APOBEC3B Promotes SARS-CoV-2 Through Activation of PKR/eIF2⍺ and AMPD2 Dysregulation
Abstract
1. Introduction
2. Results
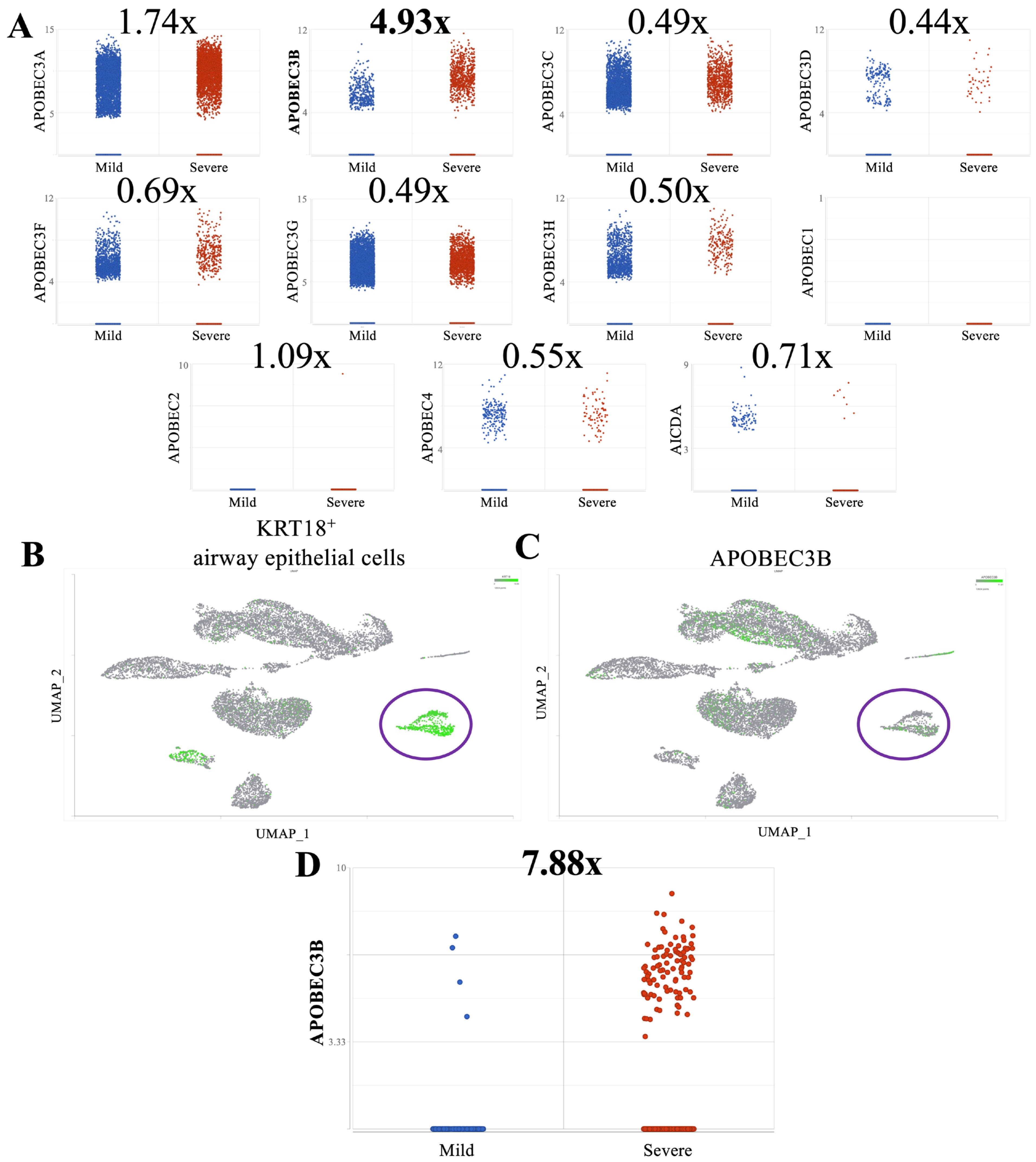
2.1. A3B Is Overexpressed in Broncho-Alveolar Lavage Fluid from Patients with Severe SARS-CoV-2 Infection
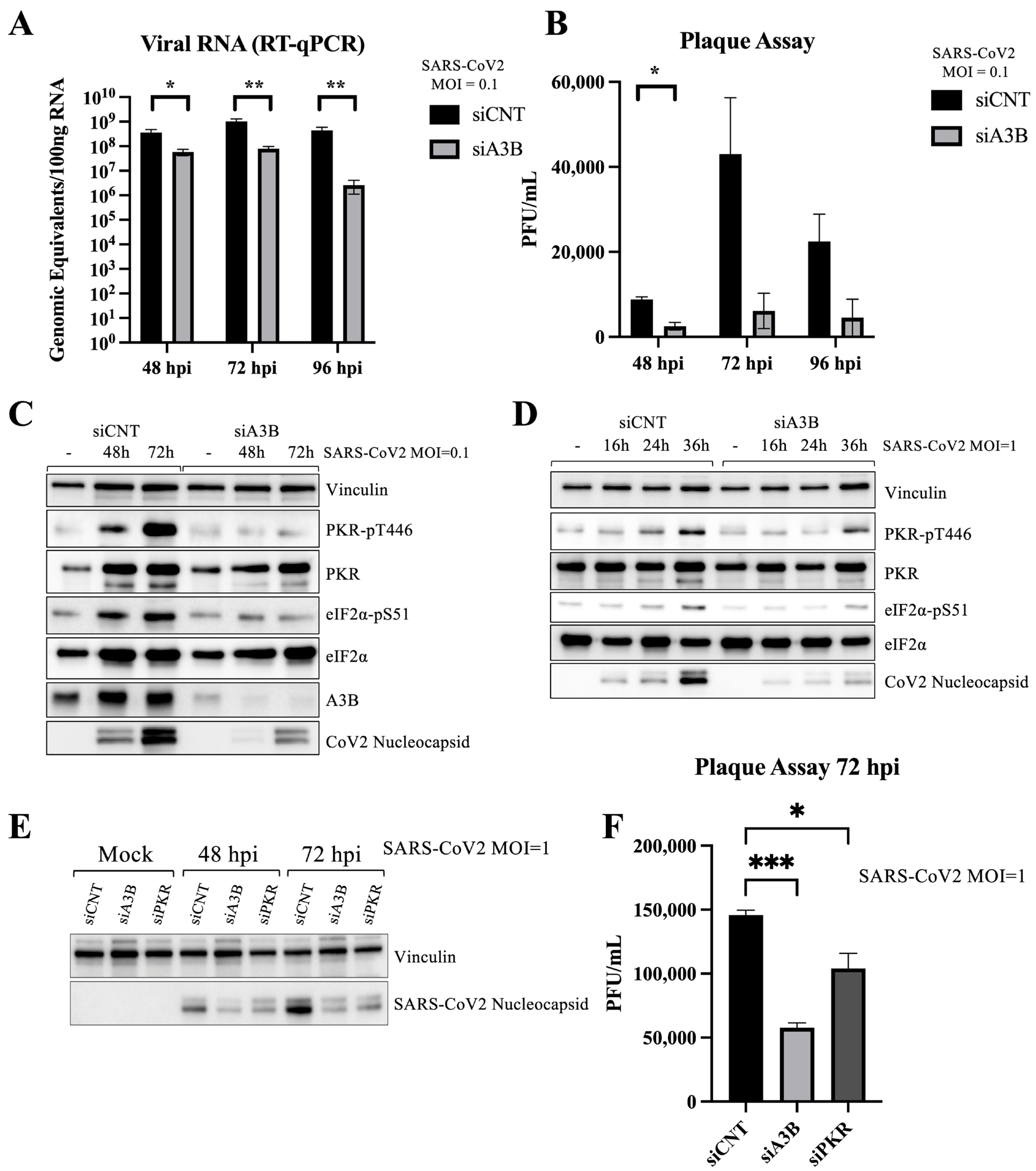
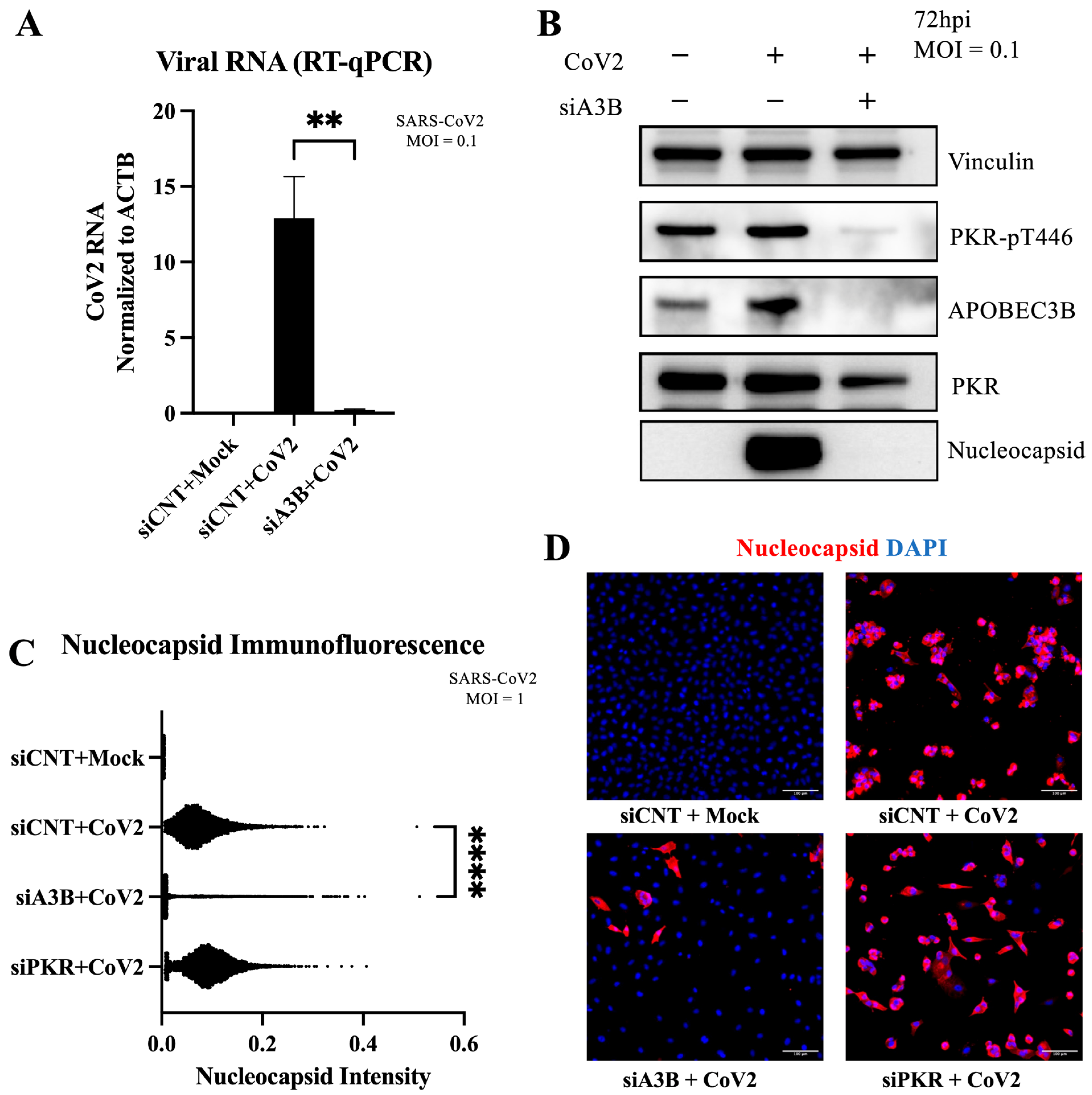
2.2. A3B Knockdown Significantly Reduces SARS-CoV-2 Infectivity in Caco-2 Cells
2.3. A3B Knockdown Reduces SARS-CoV-2 Infectivity in Caco-2 via Attenuation of PKR/eIF2⍺ Pathway
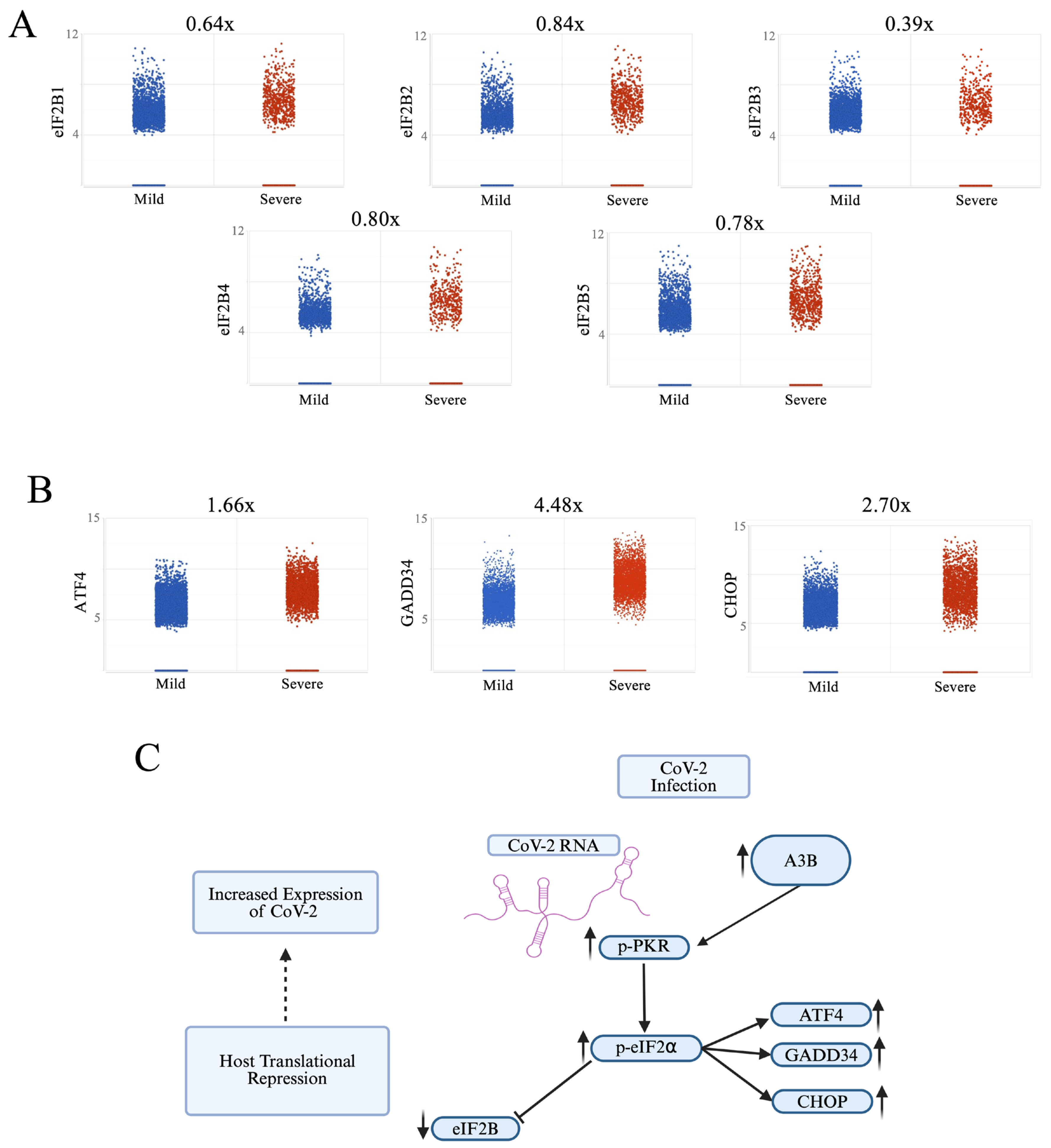
2.4. Severe COVID-19 Patient BALF Cells Show Signs of p-PKR/p-eIF2⍺ Pathway Activation
2.5. A3B Knockdown Reduces SARS-CoV-2 Infectivity in A549-ACE2 Independently of PKR Activation
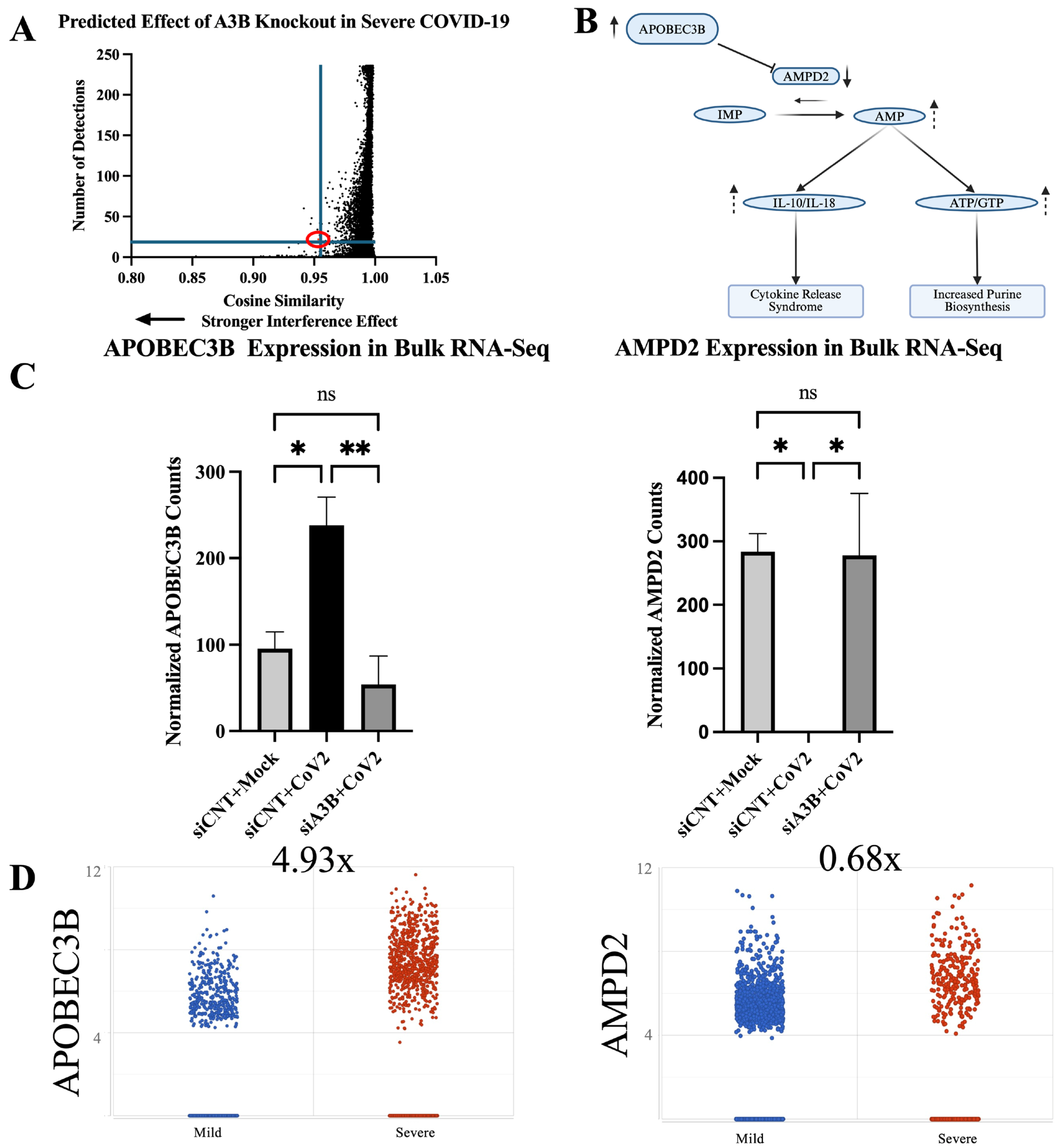
2.6. Geneformer Predicts A3B Knockout Dysregulates AMPD2 in the Context of Severe SARS-CoV-2 Infection
2.7. AMPD2 Is Downregulated in Severe COVID-19 Infection
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. RNA Interference
4.3. SARS-CoV-2 Virus Infections
4.4. Reverse Transcription-Quantitative Polymerase Chain Reaction
4.5. Western Blots
4.6. Immunofluorescence
4.7. Bulk RNA Sequencing
4.8. Bulk RNA Sequencing Analysis
4.9. Single-Cell RNA Sequencing Analysis and Cell Type Identification
4.10. Geneformer
4.11. Antibodies
4.12. Statistical Analysis and Plotting
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Salter, J.D.; Bennett, R.P.; Smith, H.C. The APOBEC protein family: United by structure, divergent in function. Trends Biochem. Sci. 2016, 41, 578–594. [Google Scholar] [CrossRef] [PubMed]
- Jarmuz, A.; Chester, A.; Bayliss, J.; Gisbourne, J.; Dunham, I.; Scott, J.; Navaratnam, N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 2002, 79, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 2006, 80, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.C.; Bennett, R.P.; Kizilyer, A.; McDougall, W.M.; Prohaska, K.M. Functions and Regulation of the APOBEC Family of Proteins; Elsevier: Amsterdam, The Netherlands, 2012; pp. 258–268. [Google Scholar]
