The Differences in the Evolutionary Dynamics of MERS and SARS Coronaviruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of Virus Genomes and Sequence Treatment
2.2. DNA Extraction, PCR, and Sequencing the DPP4 Gene
3. Results
3.1. The Evolutionary Characteristics of SARS-CoVs
3.2. The Evolutionary Characteristics of MERS-CoV
3.3. DPP4 Did Not Show Any Difference Between Arabs and Africans
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses 2019, 11, 59. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef]
- Drosten, C.; Gunther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.; et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.L.; Shi, W.F.; Zhang, W.; Zhu, Y.; Zhang, Y.W.; Xie, Q.M.; Mani, S.; et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255–258. [Google Scholar] [CrossRef]
- Gong, L.; Li, J.; Zhou, Q.; Xu, Z.; Chen, L.; Zhang, Y.; Xue, C.; Wen, Z.; Cao, Y. A new bat-HKU2-like coronavirus in swine, China, 2017. Emerg. Infect. Dis. 2017, 23, 1607–1609. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-Like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Guan, Y.J.; et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef]
- Song, H.D.; Tu, C.C.; Zhang, G.W.; Wang, S.Y.; Zheng, K.; Lei, L.C.; Chen, Q.X.; Gao, Y.W.; Zhou, H.Q.; Xiang, H.; et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA 2005, 102, 2430–2435. [Google Scholar] [CrossRef]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Sabir, J.S.M.; Lam, T.T.Y.; Ahmed, M.M.M.; Li, L.; Shen, Y.; Abo-Aba, S.E.M.; Qureshi, M.I.; Abu-Zeid, M.; Zhang, Y.; Khiyami, M.A.; et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 2016, 351, 81–84. [Google Scholar] [CrossRef]
- Anthony, S.J.; Gilardi, K.; Menachery, V.D.; Goldstein, T.; Ssebide, B.; Mbabazi, R.; Navarrete-Macias, I.; Liang, E.; Wells, H.; Hicks, A.; et al. Further evidence for bats as the evolutionary source of Middle East Respiratory Syndrome coronavirus. mBio 2017, 8, e00373-17. [Google Scholar] [CrossRef]
- Fehr, A.R.; Channappanavar, R.; Perlman, S. Middle East Respiratory Syndrome: Emergence of a Pathogenic Human Coronavirus. Annu. Rev. Med. 2017, 68, 387–399. [Google Scholar] [CrossRef]
- Millet, J.K.; Jaimes, J.A.; Whittaker, G.R. Molecular diversity of coronavirus host cell entry receptors. FEMS Microbiol. Rev. 2021, 45, fuaa057. [Google Scholar] [CrossRef]
