Pseudomonas Phage Banzai: Genomic and Functional Analysis of Novel Pbunavirus with Lytic Activity Against Pseudomonas aeruginosa
Abstract
1. Introduction
2. Materials and Methods
2.1. Phage Isolation and Purification
2.2. Experimental Procedures: Phage Adsorption and One-Step Growth
2.3. Experimental Procedures: Phage Stability Under Different Conditions
2.4. Phage Host Range Determination
2.5. Transmission Electron Microscopy
2.6. MIC Analisys
2.7. Testing Phage Activity In Vivo Using Galleria mellonella Model
- Prepare 15 Petri dishes and label them accordingly (3 Petri dishes per group; each Petri dish will contain 5 larvae).
- Prepare 15 tubes with 10 μL of 0.9% NaCl solution, 30 tubes with 5 μL of P. aeruginosa solution, and 15 tubes with 5 μL of phage Banzai (2 × 109 PFU/mL concentration was used in this study).
- Grab a larva randomly from the glass jar with the tweezers and put it on the Petri dish.
- Now take the insulin syringe and fill it with the necessary solution, then apply the needle into the last proleg of the larva. Press the plunger to inject the volume. Gently remove the pressure and subsequently the needle from the larva.
- Note: For the third group, firstly fill the syringe with 5 μL P. aeruginosa solution and then add 5 μL phage solution.
- Place it in one of the previously prepared Petri dishes.
- Repeat this procedure now for all larvae.
- After these steps we will have for each group three Petri dishes with five larvae each.
- Place the Petri dishes now into a cardboard box containing air holes and incubate the larvae in the dark at 37 °C.
- Monitor the larvae after 24 and 48 h, respectively.
- Evaluate the efficiency of the phage by scoring for alive and dead larvae.
- Analyze the data to produce a survival curve, which reflects the killing activity of phage Banzai in vivo.
2.8. Bioinformatic Analyses
3. Results
3.1. Latency Period and Burst Size Determination
3.2. Phage Stability Under Different Conditions
3.3. Phage Host Range
3.4. MIC Analisys
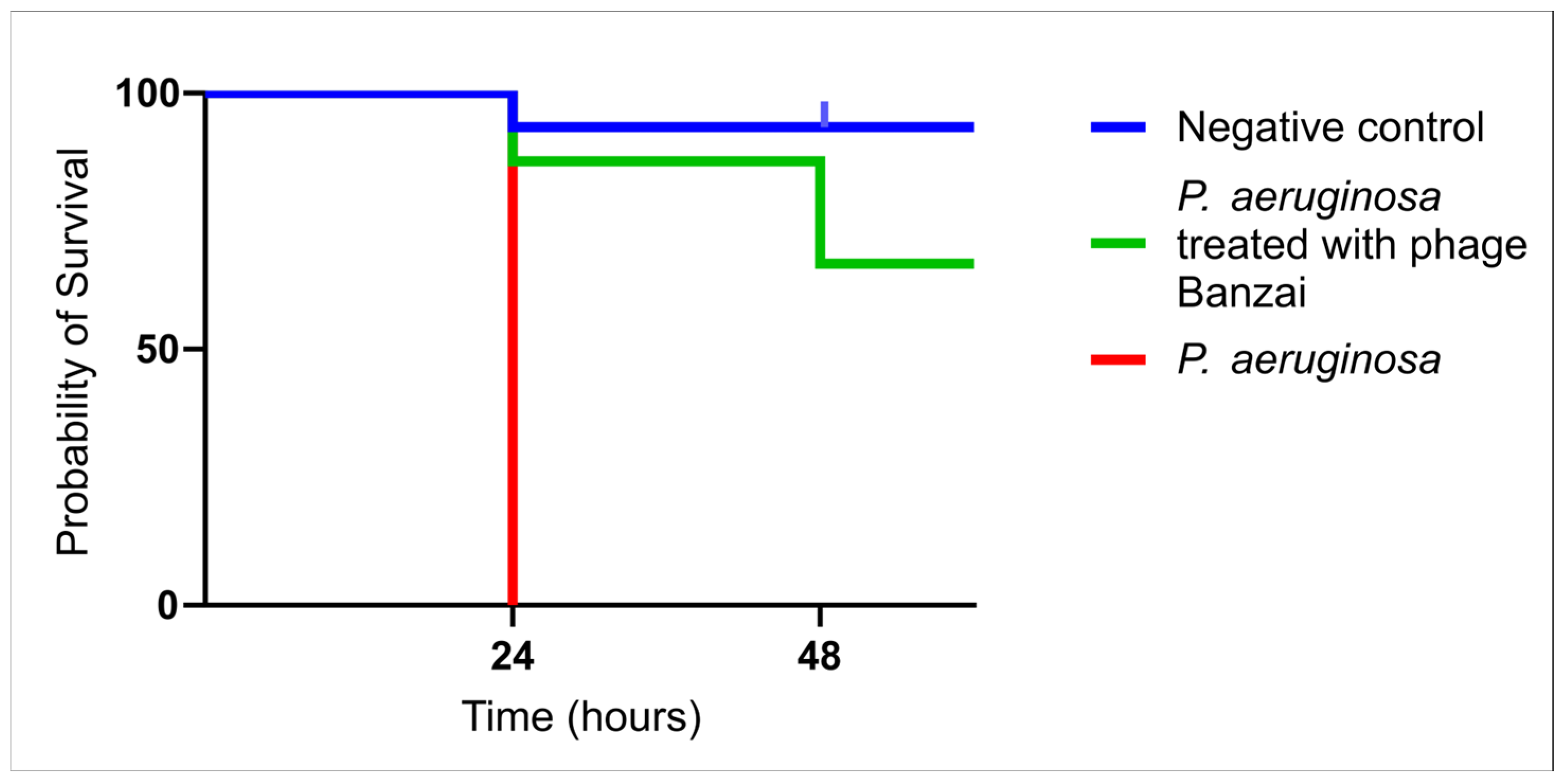
3.5. Survival Curve of G. mellonella Treated with Phage Banzai
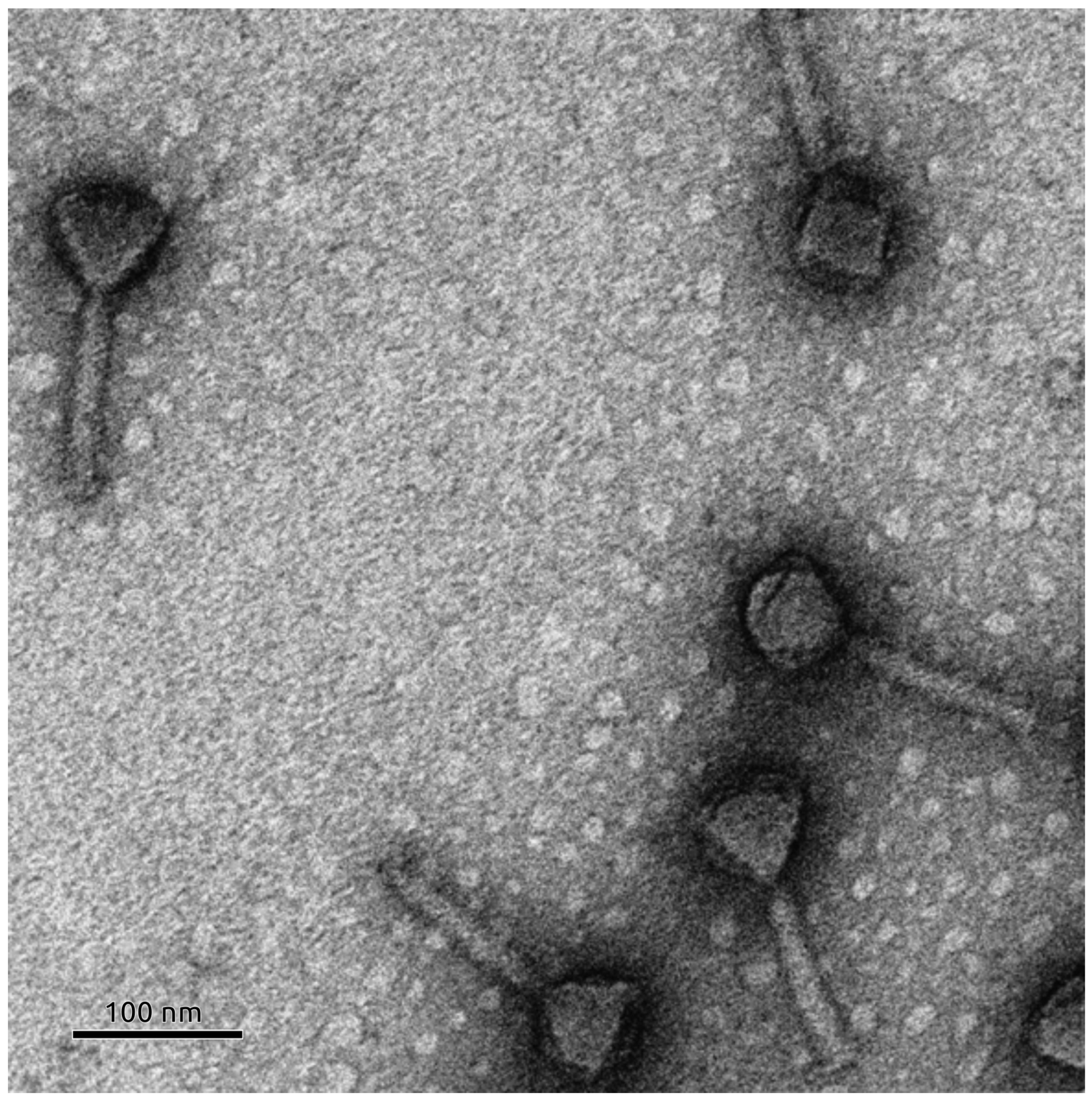
3.6. Phage Morphology
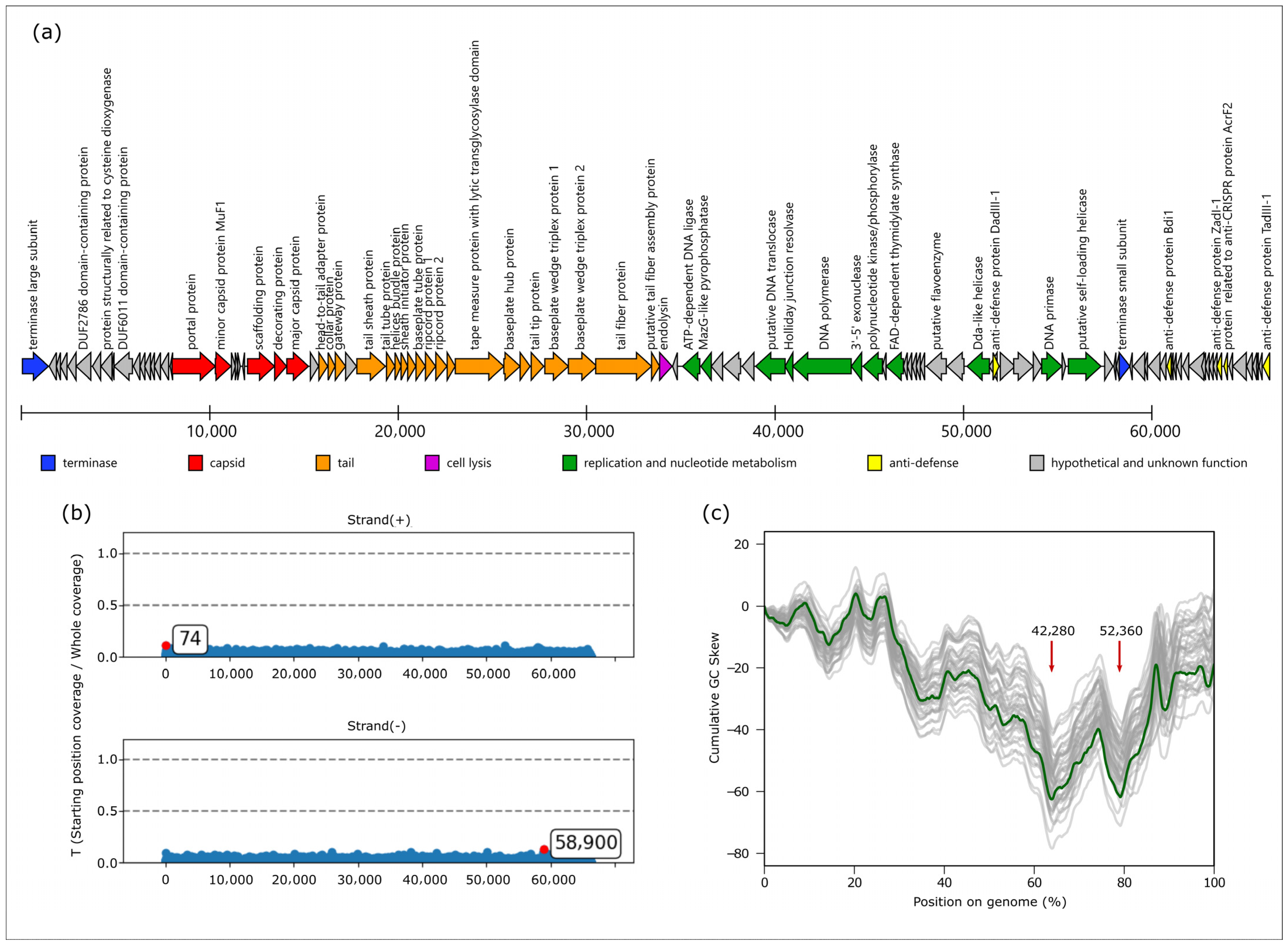
3.7. Genome and Proteome Characterization of Phage Banzai
4. Discussion
4.1. Therapeutic Potential of Phage Banzai for Pseudomonas Infections
4.2. General Virion and Genome Features
4.3. Genome Packaging
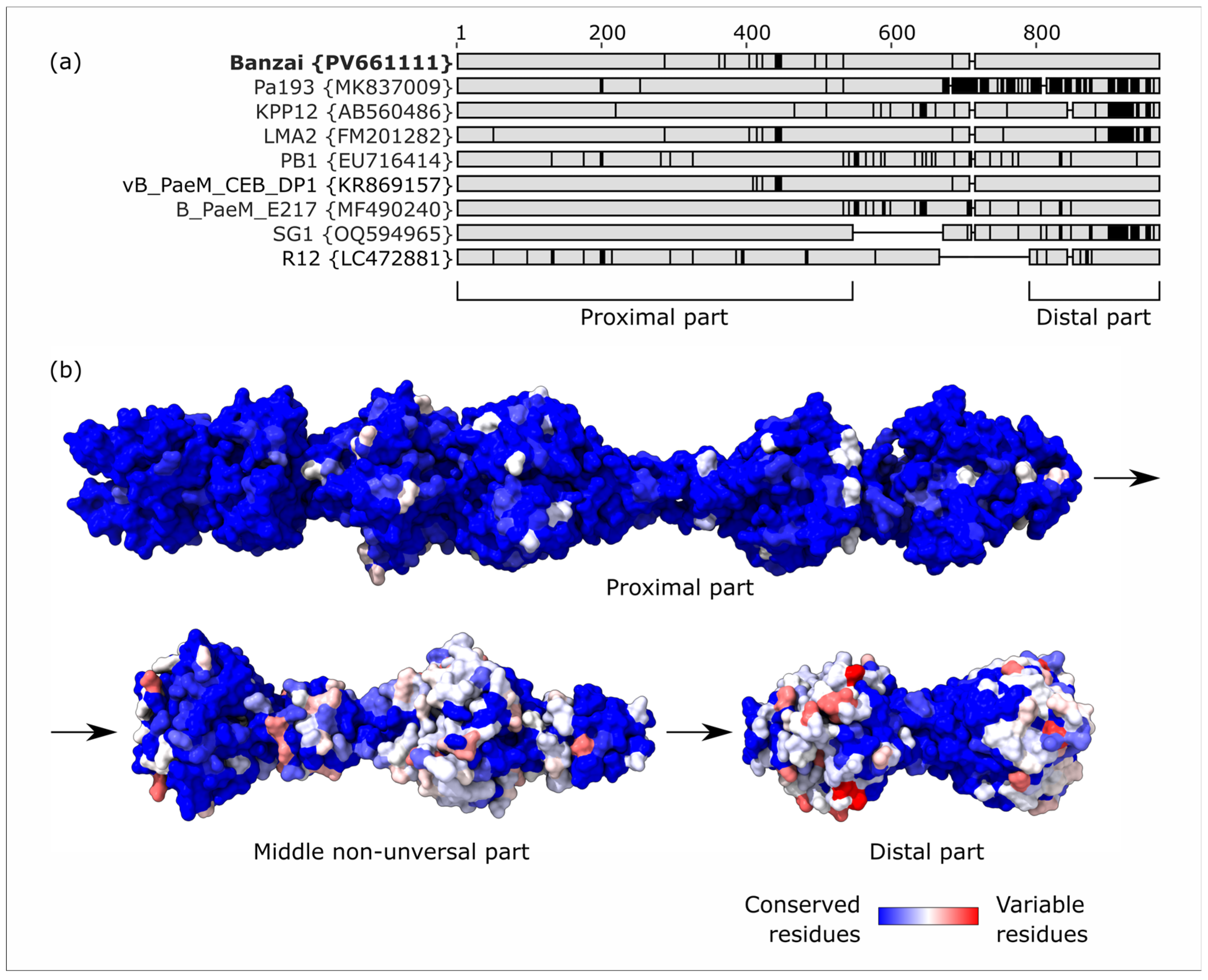
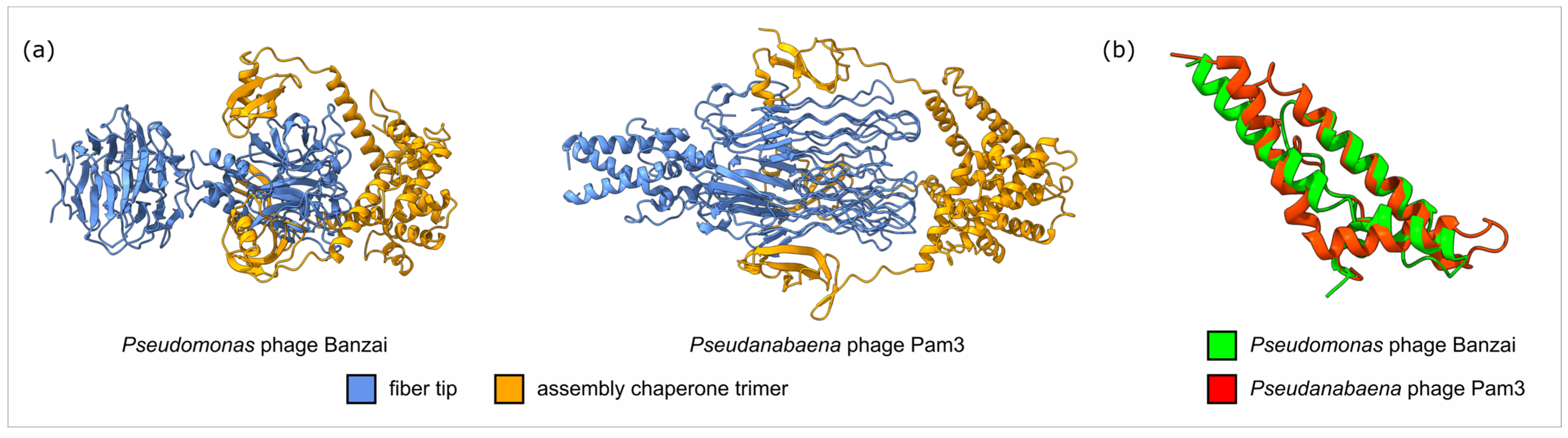
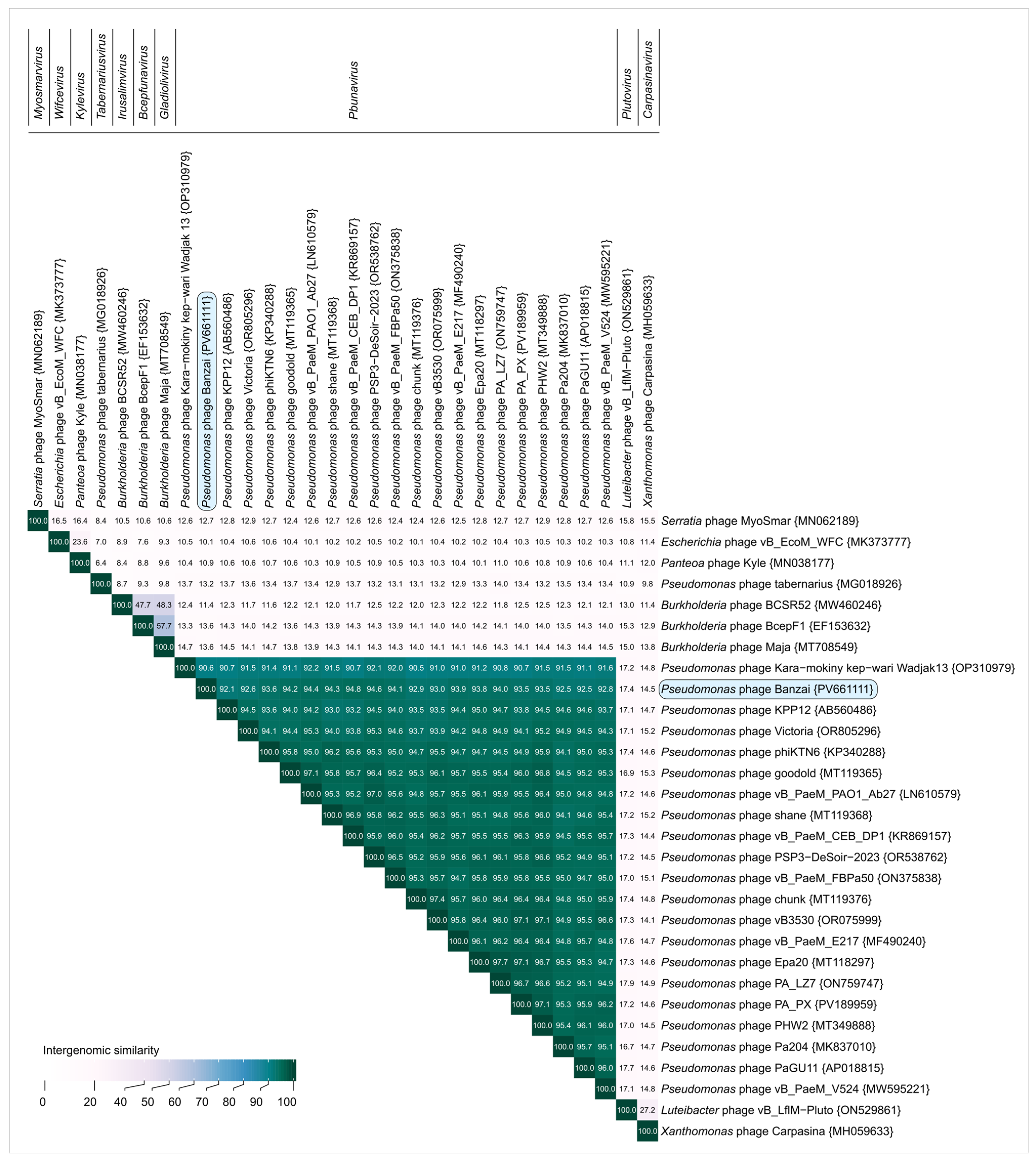
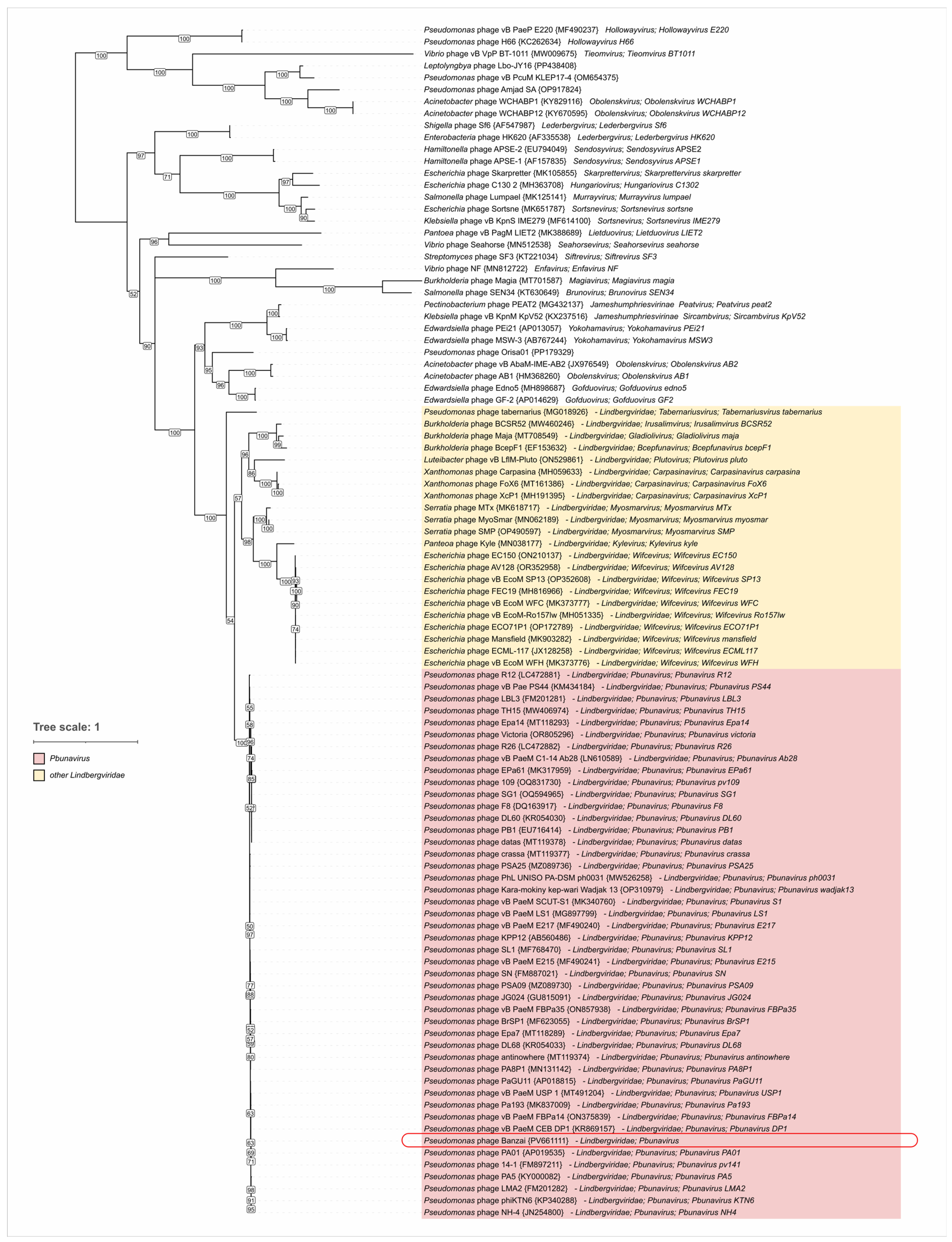
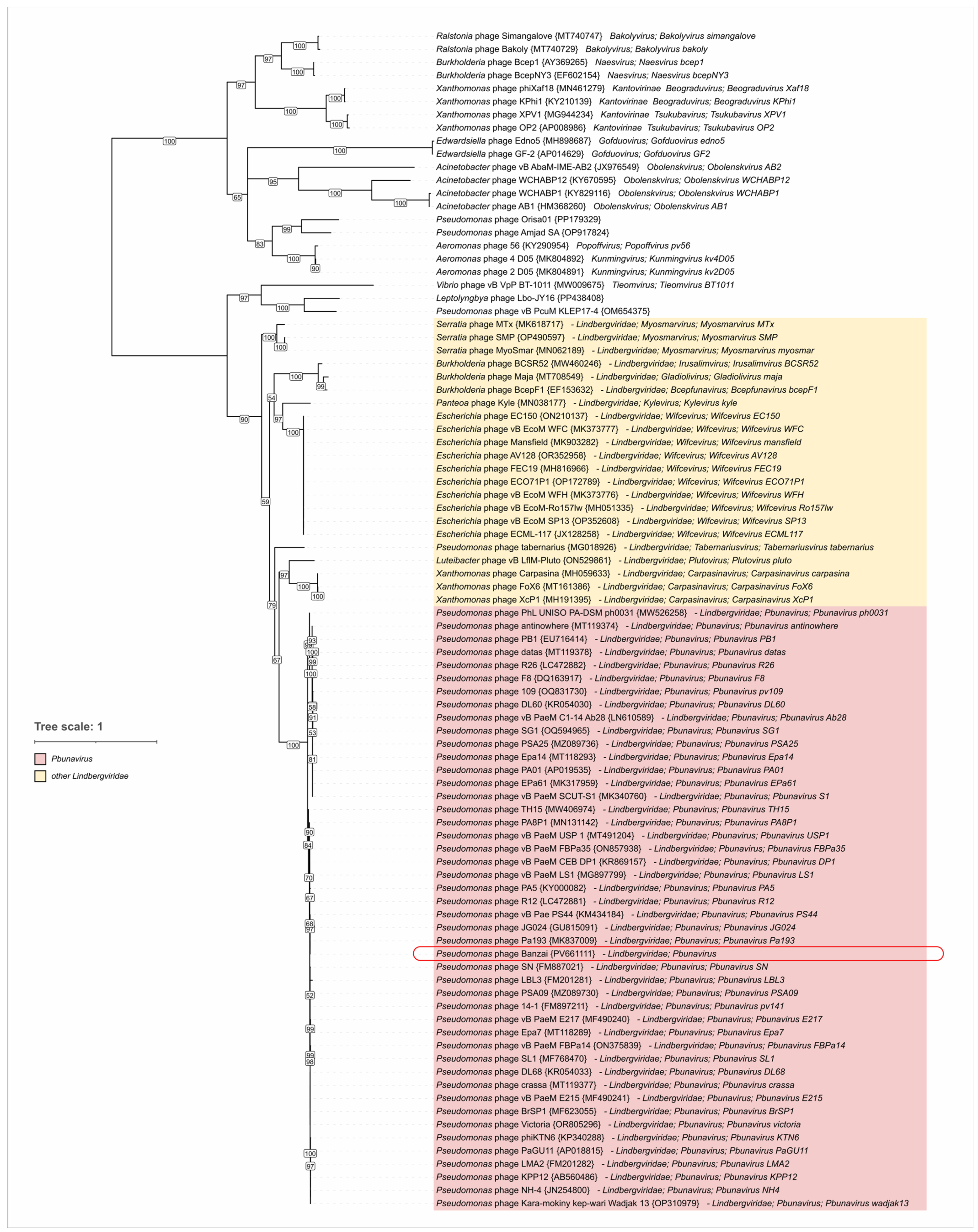
4.4. Lateral Gene Transfer and Genomic Evolution Within Pbunavirus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| bp | base pair |
| LGT | lateral gene transfer |
| MCP | major capsid protein |
| TLS | terminase large subunit |
References