- Navarro, F.; Bollman, B.; Chen, H.; König, R.; Yu, Q.; Chiles, K.; Landau, N.R. Complementary function of the two catalytic domains of APOBEC3G. Virology 2005, 333, 374–386. [Google Scholar] [CrossRef]
- Yang, H.; Ito, F.; Wolfe, A.D.; Li, S.; Mohammadzadeh, N.; Love, R.P.; Yan, M.; Zirkle, B.; Gaba, A.; Chelico, L. Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G. Nat. Commun. 2020, 11, 632. [Google Scholar] [CrossRef]
- Harris, R.S.; Liddament, M.T. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004, 4, 868–877. [Google Scholar] [CrossRef]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Belanger, K.; Savoie, M.; Rosales Gerpe, M.C.; Couture, J.-F.; Langlois, M.-A. Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses. Nucleic Acids Res. 2013, 41, 7438–7452. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Dudley, J.P. APOBECs and virus restriction. Virology 2015, 479, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Ooms, M.; Krikoni, A.; Kress, A.K.; Simon, V.; Münk, C. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J. Virol. 2012, 86, 6097–6108. [Google Scholar] [CrossRef] [PubMed]
- Beggel, B.; Münk, C.; Däumer, M.; Hauck, K.; Häussinger, D.; Lengauer, T.; Erhardt, A. Full genome ultra-deep pyrosequencing associates G-to-A hypermutation of the hepatitis B virus genome with the natural progression of hepatitis B. J. Viral Hepat. 2013, 20, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Lucifora, J.; Xia, Y.; Reisinger, F.; Zhang, K.; Stadler, D.; Cheng, X.; Sprinzl, M.F.; Koppensteiner, H.; Makowska, Z.; Volz, T. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014, 343, 1221–1228. [Google Scholar] [CrossRef] [PubMed]
- Narvaiza, I.; Linfesty, D.C.; Greener, B.N.; Hakata, Y.; Pintel, D.J.; Logue, E.; Landau, N.R.; Weitzman, M.D. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009, 5, e1000439. [Google Scholar] [CrossRef]