- Tolentino, J.E.; Lytras, S.; Ito, J.; Holmes, E.C.; Sato, K. Recombination as an evolutionary driver of MERS-related coronavirus emergence. Lancet Infect. Dis. 2024, 24, e546. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shen, L.; Gu, X. Evolutionary Dynamics of MERS-CoV: Potential Recombination, Positive Selection and Transmission. Sci. Rep. 2016, 6, 25049. [Google Scholar] [CrossRef] [PubMed]
- Tolentino, J.E.; Lytras, S.; Ito, J.; Sato, K. Recombination analysis on the receptor switching event of MERS-CoV and its close relatives: Implications for the emergence of MERS-CoV. Virol. J. 2024, 21, 84. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.B.; Liu, C.; Park, Y.J.; Tang, J.; Chen, J.; Xiong, Q.; Lee, J.; Stewart, C.; Asarnow, D.; Brown, J.; et al. Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses. Cell 2025, 188, 1693–1710.e1618. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wu, F. Global surveillance and countermeasures for ACE2-using MERS-related coronaviruses with spillover risk. Cell 2025, 188, 1465–1468. [Google Scholar] [CrossRef] [PubMed]
- Peeri, N.C.; Shrestha, N.; Rahman, M.S.; Zaki, R.; Tan, Z.; Bibi, S.; Baghbanzadeh, M.; Aghamohammadi, N.; Zhang, W.; Haque, U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: What lessons have we learned? Int. J. Epidemiol. 2020, 49, 717–726. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al-Ahmed, S.H.; Sah, R.; Alqumber, M.A.; Haque, S.; Patel, S.K.; Pathak, M.; Tiwari, R.; Yatoo, M.I.; Haq, A.U.; et al. MERS-CoV: Epidemiology, molecular dynamics, therapeutics, and future challenges. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 8. [Google Scholar] [CrossRef] [PubMed]
- Kan, B.; Wang, M.; Jing, H.; Xu, H.; Jiang, X.; Yan, M.; Liang, W.; Zheng, H.; Wan, K.; Liu, Q.; et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005, 79, 11892–11900. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Muller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 2010, 26, 1899–1900. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Martin, D.; Rybicki, E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef]
- Martin, D.P.; Posada, D.; Crandall, K.A.; Williamson, C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retroviruses 2005, 21, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Lam, H.M.; Ratmann, O.; Boni, M.F. Improved Algorithmic Complexity for the 3SEQ Recombination Detection Algorithm. Mol. Biol. Evol. 2018, 35, 247–251. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Frost, S.D. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky Pond, S.L.; Scheffler, K. FUBAR: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Zhang, X.S.; Pebody, R.; Charlett, A.; de Angelis, D.; Birrell, P.; Kang, H.; Baguelin, M.; Choi, Y.H. Estimating and modelling the transmissibility of Middle East Respiratory Syndrome CoronaVirus during the 2015 outbreak in the Republic of Korea. Influenza Other Respir. Viruses 2017, 11, 434–444. [Google Scholar] [CrossRef]