- Naureen, Z.; Dautaj, A.; Anpilogov, K.; Camilleri, G.; Dhuli, K.; Tanzi, B.; Maltese, P.E.; Cristofoli, F.; De Antoni, L.; Beccari, T.; et al. Bacteriophages Presence in Nature and Their Role in the Natural Selection of Bacterial Populations. Acta Bio Med. Atenei Parm. 2020, 91, e2020024. [Google Scholar] [CrossRef]
- Kutter, E.; Sulakvelidze, A. Bacteriophages: Biology and Applications; CRC Press: Boca Raton, FL, USA, 2004; ISBN 0-429-20872-3. [Google Scholar]
- Keen, E.C. A Century of Phage Research: Bacteriophages and the Shaping of Modern Biology. BioEssays News Rev. Mol. Cell. Dev. Biol. 2015, 37, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Campbell, A. The Future of Bacteriophage Biology. Nat. Rev. Genet. 2003, 4, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of Antibiotic Resistance Pseudomonas aeruginosa in Intensive Care Unit; a Critical Review. Genes Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, Virulence Factors, Antibiotic Resistance, Interaction with Host, Technology Advances and Emerging Therapeutics. Signal Transduct. Target. Ther. 2022, 7, 1–27. [Google Scholar] [CrossRef]
- Ghasemian, S.; Karami-Zarandi, M.; Heidari, H.; Khoshnood, S.; Kouhsari, E.; Ghafourian, S.; Maleki, A.; Kazemian, H. Molecular Characterizations of Antibiotic Resistance, Biofilm Formation, and Virulence Determinants of Pseudomonas aeruginosa Isolated from Burn Wound Infection. J. Clin. Lab. Anal. 2023, 37, e24850. [Google Scholar] [CrossRef]
- Kerr, K.G.; Snelling, A.M. Pseudomonas aeruginosa: A Formidable and Ever-Present Adversary. J. Hosp. Infect. 2009, 73, 338–344. [Google Scholar] [CrossRef]
- Sala, A.; Di Ianni, F.; Pelizzone, I.; Bertocchi, M.; Santospirito, D.; Rogato, F.; Flisi, S.; Spadini, C.; Iemmi, T.; Moggia, E.; et al. The Prevalence of Pseudomonas aeruginosa and Multidrug Resistant Pseudomonas aeruginosa in Healthy Captive Ophidian. PeerJ 2019, 7, e6706. [Google Scholar] [CrossRef]
- Wright, A.; Hawkins, C.H.; Anggård, E.E.; Harper, D.R. A Controlled Clinical Trial of a Therapeutic Bacteriophage Preparation in Chronic Otitis Due to Antibiotic-Resistant Pseudomonas aeruginosa; a Preliminary Report of Efficacy. Clin. Otolaryngol. Off. J. ENT-UK Off. J. Neth. Soc. Oto-Rhino-Laryngol. Cervico-Facial Surg. 2009, 34, 349–357. [Google Scholar] [CrossRef]
- Mittal, R.; Aggarwal, S.; Sharma, S.; Chhibber, S.; Harjai, K. Urinary Tract Infections Caused by Pseudomonas aeruginosa: A Minireview. J. Infect. Public Health 2009, 2, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Li, L.; Li, M.; Liang, H.; He, Y.; Li, S. Risk Factors and Outcomes of Patients with Carbapenem-Resistant Pseudomonas aeruginosa Bloodstream Infection. Infect. Drug Resist. 2023, 16, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Paprocka, P.; Durnaś, B.; Mańkowska, A.; Król, G.; Wollny, T.; Bucki, R. Pseudomonas aeruginosa Infections in Cancer Patients. Pathogens 2022, 11, 679. [Google Scholar] [CrossRef]