- Chen, H.; Lilley, C.E.; Yu, Q.; Lee, D.V.; Chou, J.; Narvaiza, I.; Landau, N.R.; Weitzman, M.D. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006, 16, 480–485. [Google Scholar] [CrossRef]
- Zhu, B.; Xiao, Y.; Yeager, M.; Clifford, G.; Wentzensen, N.; Cullen, M.; Boland, J.F.; Bass, S.; Steinberg, M.K.; Raine-Bennett, T. Mutations in the HPV16 genome induced by APOBEC3 are associated with viral clearance. Nat. Commun. 2020, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.J.; Xu, T.; Guo, K.; Griffin, L.M.; Westrich, J.A.; Lee, D.; Lambert, P.F.; Santiago, M.L.; Pyeon, D. APOBEC3A functions as a restriction factor of human papillomavirus. J. Virol. 2015, 89, 688–702. [Google Scholar] [CrossRef]
- Cheng, A.Z.; Yockteng-Melgar, J.; Jarvis, M.C.; Malik-Soni, N.; Borozan, I.; Carpenter, M.A.; McCann, J.L.; Ebrahimi, D.; Shaban, N.M.; Marcon, E. Epstein–Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity. Nat. Microbiol. 2019, 4, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Bekerman, E.; Jeon, D.; Ardolino, M.; Coscoy, L. A role for host activation-induced cytidine deaminase in innate immune defense against KSHV. PLoS Pathog. 2013, 9, e1003748. [Google Scholar] [CrossRef]
- Weisblum, Y.; Oiknine-Djian, E.; Zakay-Rones, Z.; Vorontsov, O.; Haimov-Kochman, R.; Nevo, Y.; Stockheim, D.; Yagel, S.; Panet, A.; Wolf, D.G. APOBEC3A is upregulated by human cytomegalovirus (HCMV) in the maternal-fetal interface, acting as an innate anti-HCMV effector. J. Virol. 2017, 91, e01296-17. [Google Scholar] [CrossRef]
- Pautasso, S.; Galitska, G.; Dell’Oste, V.; Biolatti, M.; Cagliani, R.; Forni, D.; De Andrea, M.; Gariglio, M.; Sironi, M.; Landolfo, S. Strategy of human cytomegalovirus to escape interferon beta-induced APOBEC3G editing activity. J. Virol. 2018, 92, e01224-18. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.A.; Holland, T.C.; Bhagwat, A.S. Human herpes simplex virus-1 depletes APOBEC3A from nuclei. Virology 2019, 537, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Moraes, S.N.; Becker, J.T.; Moghadasi, S.A.; Shaban, N.M.; Auerbach, A.A.; Cheng, A.Z.; Harris, R.S. Evidence linking APOBEC3B genesis and evolution of innate immune antagonism by gamma-herpesvirus ribonucleotide reductases. eLife 2022, 11, e83893. [Google Scholar] [CrossRef]