- Wong, A.C.P.; Li, X.; Lau, S.K.P.; Woo, P.C.Y. Global epidemiology of bat coronaviruses. Viruses 2019, 11, 174. [Google Scholar] [CrossRef]
- Wang, L.F.; Anderson, D.E. Viruses in bats and potential spillover to animals and humans. Curr. Opin. Virol. 2019, 34, 79–89. [Google Scholar] [CrossRef]
- Stein, R.A. Super-spreaders in infectious diseases. Int. J. Infect. Dis. 2011, 15, e510–e513. [Google Scholar] [CrossRef]
- Lambrou, A.S.; South, E.; Midgley, C.M.; Harrington, C.; Wang, L.; Cubenas, C.; Lowe, D.; Abedi, G.R.; Jones, C.; Hughes, L.J.; et al. Update on the Epidemiology of Middle East Respiratory Syndrome Coronavirus-Worldwide, 2017–2023. MMWR Morb. Mortal. Wkly. Rep. 2025, 74, 313–320. [Google Scholar] [CrossRef]
- Adney, D.R.; Wang, L.; van Doremalen, N.; Shi, W.; Zhang, Y.; Kong, W.P.; Miller, M.R.; Bushmaker, T.; Scott, D.; de Wit, E.; et al. Efficacy of an Adjuvanted Middle East Respiratory Syndrome Coronavirus Spike Protein Vaccine in Dromedary Camels and Alpacas. Viruses 2019, 11, 212. [Google Scholar] [CrossRef]
- Sugimoto, S.; Kakizaki, M.; Kawase, M.; Kawachi, K.; Ujike, M.; Kamitani, W.; Sentsui, H.; Shirato, K. Single Amino Acid Substitution in the Receptor Binding Domain of Spike Protein Is Sufficient To Convert the Neutralization Profile between Ethiopian and Middle Eastern Isolates of Middle East Respiratory Coronavirus. Microbiol. Spectr. 2023, 11, e0459022. [Google Scholar] [CrossRef] [PubMed]
- Hassell, J.M.; Zimmerman, D.; Fevre, E.M.; Zinsstag, J.; Bukachi, S.; Barry, M.; Muturi, M.; Bett, B.; Jensen, N.; Ali, S.; et al. Africa’s Nomadic Pastoralists and Their Animals Are an Invisible Frontier in Pandemic Surveillance. Am. J. Trop. Med. Hyg. 2020, 103, 1777–1779. [Google Scholar] [CrossRef]
- Alagaili, A.N.; Briese, T.; Mishra, N.; Kapoor, V.; Sameroff, S.C.; Burbelo, P.D.; de Wit, E.; Munster, V.J.; Hensley, L.E.; Zalmout, I.S.; et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 2014, 5, e00884-14. [Google Scholar] [CrossRef]
- Corman, V.M.; Jores, J.; Meyer, B.; Younan, M.; Liljander, A.; Said, M.Y.; Gluecks, I.; Lattwein, E.; Bosch, B.J.; Drexler, J.F.; et al. Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992–2013. Emerg. Infect. Dis. 2014, 20, 1319–1322. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Sabir, J.S.M.; Irwin, D.M.; Shen, Y. Vaccine against Middle East respiratory syndrome coronavirus. Lancet Infect. Dis. 2019, 19, 1053–1054. [Google Scholar] [CrossRef] [PubMed]
