea, T.J.; Cryan, P.M.; Gilbert, A.T.; Pulliam, J.R.; Mills, J.N.; Timonin, M.E.; Willis, C.K.; Cunningham, A.A.; et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc. Biol. Sci. 2013, 280, 20122753. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Luo, C.F.; Xiang, R.; Min, J.M.; Shao, Z.T.; Zhao, Y.L.; Chen, L.; Huang, L.; Zhang, Y.; Liu, S.S.; et al. Host taxonomy and environment shapes insectivore viromes and viral spillover risks in Southwestern China. Microbiome 2025, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Jern, P.; Greenwood, A.D. Wildlife endogenous retroviruses: Colonization, consequences, and cooption. Trends Genet. 2024, 40, 149–159. [Google Scholar] [CrossRef]
- Mager, D.L.; Stoye, J.P. Mammalian Endogenous Retroviruses. Microbiol. Spectr. 2015, 3, MDNA3–0009-2014. [Google Scholar] [CrossRef]
- Hayward, J.A.; Tachedjian, G. Retroviruses of Bats: A Threat Waiting in the Wings? mBio 2021, 12, e0194121. [Google Scholar] [CrossRef] [PubMed]
- Escalera-Zamudio, M.; Mendoza, M.L.; Heeger, F.; Loza-Rubio, E.; Rojas-Anaya, E.; Méndez-Ojeda, M.L.; Taboada, B.; Mazzoni, C.J.; Arias, C.F.; Greenwood, A.D. A novel endogenous betaretrovirus in the common vampire bat (Desmodus rotundus) suggests multiple independent infection and cross-species transmission events. J. Virol. 2015, 89, 5180–5184. [Google Scholar] [CrossRef]
- Albuquerque, N.K.; Silva, S.P.; Aragão, C.F.; Cunha, T.C.A.S.; Paiva, F.A.S.; Coelho, T.F.S.B.; Cruz, A.C.R. Virome analysis of Desmodus rotundus tissue samples from the Amazon region. BMC Genomics 2024, 25, 34. [Google Scholar] [CrossRef]
- Hayward, J.A.; Tachedjian, M.; Cui, J.; Field, H.; Holmes, E.C.; Wang, L.F.; Tachedjian, G. Identification of diverse full-length endogenous betaretroviruses in megabats and microbats. Retrovirology 2013, 10, 35. [Google Scholar] [CrossRef]
- Chung, H.C.; Kim, S.J.; Hwang, S.J.; Jeon, Y.S.; Song, M.S.; Ko, S.H.; Lee, J.; Choi, Y.; Chung, C.U.; Lee, J.M. Identification and characterization of recent retrovirus in Rhinolophus ferrumequinum bats. Microbiol. Spectr. 2024, 12, e0432323. [Google Scholar] [CrossRef] [PubMed]
- Bradfute, S.B.; Calisher, C.H.; Klempa, B.; Klingström, J.; Kuhn, J.H.; Laenen, L.; Tischler, N.D.; Maes, P. ICTV Virus Taxonomy Profile: Hantaviridae 2024. J. Gen. Virol. 2024, 105, 001975. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses. Family: Hantaviridae. Genus: Mobatvirus. Available online: https://ictv.global/report/chapter/hantaviridae/hantaviridae/mammantavirinae/mobatvirus (accessed on 5 August 2025).
- Barbosa Dos Santos, M.; Koide Albuquerque, N.; Patroca da Silva, S.; Silva da Silva, F.; Damous Dias, D.; Brito Mendes, S.; Fernandes Souza Barbosa Coelho, T.; Barros, M.C.; Ribeiro Cruz, A.C. A novel hantavirus identified in bats (Carollia perspicillata) in Brazil. Sci. Rep. 2024, 14, 6346. [Google Scholar] [CrossRef]
- Yamada, K.; Kikuchi, F.; Dunnum, J.L.; Gutiérrez-Moreno, P.; Armién, B.; Pérez-Callejas, M.; Land, D.; Colella, J.P.; Mizutani, T.; Maeda, K.; et al. Genetically distinct hantaviruses in two bat species in Panamá. iScience 2025, 28, 112749. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Pathogens Prioritization: A Scientific Framework for Epidemic and Pandemic Research Preparedness. Available online: https://www.who.int/publications/m/item/pathogens-prioritization-a-scientific-framework-for-epidemic-and-pandemic-research-preparedness (accessed on 5 August 2025).