- Peretti, A.; Geoghegan, E.M.; Pastrana, D.V.; Smola, S.; Feld, P.; Sauter, M.; Lohse, S.; Ramesh, M.; Lim, E.S.; Wang, D. Characterization of BK polyomaviruses from kidney transplant recipients suggests a role for APOBEC3 in driving in-host virus evolution. Cell Host Microbe 2018, 23, 628–635.e7. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, S.; Rosenthal, R.; Kanu, N.; McGranahan, N.; Bartek, J.; Quezada, S.; Hare, J.; Harris, R.; Swanton, C. Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann. Oncol. 2018, 29, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Calabrese, P.; Wang, S.; Qin, C.; Rao, Y.; Feng, P.; Chen, X.S. The roles of APOBEC-mediated RNA editing in SARS-CoV-2 mutations, replication and fitness. Sci. Rep. 2022, 12, 14972. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, L.; Oh, S.; Ortega, P.; Bouin, A.; Bournique, E.; Sanchez, A.; Martensen, P.M.; Auerbach, A.A.; Becker, J.T.; Seldin, M. APOBEC3B drives PKR-mediated translation shutdown and protects stress granules in response to viral infection. Nat. Commun. 2023, 14, 820. [Google Scholar] [CrossRef]
- Pavitt, G.D. Regulation of translation initiation factor eIF2B at the hub of the integrated stress response. Wiley Interdiscip. Rev. RNA 2018, 9, e1491. [Google Scholar] [CrossRef]
- McCormick, C.; Khaperskyy, D.A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017, 17, 647–660. [Google Scholar] [CrossRef]
- Wei, L.-H.; Sun, Y.; Guo, J.U. Genome-wide CRISPR screens identify noncanonical translation factor eIF2A as an enhancer of SARS-CoV-2 programmed—1 ribosomal frameshifting. Cell Rep. 2023, 42, 112987. [Google Scholar] [CrossRef]