- Reusken, C.B.; Haagmans, B.L.; Muller, M.A.; Gutierrez, C.; Godeke, G.J.; Meyer, B.; Muth, D.; Raj, V.S.; Smits-De Vries, L.; Corman, V.M.; et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: A comparative serological study. Lancet Infect. Dis. 2013, 13, 859–866. [Google Scholar] [CrossRef]
- Meyer, B.; Müller, M.A.; Corman, V.M.; Reusken, C.B.E.M.; Ritz, D.; Godeke, G.-J.; Lattwein, E.; Kallies, S.; Siemens, A.; van Beek, J.; et al. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg. Infect. Dis. 2014, 20, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.M.; Muhlemann, B.; Al-Subhi, T.L.; Rodon, J.; El-Kafrawy, S.A.; Memish, Z.; Melchert, J.; Bleicker, T.; Mauno, T.; Perlman, S.; et al. Ongoing Evolution of Middle East Respiratory Syndrome Coronavirus, Saudi Arabia, 2023–2024. Emerg. Infect. Dis. 2025, 31, 57–65. [Google Scholar] [CrossRef]
- Salomon, I. Saudi Arabia’s Middle East respiratory syndrome Coronavirus (MERS-CoV) outbreak: Consequences, reactions, and takeaways. Ann. Med. Surg. 2024, 86, 4668–4674. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Yuan, Y.; Cao, D.; Zhang, Y.; Ma, J.; Qi, J.; Wang, Q.; Lu, G.; Wu, Y.; Yan, J.; Shi, Y.; et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017, 8, 15092. [Google Scholar] [CrossRef]
- Millet, J.K.; Goldstein, M.E.; Labitt, R.N.; Hsu, H.L.; Daniel, S.; Whittaker, G.R. A camel-derived MERS-CoV with a variant spike protein cleavage site and distinct fusion activation properties. Emerg. Microbes Infect. 2016, 5, e126. [Google Scholar] [CrossRef] [PubMed]
- Hulswit, R.J.; de Haan, C.A.; Bosch, B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016, 96, 29–57. [Google Scholar] [CrossRef]
- Consortium, C.S.M.E. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 2004, 303, 1666–1669. [Google Scholar] [CrossRef]
- Adney, D.R.; van Doremalen, N.; Brown, V.R.; Bushmaker, T.; Scott, D.; de Wit, E.; Bowen, R.A.; Munster, V.J. Replication and shedding of MERS-CoV in upper respiratory tract of inoculated dromedary camels. Emerg. Infect. Dis. 2014, 20, 1999–2005. [Google Scholar] [CrossRef]
- Islam, A.; Ferdous, J.; Islam, S.; Sayeed, M.A.; Dutta Choudhury, S.; Saha, O.; Hassan, M.M.; Shirin, T. Evolutionary Dynamics and Epidemiology of Endemic and Emerging Coronaviruses in Humans, Domestic Animals, and Wildlife. Viruses 2021, 13, 1908. [Google Scholar] [CrossRef]