- Barquez, R.; Perez, S.; Miller, B.; Diaz, M. The IUCN Red List of Threatened Species 2015. Carollia perspicillata. Available online: https://www.iucnredlist.org/species/3905/22133716 (accessed on 5 August 2025).
- Sampaio, E.; Lim, B.; Peters, S. The IUCN Red List of Threatened Species 2016. Carollia brevicauda. Available online: https://www.iucnredlist.org/species/3903/22134642 (accessed on 5 August 2025).
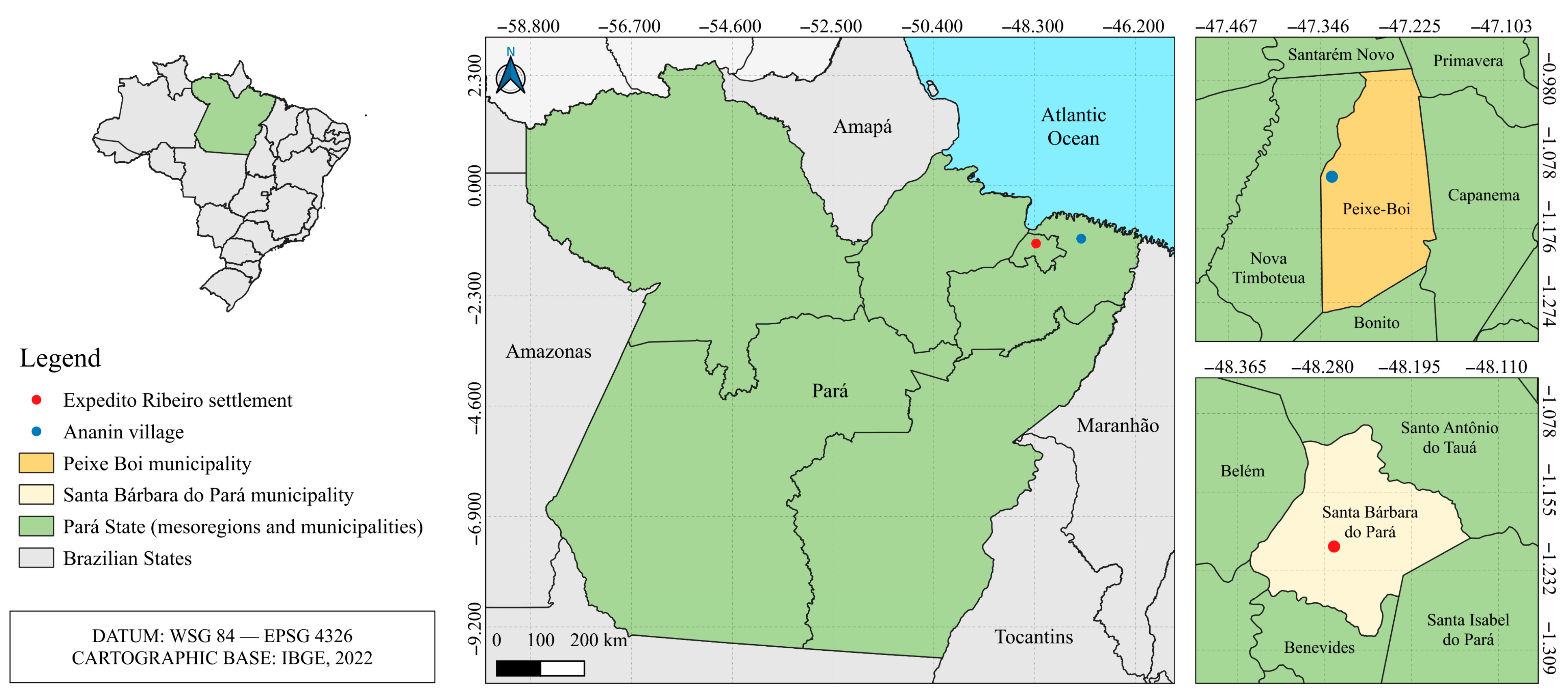


| Location | Sample ID | Tissue | Host Species | Collection Date | Total Reads | Fastp | SortMeRNA | Viral Reads |
|---|---|---|---|---|---|---|---|---|
| Peixe-Boi (Ananin village) | 01_MA7151 | Viscera | Cryptonanus agricolai | 11 September 2015 | 52,309,212 | 47,490,186 | 8,498,016 | 20,423 |
| 02_RO22873 | Viscera | Rattus rattus | 12 September 2015 | 49,198,384 | 45,713,586 | 12,068,688 | 6731 | |
| 03_MA7159 | Viscera | Marmosops pinheiroi | 14 September 2015 | 52,770,248 | 47,445,710 | 16,385,644 | 65,091 | |
| 04_MA7162 | Viscera | Metachirus myosuros | 15 September 2015 | 49,699,504 | 46,350,714 | 13,363,622 | 198,399 | |
| 05_MA7169 | Viscera | Philander opossum | 18 September 2015 | 57,013,870 | 52,138,166 | 14,284,912 | 25,126 | |
| 06_CH9918/20 | Viscera | Carollia perspicillata | 25 May 2016 | 46,011,536 | 42,632,696 | 26,247,326 | 30,878 | |
| 07_CH9918/20 | Liver | 34,922,698 | 31,903,538 | 19,319,456 | 20,866 | |||
| 08_CH9918/20 | Brain | 42,923,108 | 39,645,450 | 6,098,404 | 14,962 | |||
| 09_CH9921/24 | Viscera | Carollia perspicillata | 26 May 2016 | 10,162,764 | 10,161,884 | 9,438,986 | 7148 | |
| 10_CH9921/24 | Liver | 43,566,608 | 40,290,140 | 16,003,212 | 18,795 | |||
| 11_CH9921/24 | Brain | 33,354,502 | 30,545,230 | 11,003,166 | 29,856 | |||
| Santa Bárbara do Pará (Expedito Ribeiro settlement) | 12_CH9882 | Viscera | Carollia brevicauda | 17 October 2014 | 60,996,864 | 55,590,760 | 38,036,772 | 47,276 |