- Sanya, D.R.A.; Onésime, D.; Vizzarro, G.; Jacquier, N. Recent Advances in Therapeutic Targets Identification and Development of Treatment Strategies towards Pseudomonas aeruginosa Infections. BMC Microbiol. 2023, 23, 86. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.-I.; Kim, S.-H.; Kim, H.-B.; Park, S.-W.; Choe, Y.-J.; Oh, M.-D.; Kim, E.-C.; Choe, K.-W. Pseudomonas aeruginosa Bacteremia: Risk Factors for Mortality and Influence of Delayed Receipt of Effective Antimicrobial Therapy on Clinical Outcome. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2003, 37, 745–751. [Google Scholar] [CrossRef]
- Frem, J.A.; Doumat, G.; Kazma, J.; Gharamti, A.; Kanj, S.S.; Abou Fayad, A.G.; Matar, G.M.; Kanafani, Z.A. Clinical Predictors of Mortality in Patients with Pseudomonas aeruginosa Infection. PLoS ONE 2023, 18, e0282276. [Google Scholar] [CrossRef]
- Makky, S.; Abdelrahman, F.; Rezk, N.; Easwaran, M.; El-Shibiny, A. Phages for Treatment Pseudomonas aeruginosa Infection. Prog. Mol. Biol. Transl. Sci. 2023, 201, 1–19. [Google Scholar] [CrossRef]
- Wood, S.J.; Kuzel, T.M.; Shafikhani, S.H. Pseudomonas aeruginosa: Infections, Animal Modeling, and Therapeutics. Cells 2023, 12, 199. [Google Scholar] [CrossRef]
- Alipour-Khezri, E.; Skurnik, M.; Zarrini, G. Pseudomonas aeruginosa Bacteriophages and Their Clinical Applications. Viruses 2024, 16, 1051. [Google Scholar] [CrossRef]
- Holger, D.J.; El Ghali, A.; Bhutani, N.; Lev, K.L.; Stamper, K.; Kebriaei, R.; Kunz Coyne, A.J.; Morrisette, T.; Shah, R.; Alexander, J.; et al. Phage-Antibiotic Combinations against Multidrug-Resistant Pseudomonas aeruginosa in in Vitro Static and Dynamic Biofilm Models. Antimicrob. Agents Chemother. 2023, 67, e0057823. [Google Scholar] [CrossRef]
- Kovacs, C.J.; Rapp, E.M.; Rankin, W.R.; McKenzie, S.M.; Brasko, B.K.; Hebert, K.E.; Bachert, B.A.; Kick, A.R.; Burpo, F.J.; Barnhill, J.C. Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics. Viruses 2024, 16, 1000. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage Therapy: An Alternative to Antibiotics in the Age of Multi-Drug Resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Evseev, P.; Tkachenko, N.; Miroshnikov, K. Diversity of Tailed Bacteriophages Infecting Pseudomonas aeruginosa. In Proceedings of the 2024 IEEE International Multi-Conference on Engineering, Computer and Information Sciences (SIBIRCON), Novosibirsk, Russia, 30 September–2 October 2024; pp. 261–265. [Google Scholar]
- Troshin, K.; Sykilinda, N.; Shuraleva, S.; Tokmakova, A.; Tkachenko, N.; Kurochkina, L.; Miroshnikov, K.; Suzina, N.; Brzhozovskaya, E.; Petrova, K.; et al. Pseudomonas Phage Lydia and the Evolution of the Mesyanzhinovviridae Family. Viruses 2025, 17, 369. [Google Scholar] [CrossRef] [PubMed]