- Reusken, C.B.; Raj, V.S.; Koopmans, M.P.; Haagmans, B.L. Cross host transmission in the emergence of MERS coronavirus. Curr. Opin. Virol. 2016, 16, 55–62. [Google Scholar] [CrossRef]
- Chu, D.K.; Poon, L.L.; Gomaa, M.M.; Shehata, M.M.; Perera, R.A.; Abu Zeid, D.; El Rifay, A.S.; Siu, L.Y.; Guan, Y.; Webby, R.J.; et al. MERS coronaviruses in dromedary camels, Egypt. Emerg. Infect. Dis. 2014, 20, 1049–1053. [Google Scholar] [CrossRef]
- Chu, D.K.; Oladipo, J.O.; Perera, R.A.; Kuranga, S.A.; Chan, S.M.; Poon, L.L.; Peiris, M. Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Nigeria, 2015. Eurosurveillance 2015, 20, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Miguel, E.; Chevalier, V.; Ayelet, G.; Ben Bencheikh, M.N.; Boussini, H.; Chu, D.K.; El Berbri, I.; Fassi-Fihri, O.; Faye, B.; Fekadu, G.; et al. Risk factors for MERS coronavirus infection in dromedary camels in Burkina Faso, Ethiopia, and Morocco, 2015. Eurosurveillance 2017, 22, 30498. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.W.; Hui, K.P.Y.; Perera, R.A.P.M.; Miguel, E.; Niemeyer, D.; Zhao, J.; Channappanavar, R.; Dudas, G.; Oladipo, J.O.; Traoré, A.; et al. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc. Natl. Acad. Sci. USA 2018, 115, 3144–3149. [Google Scholar] [CrossRef]
- Kiambi, S.; Corman, V.M.; Sitawa, R.; Githinji, J.; Ngoci, J.; Ozomata, A.S.; Gardner, E.; von Dobschuetz, S.; Morzaria, S.; Kimutai, J.; et al. Detection of distinct MERS-Coronavirus strains in dromedary camels from Kenya, 2017. Emerg. Microbes Infect. 2018, 7, 195. [Google Scholar] [CrossRef] [PubMed]
- Ommeh, S.; Zhang, W.; Zohaib, A.; Chen, J.; Zhang, H.; Hu, B.; Ge, X.Y.; Yang, X.L.; Masika, M.; Obanda, V.; et al. Genetic evidence of Middle East Respiratory Syndrome coronavirus (MERS-Cov) and widespread seroprevalence among camels in Kenya. Virol. Sin. 2018, 33, 484–492. [Google Scholar] [CrossRef]
- Karani, A.; Ombok, C.; Situma, S.; Breiman, R.; Mureithi, M.; Jaoko, W.; Njenga, M.K.; Ngere, I. Low-Level Zoonotic Transmission of Clade C MERS-CoV in Africa: Insights from Scoping Review and Cohort Studies in Hospital and Community Settings. Viruses 2025, 17, 125. [Google Scholar] [CrossRef] [PubMed]
- Mackay, I.M.; Arden, K.E. MERS coronavirus: Diagnostics, epidemiology and transmission. Virol. J. 2015, 12, 222. [Google Scholar] [CrossRef]
- Barlan, A.; Zhao, J.; Sarkar, M.K.; Li, K.; McCray, P.B., Jr.; Perlman, S.; Gallagher, T. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J. Virol. 2014, 88, 4953–4961. [Google Scholar] [CrossRef]