- Slobodin, B.; Sehrawat, U.; Lev, A.; Hayat, D.; Zuckerman, B.; Fraticelli, D.; Ogran, A.; Ben-Shmuel, A.; Bar-David, E.; Levy, H. Cap-independent translation and a precisely located RNA sequence enable SARS-CoV-2 to control host translation and escape anti-viral response. Nucleic Acids Res. 2022, 50, 8080–8092. [Google Scholar] [CrossRef] [PubMed]
- Renner, D.M.; Parenti, N.A.; Bracci, N.; Weiss, S.R. Betacoronaviruses Differentially Activate the Integrated Stress Response to Optimize Viral Replication in Lung-Derived Cell Lines. Viruses 2025, 17, 120. [Google Scholar] [CrossRef]
- Ricciardi-Jorge, T.; da Rocha, E.L.; Gonzalez-Kozlova, E.; Rodrigues-Luiz, G.F.; Ferguson, B.J.; Sweeney, T.; Irigoyen, N.; Mansur, D.S. PKR-mediated stress response enhances dengue and Zika virus replication. Mbio 2023, 14, e0093423. [Google Scholar] [CrossRef]
- Qin, C.; Rao, Y.; Yuan, H.; Wang, T.-Y.; Zhao, J.; Espinosa, B.; Liu, Y.; Zhang, S.; Savas, A.C.; Liu, Q. SARS-CoV-2 couples evasion of inflammatory response to activated nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2122897119. [Google Scholar] [CrossRef]
- Xiao, N.; Nie, M.; Pang, H.; Wang, B.; Hu, J.; Meng, X.; Li, K.; Ran, X.; Long, Q.; Deng, H. Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 2021, 12, 1618. [Google Scholar] [CrossRef]
- Theodoris, C.V.; Xiao, L.; Chopra, A.; Chaffin, M.D.; Al Sayed, Z.R.; Hill, M.C.; Mantineo, H.; Brydon, E.M.; Zeng, Z.; Liu, X.S. Transfer learning enables predictions in network biology. Nature 2023, 618, 616–624. [Google Scholar] [CrossRef]
- Chen, H.; Venkatesh, M.S.; Ortega, J.G.; Mahesh, S.V.; Nandi, T.N.; Madduri, R.K.; Pelka, K.; Theodoris, C.V. Quantized multi-task learning for context-specific representations of gene network dynamics. bioRxiv 2024. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef] [PubMed]
- Zupin, L.; Fontana, F.; Clemente, L.; Ruscio, M.; Ricci, G.; Crovella, S. Effect of short time of SARS-CoV-2 infection in Caco-2 cells. Viruses 2022, 14, 704. [Google Scholar] [CrossRef] [PubMed]