- Ilyina, V.; Gatina, A.; Trizna, E.; Siniagina, M.; Yadykova, L.; Ivannikova, A.; Ozhegov, G.; Zhuravleva, D.; Fedorova, M.; Gorshkova, A.; et al. New Bacteriophage Pseudomonas Phage Ka2 from a Tributary Stream of Lake Baikal. Viruses 2025, 17, 189. [Google Scholar] [CrossRef]
- Garbe, J.; Wesche, A.; Bunk, B.; Kazmierczak, M.; Selezska, K.; Rohde, C.; Sikorski, J.; Rohde, M.; Jahn, D.; Schobert, M. Characterization of JG024, a Pseudomonas aeruginosa PB1-like Broad Host Range Phage under Simulated Infection Conditions. BMC Microbiol. 2010, 10, 301. [Google Scholar] [CrossRef]
- Fujiki, J.; Furusawa, T.; Munby, M.; Kawaguchi, C.; Matsuda, Y.; Shiokura, Y.; Nakamura, K.; Nakamura, T.; Sasaki, M.; Usui, M.; et al. Susceptibility of Pseudomonas aeruginosa Veterinary Isolates to Pbunavirus PB1-like Phages. Microbiol. Immunol. 2020, 64, 778–782. [Google Scholar] [CrossRef]
- Nakamura, K.; Fujiki, J.; Nakamura, T.; Furusawa, T.; Gondaira, S.; Usui, M.; Higuchi, H.; Tamura, Y.; Iwano, H. Fluctuating Bacteriophage-Induced galU Deficiency Region Is Involved in Trade-off Effects on the Phage and Fluoroquinolone Sensitivity in Pseudomonas aeruginosa. Virus Res. 2021, 306, 198596. [Google Scholar] [CrossRef]
- Wang, X.; Tang, J.; Dang, W.; Xie, Z.; Zhang, F.; Hao, X.; Sun, S.; Liu, X.; Luo, Y.; Li, M.; et al. Isolation and Characterization of Three Pseudomonas aeruginosa Viruses with Therapeutic Potential. Microbiol. Spectr. 2023, 11, e04636. [Google Scholar] [CrossRef]
- Weiner, I.; Kahan-Hanum, M.; Buchstab, N.; Zelcbuch, L.; Navok, S.; Sherman, I.; Nicenboim, J.; Axelrod, T.; Berko-Ashur, D.; Olshina, M.; et al. Phage Therapy with Nebulized Cocktail BX004-A for Chronic Pseudomonas aeruginosa Infections in Cystic Fibrosis: A Randomized First-in-Human Trial. Nat. Commun. 2025, 16, 5579. [Google Scholar] [CrossRef]
- Iglesias, S.M.; Hou, C.-F.D.; Reid, J.; Schauer, E.; Geier, R.; Soriaga, A.; Sim, L.; Gao, L.; Whitelegge, J.; Kyme, P.; et al. Cryo-EM Analysis of Pseudomonas Phage Pa193 Structural Components. Commun. Biol. 2024, 7, 1275. [Google Scholar] [CrossRef]
- Li, F.; Hou, C.-F.D.; Lokareddy, R.K.; Yang, R.; Forti, F.; Briani, F.; Cingolani, G. High-Resolution Cryo-EM Structure of the Pseudomonas Bacteriophage E217. Nat. Commun. 2023, 14, 4052. [Google Scholar] [CrossRef]
- Koch, G.; Nadal-Jimenez, P.; Cool, R.H.; Quax, W.J. Assessing Pseudomonas Virulence with Nonmammalian Host: Galleria Mellonella. Methods Mol. Biol. Clifton NJ 2014, 1149, 681–688. [Google Scholar] [CrossRef]
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Middelboe, M.; Chan, A.; Bertelsen, S.K. Isolation and Life Cycle Characterization of Lytic Viruses Infecting Heterotrophic Bacteria and Cyanobacteria. In Manual of Aquatic Viral Ecology; Wilhelm, S.W., Weinbauer, M.G., Suttle, C.A., Eds.; American Society of Limnology and Oceanography, Inc.: Waco, TX, USA, 2010; pp. 118–133. [Google Scholar]