- Sims, A.C.; Schafer, A.; Okuda, K.; Leist, S.R.; Kocher, J.F.; Cockrell, A.S.; Hawkins, P.E.; Furusho, M.; Jensen, K.L.; Kyle, J.E.; et al. Dysregulation of lung epithelial cell homeostasis and immunity contributes to Middle East respiratory syndrome coronavirus disease severity. mSphere 2025, 10, e0095124. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Das, A.; Saha, A. Regulation of miRNA in Cytokine Storm (CS) of COVID-19 and Other Viral Infection: An Exhaustive Review. Rev. Med. Virol. 2025, 35, e70026. [Google Scholar] [CrossRef]
- Dobrijevic, Z.; Gligorijevic, N.; Sunderic, M.; Penezic, A.; Miljus, G.; Tomic, S.; Nedic, O. The association of human leucocyte antigen (HLA) alleles with COVID-19 severity: A systematic review and meta-analysis. Rev. Med. Virol. 2023, 33, e2378. [Google Scholar] [CrossRef]
- Tanaka, K.; Meguro, A.; Hara, Y.; Endo, L.; Izawa, A.; Muraoka, S.; Kaneko, A.; Somekawa, K.; Hirata, M.; Otsu, Y.; et al. HLA-DQA1*01:03 and DQB1*06:01 are risk factors for severe COVID-19 pneumonia. HLA 2024, 104, e15609. [Google Scholar] [CrossRef]
- Hajeer, A.H.; Balkhy, H.; Johani, S.; Yousef, M.Z.; Arabi, Y. Association of human leukocyte antigen class II alleles with severe Middle East respiratory syndrome-coronavirus infection. Ann. Thorac. Med. 2016, 11, 211–213. [Google Scholar] [CrossRef] [PubMed]
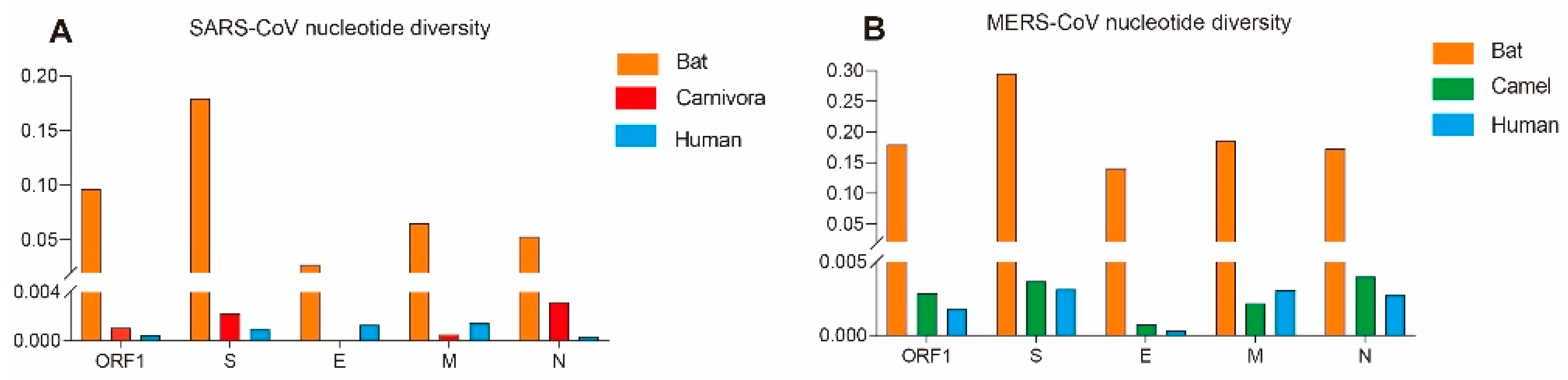
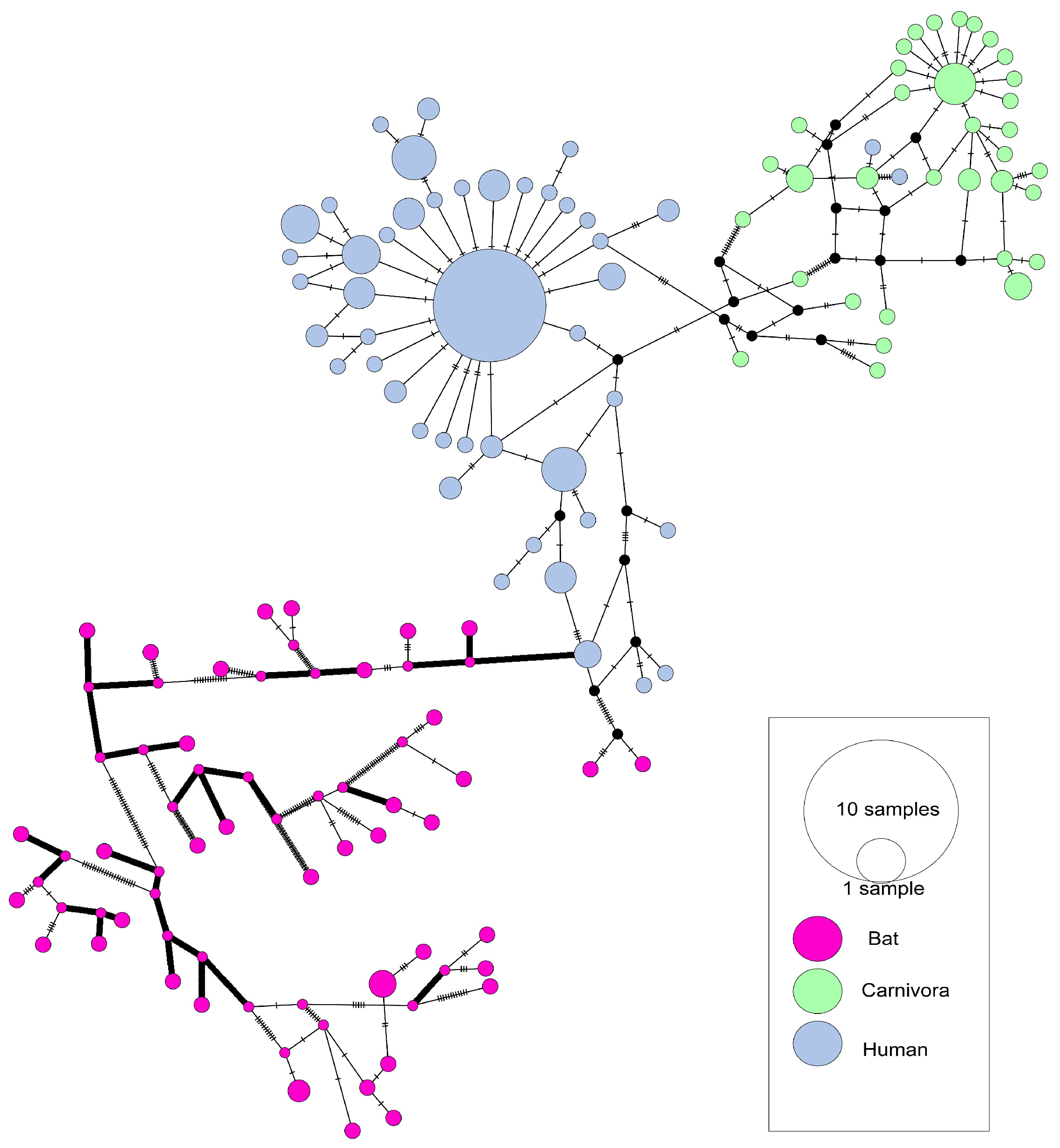
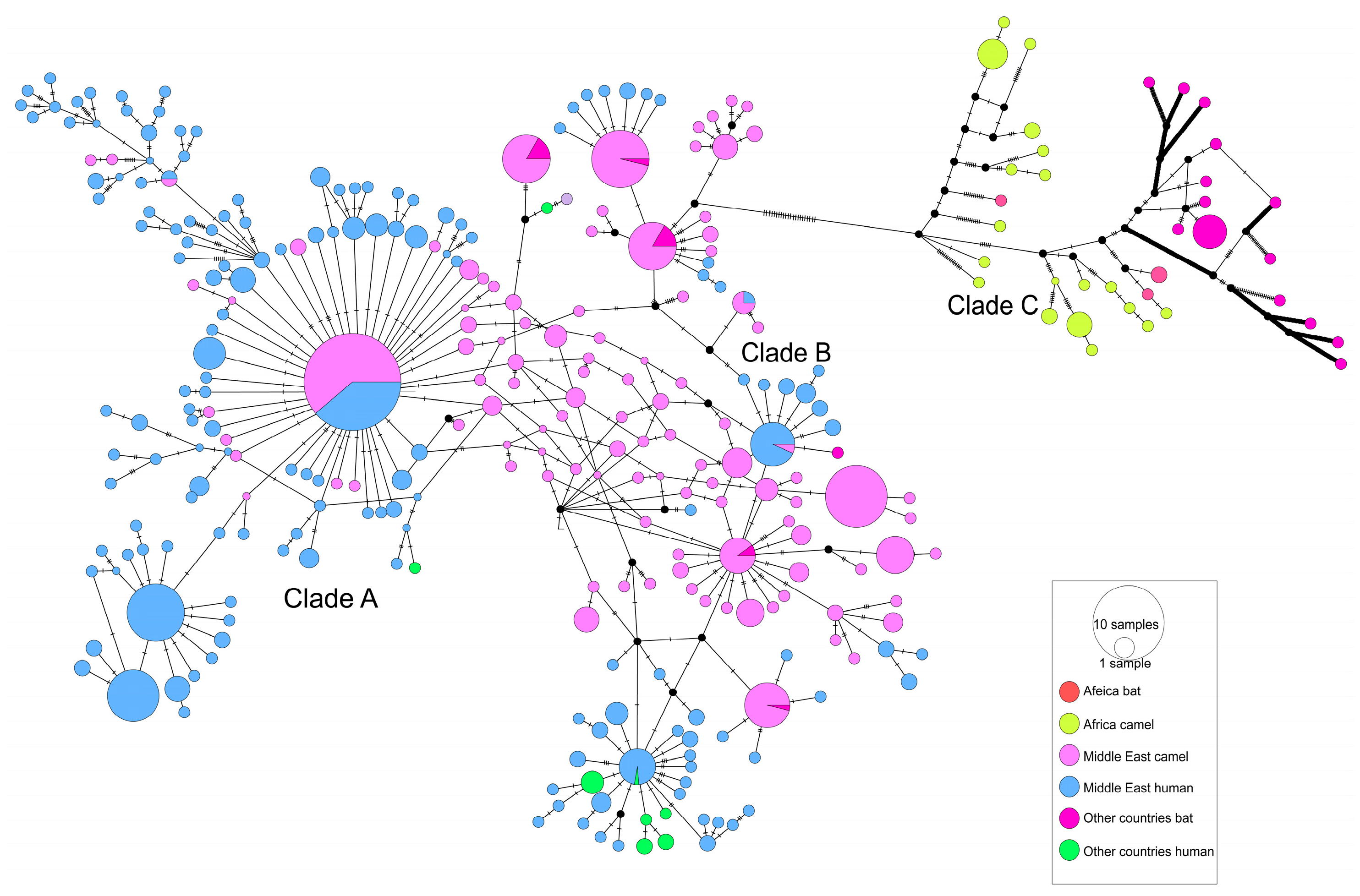
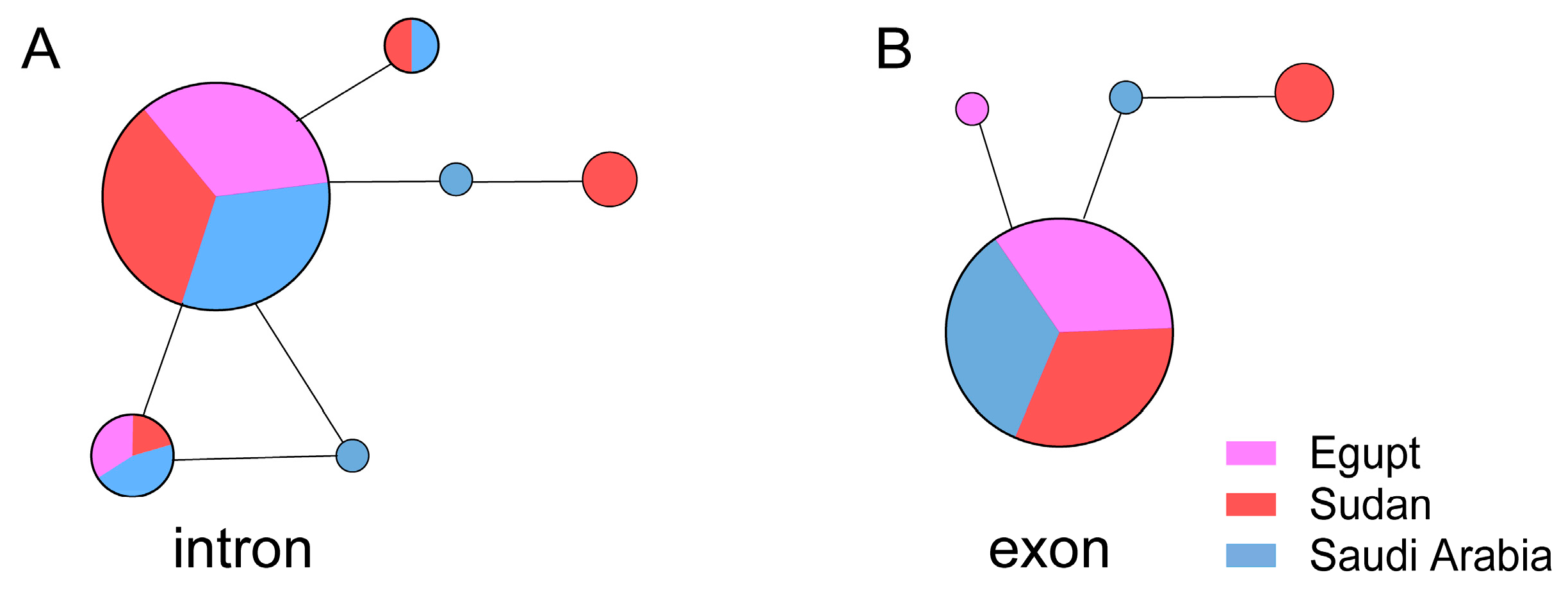
| Gene | Host | Nucleotide Diversity (π) | Neutrality Analyses | ||
|---|---|---|---|---|---|
| Fu and Li’s D * | Fu and Li’s D * | ||||
| SARS-CoV | ORF1 | Human | 0.00048 | −2.63679 ** | −5.05802 ** |
| Bat | 0.09634 | −0.72289 | −0.85435 | ||
| Carnivora | 0.00107 | −1.45552 | −1.05296 | ||
| S | Human | 0.00094 | −2.29683 ** | −4.31580 ** | |
| Bat | 0.17954 | 0.35301 | −0.36182 | ||
| Carnivora | 0.00225 | −0.10083 | −1.05012 | ||
| E | Human | 0.00132 | −1.63734 * | −0.82508 | |
| Bat | 0.02742 | −1.05697 | −1.11301 | ||
| Carnivora | / | / | / | ||
| M | Human | 0.00151 | −1.99699 * | −3.68341 ** | |
| Bat | 0.06514 | −1.22163 | −1.01881 | ||
| Carnivora | 0.00054 | −1.03789 | −0.50381 | ||
| N | Human | 0.00037 | −2.23097 ** | −4.79424 ** | |
| Bat | 0.05313 | −1.43029 | −1.77074 ** | ||
| Carnivora | 0.00320 | −0.63115 | 0.04240 | ||
| MERS-CoV | ORF1 | Human | 0.00180 | −2.32084 ** | −8.53056 ** |