- Pires De Souza, G.A.; Le Bideau, M.; Boschi, C.; Wurtz, N.; Colson, P.; Aherfi, S.; Devaux, C.; La Scola, B. Choosing a cellular model to study SARS-CoV-2. Front. Cell. Infect. Microbiol. 2022, 12, 1003608. [Google Scholar] [CrossRef]
- Garcia, M.; Meurs, E.; Esteban, M. The dsRNA protein kinase PKR: Virus and cell control. Biochimie 2007, 89, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.; Liu, Y.; Dakshanamurthy, S. Single-cell and bulk RNASeq profiling of COVID-19 patients reveal immune and inflammatory mechanisms of infection-induced organ damage. Viruses 2021, 13, 2418. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xu, X.; Fan, J.; Chen, H.; Zhao, Y.; Huang, W.; Liu, W.; Zhang, Z.; Cui, Q.; Li, Q. APOBEC3-related mutations in the spike protein-encoding region facilitate SARS-CoV-2 evolution. Heliyon 2024, 10, e32139. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; He, B.; Carver, K.; Vanheyningen, D.; Parkin, B.; Garmire, L.X.; Olszewski, M.A.; Deng, J.C. Heterogeneity of neutrophils and inflammatory responses in patients with COVID-19 and healthy controls. Front. Immunol. 2022, 13, 970287. [Google Scholar] [CrossRef]
- Ehlers, L.; Kuppe, A.; Damerau, A.; Wilantri, S.; Kirchner, M.; Mertins, P.; Strehl, C.; Buttgereit, F.; Gaber, T. Surface AMP deaminase 2 as a novel regulator modifying extracellular adenine nucleotide metabolism. FASEB J. 2021, 35, e21684. [Google Scholar] [CrossRef]
- Leonard, B.; McCann, J.L.; Starrett, G.J.; Kosyakovsky, L.; Luengas, E.M.; Molan, A.M.; Burns, M.B.; McDougle, R.M.; Parker, P.J.; Brown, W.L. The PKC/NF-κB signaling pathway induces APOBEC3B expression in multiple human cancers. Cancer Res. 2015, 75, 4538–4547. [Google Scholar] [CrossRef] [PubMed]
- Faure-Dupuy, S.; Riedl, T.; Rolland, M.; Hizir, Z.; Reisinger, F.; Neuhaus, K.; Schuehle, S.; Remouchamps, C.; Gillet, N.; Schönung, M. Control of APOBEC3B induction and cccDNA decay by NF-κB and miR-138-5p. JHEP Rep. 2021, 3, 100354. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-W.; Zhou, S.-F. Inflammation, Cytokines, the IL-17/IL-6/STAT3/NF-κB Axis, and Tumorigenesis; Taylor & Francis: Abingdon, UK, 2015; pp. 2941–2946. [Google Scholar]