- Lerdsittikul, V.; Thongdee, M.; Chaiwattanarungruengpaisan, S.; Atithep, T.; Apiratwarrasakul, S.; Withatanung, P.; Clokie, M.R.J.; Korbsrisate, S. A Novel Virulent Litunavirus Phage Possesses Therapeutic Value against Multidrug Resistant Pseudomonas aeruginosa. Sci. Rep. 2022, 12, 21193. [Google Scholar] [CrossRef] [PubMed]
- Wdowiak, M.; Paczesny, J.; Raza, S. Enhancing the Stability of Bacteriophages Using Physical, Chemical, and Nano-Based Approaches: A Review. Pharmaceutics 2022, 14, 1936. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Gao, Y.; Xue, Y.; Liu, Y.; Zeng, X.; Cheng, Y.; Ma, J.; Wang, H.; Sun, J.; Wang, Z.; et al. Bacteriophage Cocktails Protect Dairy Cows Against Mastitis Caused By Drug Resistant Escherichia coli Infection. Front. Cell. Infect. Microbiol. 2021, 11, 690377. [Google Scholar] [CrossRef]
- Bocharova, Y.; Chebotar, I.; Savinova, T.; Lyamin, A.; Kondratenko, O.; Polikarpova, S.; Fedorova, N.; Semykin, S.; Korostin, D.; Chaplin, A.; et al. Clonal Diversity, Antimicrobial Resistance, and Genome Features among Non-Fermenting Gram-Negative Bacteria Isolated from Patients with Cystic Fibrosis in Russia. Diagn. Microbiol. Infect. Dis. 2023, 108, 116102. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast One-Pass FASTQ Data Preprocessing, Quality Control, and Deduplication Using Fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. Pharokka: A Fast Scalable Bacteriophage Annotation Tool. Bioinform. Oxf. Engl. 2023, 39, btac776. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at Its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- van Kempen, M.; Kim, S.S.; Tumescheit, C.; Mirdita, M.; Lee, J.; Gilchrist, C.L.M.; Söding, J.; Steinegger, M. Fast and Accurate Protein Structure Search with Foldseek. Nat. Biotechnol. 2024, 42, 243–246. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle5: High-Accuracy Alignment Ensembles Enable Unbiased Assessments of Sequence Homology and Phylogeny. Nat. Commun. 2022, 13, 6968. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC-A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Petit, R.A., III. Pasty: A Tool Easily Taken Advantage of for in Silico Serogrouping of Pseudomonas aeruginosa Isolates. Available online: https://ictv.global/taxonomy (accessed on 10 July 2025).
- Anbo, M.; Jelsbak, L. A Bittersweet Fate: Detection of Serotype Switching in Pseudomonas aeruginosa. Microb. Genom. 2023, 9, mgen000919. [Google Scholar] [CrossRef]
- Tesson, F.; Hervé, A.; Mordret, E.; Touchon, M.; d’Humières, C.; Cury, J.; Bernheim, A. Systematic and Quantitative View of the Antiviral Arsenal of Prokaryotes. Nat. Commun. 2022, 13, 2561. [Google Scholar] [CrossRef]
- Yee, W.-X.; Lee, Y.-J.; Klein, T.A.; Wirganowicz, A.; Gabagat, A.E.; Csörgő, B.; Makarova, K.S.; Koonin, E.V.; Weigele, P.R.; Bondy-Denomy, J. END Nucleases: Antiphage Defense Systems Targeting Multiple Hypermodified Phage Genomes. bioRxiv 2025. [Google Scholar] [CrossRef]