| Camel | 0.00286 | −2.03977 * | −6.02846 ** | ||
| Bat | 0.17953 | −0.04466 | 0.68164 | ||
| S | Human | 0.00316 | −2.23642 ** | −7.86620 ** | |
| Camel | 0.00369 | −2.12122 ** | −5.43913 ** | ||
| Bat | 0.29554 | −0.13075 | 0.44654 | ||
| E | Human | 0.00033 | −2.06853 * | −5.60248 ** | |
| Camel | 0.00078 | −1.89892 * | −3.04304 * | ||
| Bat | 0.14117 | 0.35028 | 0.68165 | ||
| M | Human | 0.00307 | −1.65090 | −5.28654 ** | |
| Camel | 0.00219 | −2.08234 * | −3.89494 ** | ||
| Bat | 0.18639 | 0.06353 | 0.68401 | ||
| N | Human | 0.00276 | −2.15013 ** | −4.22054 ** | |
| Camel | 0.00401 | −2.10318 * | −4.26757 ** | ||
| Bat | 0.17372 | −0.07776 | 0.54725 | ||
| Gene | Positive Selection Pressure Sites Identified by Different Methods | |||||
|---|---|---|---|---|---|---|
| M8 vs. M8a | FEL | SLAC | FUBAR | MEME | PSC * | |
| Bat S | 5, 8, 20, 132, 133, 135, 136, 138, 154, 182, 195, 411, 412, 422, 439, 481, 503, 589, 689 | 166, 540, 596, 633 | None | 451, 540, 648 | 7, 20, 24, 43, 81, 82, 83, 89, 91, 101, 130, 156, 157, 160, 166, 174, 178, 185, 190, 207, 210, 231, 242, 252, 314, 410, 411, 451, 454, 464, 465, 477, 501, 502, 509, 516, 540, 562, 596, 633, 648, 689, 693, 753, 876, 935, 1117, 1250, 1253 | 540 |
| Bat E | None | None | None | None | None | - |
| Bat M | 14 | 97 | None | None | 97 | - |
| Bat N | 8, 22, 268, 410 | 8, 22, 25, 34, 81, 121, 268, 410 | None | 8, 25, 81, 410 | 8, 22, 25, 34, 81, 121, 196, 268, 297, 408, 410 | 8, 22, 25, 81, 268, 410 |
| Carnivora S | 77, 108, 113, 139, 147, 194, 227, 239, 243, 244, 261, 294, 336, 344, 360, 461, 470, 477, 478, 556, 575, 578, 605, 607, 611, 630, 642, 645, 648, 663, 699, 701, 741, 752, 763, 776, 819, 837, 842, 892, 898, 1050, 1078, 1161, 1217, | 77, 147, 227, 479, 609, 743, 894, 1080 | None | 147, 227, 244, 344, 360, 440, 462, 479, 480, 609, 613, 743, 1052, 1080, 1219 | 147, 227, 479, 609, | 147, 227, 462, 479, 609 |
| Carnivoral E | None | None | None | None | None | - |
| Carnivora M | None | None | None | None | None | - |
| Carnivora N | 384 | None | None | 384 | None | - |
| Human S | 2, 5, 12, 49, 75, 77, 78, 138, 139, 144, 147, 238, 243, 310, 343, 349, 352, 359, 383, 424, 435, 441, 462, 471, 479, 486, 500, 576, 599, 604, 607, 608, 612, 622, 651, 664, 665, 742, 764, 777, 793, 855, 859, 860, 862, 1000, 1131, 1147, 1162, 1168, 1182, 1207, 1222, 1246 | None | None | 12, 138, 311, 608, 609, 1148, 1163, 1208 | 138 | 138 |
| Human E | 5, 6, 23, 29 | None | None | None | None | - |
| Human M | 5, 11, 27, 38, 68, 73, 81, 86, 91, 99, 113, 119, 154, 210 | None | None | 11 | None | - |