- Liu, W.; Wu, J.; Yang, F.; Ma, L.; Ni, C.; Hou, X.; Wang, L.; Xu, A.; Song, J.; Deng, Y. Genetic polymorphisms predisposing the interleukin 6–induced APOBEC3B-UNG imbalance increase HCC risk via promoting the generation of APOBEC-signature HBV mutations. Clin. Cancer Res. 2019, 25, 5525–5536. [Google Scholar] [CrossRef]
- Li, S.; Bao, X.; Wang, D.; You, L.; Li, X.; Yang, H.; Bian, J.; Wang, Y.; Yang, Y. APOBEC3B and IL-6 form a positive feedback loop in hepatocellular carcinoma cells. S. China Life Sci. 2017, 60, 617–626. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jia, M.; He, Z.; Liu, X.-S. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene 2018, 37, 3924–3936. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.B.; Temiz, N.A.; Harris, R.S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013, 45, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Nikkilä, J.; Kumar, R.; Campbell, J.; Brandsma, I.; Pemberton, H.N.; Wallberg, F.; Nagy, K.; Scheer, I.; Vertessy, B.G.; Serebrenik, A.A. Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells. Br. J. Cancer 2017, 117, 113–123. [Google Scholar] [CrossRef]
- Taylor, B.J.; Nik-Zainal, S.; Wu, Y.L.; Stebbings, L.A.; Raine, K.; Campbell, P.J.; Rada, C.; Stratton, M.R.; Neuberger, M.S. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2013, 2, e00534. [Google Scholar] [CrossRef] [PubMed]
- Fixman, B.; Díaz-Gay, M.; Qiu, C.; Margaryan, T.; Lee, B.; Chen, X.S. Validation of the APOBEC3A-Mediated RNA Single Base Substitution Signature and Proposal of Novel APOBEC1, APOBEC3B, and APOBEC3G RNA Signatures. J. Mol. Biol. 2024, 436, 168854. [Google Scholar] [CrossRef] [PubMed]
- Fixman, B.B. APOBEC RNA Mutational Signatures and the Role of APOBEC3B in SARS-CoV-2 Infection. Ph.D. Thesis, University of Southern California, Los Angeles, CA, USA, 2025. [Google Scholar]
- Partek. Partek™ Flow™, version 10.0. Computer Software. Illumina: Tokyo, Japan, 2024.