- Ipoutcha, T.; Racharaks, R.; Huttelmaier, S.; Wilson, C.J.; Ozer, E.A.; Hartmann, E.M. A Synthetic Biology Approach to Assemble and Reboot Clinically Relevant Pseudomonas aeruginosa Tailed Phages. Microbiol. Spectr. 2024, 12, e0289723. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.-C.; Bikard, D.; Monot, M. PhageTerm: A Tool for Fast and Accurate Determination of Phage Termini and Packaging Mechanism Using next-Generation Sequencing Data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, A. Analyzing Genomes with Cumulative Skew Diagrams. Nucleic Acids Res. 1998, 26, 2286–2290. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.L.; Lou, Y.C.; Sachdeva, R.; Al-Shayeb, B.; Penev, P.I.; Jaffe, A.L.; Lei, S.; Santini, J.M.; Banfield, J.F. Widespread Stop-Codon Recoding in Bacteriophages May Regulate Translation of Lytic Genes. Nat. Microbiol. 2022, 7, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Jamet, A.; Touchon, M.; Ribeiro-Gonçalves, B.; Carriço, J.A.; Charbit, A.; Nassif, X.; Ramirez, M.; Rocha, E.P.C. A Widespread Family of Polymorphic Toxins Encoded by Temperate Phages. BMC Biol. 2017, 15, 75. [Google Scholar] [CrossRef]
- Godinho, L.M.; El Sadek Fadel, M.; Monniot, C.; Jakutyte, L.; Auzat, I.; Labarde, A.; Djacem, K.; Oliveira, L.; Carballido-Lopez, R.; Ayora, S.; et al. The Revisited Genome of Bacillus subtilis Bacteriophage SPP1. Viruses 2018, 10, 705. [Google Scholar] [CrossRef]
- Wei, Z.-L.; Yang, F.; Li, B.; Hou, P.; Kong, W.-W.; Wang, J.; Chen, Y.; Jiang, Y.-L.; Zhou, C.-Z. Structural Insights into the Chaperone-Assisted Assembly of a Simplified Tail Fiber of the Myocyanophage Pam3. Viruses 2022, 14, 2260. [Google Scholar] [CrossRef]
- North, O.I.; Davidson, A.R. Phage Proteins Required for Tail Fiber Assembly Also Bind Specifically to the Surface of Host Bacterial Strains. J. Bacteriol. 2021, 203, e00406-20. [Google Scholar] [CrossRef]
- Qiao, C.; Debiasi-Anders, G.; Mir-Sanchis, I. Staphylococcal Self-Loading Helicases Couple the Staircase Mechanism with Inter Domain High Flexibility. Nucleic Acids Res. 2022, 50, 8349–8362. [Google Scholar] [CrossRef]
- Yan, L.; Chen, Z. A Unifying Mechanism of DNA Translocation Underlying Chromatin Remodeling. Trends Biochem. Sci. 2020, 45, 217–227. [Google Scholar] [CrossRef]
- Zecchi, L.; Lo Piano, A.; Suzuki, Y.; Cañas, C.; Takeyasu, K.; Ayora, S. Characterization of the Holliday Junction Resolving Enzyme Encoded by the Bacillus subtilis Bacteriophage SPP1. PLoS ONE 2012, 7, e48440. [Google Scholar] [CrossRef]
- Johnson, G.; Banerjee, S.; Putonti, C. Diversity of Pseudomonas aeruginosa Temperate Phages. mSphere 2022, 7, e01015-21. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.R.; van den Berg, D.F.; Esser, J.Q.; van den Bossche, H.; Pozhydaieva, N.; Kalogeropoulos, K.; Brouns, S.J.J. Bacteriophage Genomes Encode Both Broad and Specific Counter-Defense Repertoires to Overcome Bacterial Defense Systems. Cell Host Microbe 2025, 33, 1161–1172.e5. [Google Scholar] [CrossRef] [PubMed]