| Human N | None | None | None | None | None | - |
| Gene | Positive Selection Pressure Sites Identified Using Different Methods | |||||
|---|---|---|---|---|---|---|
| M8 vs. M8a | FEL | SLAC | FUBAR | MEME | PSC a | |
| Bat S | None | 3, 7, 25, 225, 328, 625, 731, 777 | None | 219 | 3, 7, 25, 145, 199, 222, 225, 232, 235, 239, 591, 687, 713, 731, 772, 777, 794, 795, 908, 966, 1277, 1293 | - |
| Bat E | None | None | None | None | None | - |
| Bat M | None | None | None | None | 96 | - |
| Bat N | None | 200, 328, 378, 389, 394, 402, 403, 424, 435 | None | 200, 328, 389, 398, 424 | 111, 200, 210, 328, 367, 378, 389, 402, 403, 406, 424, 431 | 200, 328, 389, 424 |
| Camel S | 26, 459, 465, 612, 723, 1193, 1224 | 26, 28, 424, 459, 723, 1224 | 26 | 26, 28, 158, 390, 424, 710, 723, 1193, 1224 | 26, 28, 424, 459, 723, 1224 | 26, 28, 424, 459, 723, 1224 |
| Camel E | None | None | None | None | None | - |
| Camel M | None | 69, 111, 155 | None | 8, 69, 82 | 69 | 69 |
| Camel N | None | 3, 198 | None | 3, 198 | 3, 198 | 3, 198 |
| Human S | None | 424 | None | 26, 91, 95, 301, 424, 507, 509, 534, 914, 1158 | 1020 | - |
| Human E | None | None | None | None | None | - |
| Human M | None | None | None | 15, 20, 69 | None | - |
| Human N | None | None | None | 8, 126, 300 | None | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Liu, J.; Sabir, J.S.M.; Cui, X.; Shen, X.; Hajrah, N.H.; Ahmed, M.M.M.; Sabir, M.J.; Al-Zogabi, O.G.; Irwin, D.M.; et al. The Differences in the Evolutionary Dynamics of MERS and SARS Coronaviruses. Viruses 2025, 17, 1114. https://doi.org/10.3390/v17081114
Ding Y, Liu J, Sabir JSM, Cui X, Shen X, Hajrah NH, Ahmed MMM, Sabir MJ, Al-Zogabi OG, Irwin DM, et al. The Differences in the Evolutionary Dynamics of MERS and SARS Coronaviruses. Viruses. 2025; 17(8):1114. https://doi.org/10.3390/v17081114
Chicago/Turabian StyleDing, Yushan, Jiameng Liu, Jamal S. M. Sabir, Xinyuan Cui, Xuejuan Shen, Nahid H. Hajrah, Mohamed M. M. Ahmed, Meshaal J. Sabir, Onaizan Godian Al-Zogabi, David M. Irwin, and et al. 2025. "The Differences in the Evolutionary Dynamics of MERS and SARS Coronaviruses" Viruses 17, no. 8: 1114. https://doi.org/10.3390/v17081114
APA StyleDing, Y., Liu, J., Sabir, J. S. M., Cui, X., Shen, X., Hajrah, N. H., Ahmed, M. M. M., Sabir, M. J., Al-Zogabi, O. G., Irwin, D. M., & Shen, Y. (2025). The Differences in the Evolutionary Dynamics of MERS and SARS Coronaviruses. Viruses, 17(8), 1114. https://doi.org/10.3390/v17081114