| 13_RO22795 | Liver | Echimys chrysurus | 20 October 2014 | 72,620,606 | 68,639,944 | 62,188,324 | 290,584 | |
| 14_MA7042 | Viscera | Marmosops pinheiroi | 23 October 2014 | 60,191,660 | 56,574,576 | 54,504,442 | 323,359 | |
| 15_MA7042 | Liver | 27,118,572 | 25,941,438 | 21,622,680 | 92,321 | |||
| 16_MA7043 | Viscera | Philander opossum | 23 October 2014 | 41,365,578 | 37,067,938 | 36,728,954 | 238,408 | |
| 17_MA7043 | Liver | 14,035,228 | 13,518,008 | 478,446 | 230 | |||
| 18_RO22798 | Viscera | Oecomys paricola | 24 October 2014 | 48,997,760 | 41,275,094 | 27,560,462 | 50,691 | |
| 19_RO22798 | Liver | 10,378,774 | 9,977,804 | 3,811,070 | 16,257 | |||
| 20_MA7044 | Viscera | Marmosops pinheiroi | 25 October 2014 | 45,637,316 | 40,316,828 | 32,671,306 | 94,728 | |
| 21_MA7044 | Liver | 28,369,886 | 25,422,456 | 16,867,168 | 28,167 | |||
| 22_RO22828 | Viscera | Oecomys paricola | 28 October 2014 | 31,158,412 | 29,536,146 | 26,805,502 | 260,923 | |
| 23_RO22828 | Liver | 17,589,744 | 16,813,682 | 11,660,122 | 83,672 | |||
| 24_CH9911 | Viscera | Rhinophylla pumilio | 07 April 2015 | 43,168,902 | 39,104,674 | 21,264,060 | 44,178 | |
| 25_CH9911 | Brain | 33,583,454 | 30,474,710 | 4,988,994 | 9857 | |||
| 26_RO22829 | Viscera | Nectomys rattus | 08 April 2015 | 36,475,646 | 32,978,740 | 27,859,838 | 508,107 | |
| 27_CH9913 | Viscera | Carollia perspicillata | 09 April 2015 | 30,715,366 | 28,149,712 | 9,439,310 | 15,579 | |
| 28_CH9913 | Brain | 35,577,070 | 32,750,072 | 9,624,060 | 10,227 | |||
| 29_CH9914 | Viscera | Carollia perspicillata | 09 April 2015 | 34,483,028 | 31,526,694 | 5,905,172 | 2030 | |
| 30_CH9914 | Brain | 24,492,902 | 22,588,498 | 10,498,444 | 20,738 | |||
| 31_RO22830 | Viscera | Guerlinguetus aestuans | 08 April 2015 | 39,958,576 | 36,804,108 | 5,893,486 | 28,790 | |
| 32_RO22835 | Viscera | Rattus rattus | 16 April 2015 | 21,722,476 | 18,281,404 | 14,685,132 | 40,663 | |