- Rieper, F.; Wittmann, J.; Bunk, B.; Spröer, C.; Häfner, M.; Willy, C.; Müsken, M.; Ziehr, H.; Korf, I.H.E.; Jahn, D. Systematic Bacteriophage Selection for the Lysis of Multiple Pseudomonas aeruginosa Strains. Front. Cell. Infect. Microbiol. 2025, 15, 1597009. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.R.; van den Berg, D.F.; Esser, J.Q.; Muralidharan, A.; van den Bossche, H.; Bonilla, B.E.; van der Steen, B.A.; Haagsma, A.C.; Fluit, A.C.; Nobrega, F.L.; et al. Accumulation of Defense Systems in Phage-Resistant Strains of Pseudomonas aeruginosa. Sci. Adv. 2024, 10, eadj0341. [Google Scholar] [CrossRef]
- Chichkova, M.A.T. Phage-Based Approaches Targeting MDR/XDR High-Risk Clones of Pseudomonas aeruginosa; DTU Bioengineering: Lyngby, Denmark, 2023. [Google Scholar]
- Forti, F.; Bertoli, C.; Cafora, M.; Gilardi, S.; Pistocchi, A.; Briani, F. Identification and Impact on Pseudomonas aeruginosa Virulence of Mutations Conferring Resistance to a Phage Cocktail for Phage Therapy. Microbiol. Spectr. 2023, 11, e01477-23. [Google Scholar] [CrossRef]
- Lam, J.S.; Taylor, V.L.; Islam, S.T.; Hao, Y.; Kocíncová, D. Genetic and Functional Diversity of Pseudomonas aeruginosa Lipopolysaccharide. Front. Microbiol. 2011, 2, 118. [Google Scholar] [CrossRef]
- Thrane, S.W.; Taylor, V.L.; Lund, O.; Lam, J.S.; Jelsbak, L. Application of Whole-Genome Sequencing Data for O-Specific Antigen Analysis and In Silico Serotyping of Pseudomonas aeruginosa Isolates. J. Clin. Microbiol. 2016, 54, 1782–1788. [Google Scholar] [CrossRef]
- Hancock, R.E.; Mutharia, L.M.; Chan, L.; Darveau, R.P.; Speert, D.P.; Pier, G.B. Pseudomonas aeruginosa Isolates from Patients with Cystic Fibrosis: A Class of Serum-Sensitive, Nontypable Strains Deficient in Lipopolysaccharide O Side Chains. Infect. Immun. 1983, 42, 170–177. [Google Scholar] [CrossRef]
- Høiby, N.; Moser, C.; Ciofu, O. Pseudomonas aeruginosa in the Frontline of the Greatest Challenge of Biofilm Infection—Its Tolerance to Antibiotics. Microorganisms 2024, 12, 2115. [Google Scholar] [CrossRef]
- Yin, R.; Cheng, J.; Wang, J.; Li, P.; Lin, J. Treatment of Pseudomonas aeruginosa Infectious Biofilms: Challenges and Strategies. Front. Microbiol. 2022, 13, 955286. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, P.; Lin, Z.; Wang, T. Characterization of Two Pseudomonas aeruginosa Viruses vB_PaeM_SCUT-S1 and vB_PaeM_SCUT-S2. Viruses 2019, 11, 318. [Google Scholar] [CrossRef]
- Billaud, M.; Plantady, C.; Lerouge, B.; Ollivier, E.; Lossouarn, J.; Moncaut, E.; Deschamps, J.; Briandet, R.; Cleret, A.; Fevre, C.; et al. Complementary Killing Activities of Pbunavirus LS1 and Bruynoghevirus LUZ24 Phages on Planktonic and Sessile Pseudomonas aeruginosa PAO1 Derivatives. Antimicrob. Agents Chemother. 2025, e00579-25. [Google Scholar] [CrossRef]