| siRNA | Sequence | Company | Catalog Number |
|---|---|---|---|
| Control | Thermo Fisher Scientific | 4390843 | |
| APOBEC3B | CCUCAGUACCACGCAGAAATT | Thermo Fisher Scientific | s18411 |
| APOBEC3B | GAGAUUCUCAGAUACCUGATT | Thermo Fisher Scientific | s18412 |
| PKR | GGUGAAGGUAGAUCAAAGATT | Thermo Fisher Scientific | s11187 |
| PKR | GACGGAAAGACUUACGUUATT | Thermo Fisher Scientific | s11185 |
| Primer | Sequence | Company |
|---|---|---|
| Actin—Forward | CTGGCACCCAGCACAATG | IDT DNA |
| Actin—Reverse | GCCGATCCACACGGAGTACT | IDT DNA |
| CoV-2 N1—Forward | GGACCCCAAAATCAGCGAAAT | IDT DNA |
| CoV-2 N1—Reverse | TTCTGGTTACTGCCAGTTGAATCTG | IDT DNA |
| Antibody | Isotype | Company | Catalog Number |
|---|---|---|---|
| SARS-CoV-2 Nucleocapsid | Rabbit monoclonal | Cell Signaling Technology (Danvers, MA, USA) | 86326 |
| GAPDH | Rabbit polyclonal | EMD Millipore (Burlington, MA, USA) | ABS16 |
| Vinculin | Mouse monoclonal | Sigma (St. Louis, MO, USA) | V9264 |
| PKR | Mouse monoclonal | BD Biosciences (San Jose, CA, USA) | 610764 |
| PKR-pT446 | Rabbit monoclonal | Abcam (Cambridge, UK) | ab32036 |
| eIF2⍺ | Rabbit monoclonal | Cell Signaling Technology (Danvers, MA, USA) | 5324T |
| eIF2⍺-pS51 | Rabbit monoclonal | Abcam (Cambridge, UK) | 32157 |
| A3B | Rabbit monoclonal | Abcam (Cambridge, UK) | 184990 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fixman, B.; Manjunath, L.; Sell, P.; Wang, S.; Margaryan, T.; Qiu, C.; Yang, H.; Buisson, R.; Chen, X.S. APOBEC3B Promotes SARS-CoV-2 Through Activation of PKR/eIF2⍺ and AMPD2 Dysregulation. Viruses 2025, 17, 1176. https://doi.org/10.3390/v17091176
Fixman B, Manjunath L, Sell P, Wang S, Margaryan T, Qiu C, Yang H, Buisson R, Chen XS. APOBEC3B Promotes SARS-CoV-2 Through Activation of PKR/eIF2⍺ and AMPD2 Dysregulation. Viruses. 2025; 17(9):1176. https://doi.org/10.3390/v17091176
Chicago/Turabian StyleFixman, Benjamin, Lavanya Manjunath, Philip Sell, Shanshan Wang, Tamara Margaryan, Connor Qiu, Hanjing Yang, Rémi Buisson, and Xiaojiang S. Chen. 2025. "APOBEC3B Promotes SARS-CoV-2 Through Activation of PKR/eIF2⍺ and AMPD2 Dysregulation" Viruses 17, no. 9: 1176. https://doi.org/10.3390/v17091176
APA StyleFixman, B., Manjunath, L., Sell, P., Wang, S., Margaryan, T., Qiu, C., Yang, H., Buisson, R., & Chen, X. S. (2025). APOBEC3B Promotes SARS-CoV-2 Through Activation of PKR/eIF2⍺ and AMPD2 Dysregulation. Viruses, 17(9), 1176. https://doi.org/10.3390/v17091176