| 33_RO22836 | Viscera | Oecomys paricola | 16 April 2015 | 31,059,340 | 27,370,724 | 11,802,244 | 11,952 | |
| Total | - | - | 1,261,629,594 | 1,155,021,310 | 607,607,420 | 2,657,012 |
| Origin Sample | Host Species | Length (nt) | Coverage (×) | Nt Identity (%) with Reference |
|---|---|---|---|---|
| 06_CH9918/20 | C. perspicillata | 7473 | 26.5 | 74.08 |
| 09_CH9921/24 | C. perspicillata | 7748 | 15.4 | 71.99 |
| 12_CH9882 | C. brevicauda | 7805 | 54.2 | 75.36 |
| 24_CH9911 1 | R. pumilio | 2483 | 82.6 | 69.3 |
| 27_CH9913 | C. perspicillata | 7097 | 9.4 | 88.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández, L.H.A.; da Silva, F.S.; da Paz, T.Y.B.; Dias, D.D.; de Barros, B.d.C.V.; Nunes, B.T.D.; Casseb, L.M.N.; da Silva, S.P.; da Costa Vasconcelos, P.F.; Cruz, A.C.R. Virome Analysis of Small Mammals from the Brazilian Amazon. Viruses 2025, 17, 1251. https://doi.org/10.3390/v17091251
Hernández LHA, da Silva FS, da Paz TYB, Dias DD, de Barros BdCV, Nunes BTD, Casseb LMN, da Silva SP, da Costa Vasconcelos PF, Cruz ACR. Virome Analysis of Small Mammals from the Brazilian Amazon. Viruses. 2025; 17(9):1251. https://doi.org/10.3390/v17091251
Chicago/Turabian StyleHernández, Leonardo Henrique Almeida, Fábio Silva da Silva, Thito Yan Bezerra da Paz, Daniel Damous Dias, Bruno de Cássio Veloso de Barros, Bruno Tardelli Diniz Nunes, Lívia Medeiros Neves Casseb, Sandro Patroca da Silva, Pedro Fernando da Costa Vasconcelos, and Ana Cecília Ribeiro Cruz. 2025. "Virome Analysis of Small Mammals from the Brazilian Amazon" Viruses 17, no. 9: 1251. https://doi.org/10.3390/v17091251
APA StyleHernández, L. H. A., da Silva, F. S., da Paz, T. Y. B., Dias, D. D., de Barros, B. d. C. V., Nunes, B. T. D., Casseb, L. M. N., da Silva, S. P., da Costa Vasconcelos, P. F., & Cruz, A. C. R. (2025). Virome Analysis of Small Mammals from the Brazilian Amazon. Viruses, 17(9), 1251. https://doi.org/10.3390/v17091251