- Lokareddy, R.K.; Hou, C.-F.D.; Doll, S.G.; Li, F.; Gillilan, R.E.; Forti, F.; Horner, D.S.; Briani, F.; Cingolani, G. Terminase Subunits from the Pseudomonas-Phage E217. J. Mol. Biol. 2022, 434, 167799. [Google Scholar] [CrossRef] [PubMed]
- Ceyssens, P.-J.; Miroshnikov, K.; Mattheus, W.; Krylov, V.; Robben, J.; Noben, J.-P.; Vanderschraeghe, S.; Sykilinda, N.; Kropinski, A.M.; Volckaert, G.; et al. Comparative Analysis of the Widespread and Conserved PB1-like Viruses Infecting Pseudomonas aeruginosa. Environ. Microbiol. 2009, 11, 2874–2883. [Google Scholar] [CrossRef] [PubMed]
- Malki, K.; Kula, A.; Bruder, K.; Sible, E.; Hatzopoulos, T.; Steidel, S.; Watkins, S.C.; Putonti, C. Bacteriophages Isolated from Lake Michigan Demonstrate Broad Host-Range across Several Bacterial Phyla. Virol. J. 2015, 12, 164. [Google Scholar] [CrossRef] [PubMed]
- Borodovich, T.; Shkoporov, A.N.; Ross, R.P.; Hill, C. Phage-Mediated Horizontal Gene Transfer and Its Implications for the Human Gut Microbiome. Gastroenterol. Rep. 2022, 10, goac012. [Google Scholar] [CrossRef]
- Abedon, S.T. Transduction of Large Amounts of DNA. In Bacteriophages as Drivers of Evolution: An Evolutionary Ecological Perspective; Abedon, S.T., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 137–150. ISBN 978-3-030-94309-7. [Google Scholar]
- Morgan, A.F. Transduction of Pseudomonas aeruginosa with a Mutant of Bacteriophage E79. J. Bacteriol. 1979, 139, 137–140. [Google Scholar] [CrossRef]
- Yin, S.; Huang, G.; Zhang, Y.; Jiang, B.; Yang, Z.; Dong, Z.; You, B.; Yuan, Z.; Hu, F.; Zhao, Y.; et al. Phage Abp1 Rescues Human Cells and Mice from Infection by Pan-Drug Resistant Acinetobacter baumannii. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 44, 2337–2345. [Google Scholar] [CrossRef]
- Popova, A.V.; Zhilenkov, E.L.; Myakinina, V.P.; Krasilnikova, V.M.; Volozhantsev, N.V. Isolation and Characterization of Wide Host Range Lytic Bacteriophage AP22 Infecting Acinetobacter baumannii. FEMS Microbiol. Lett. 2012, 332, 40–46. [Google Scholar] [CrossRef]
- Gunalan, A.; Sarumathi, D.; Sastry, A.S.; Ramanathan, V.; Rajaa, S.; Sistla, S. Effect of Combined Colistin and Meropenem against Meropenem Resistant Acinetobacter baumannii and Pseudomonas aeruginosa by Checkerboard Method: A Cross Sectional Analytical Study. Indian J. Pharmacol. 2021, 53, 207. [Google Scholar] [CrossRef] [PubMed]
- Ahuatzin-Flores, O.E.; Torres, E.; Chávez-Bravo, E. Acinetobacter baumannii, a Multidrug-Resistant Opportunistic Pathogen in New Habitats: A Systematic Review. Microorganisms 2024, 12, 644. [Google Scholar] [CrossRef] [PubMed]
- Crone, S.; Vives-Flórez, M.; Kvich, L.; Saunders, A.M.; Malone, M.; Nicolaisen, M.H.; Martínez-García, E.; Rojas-Acosta, C.; Catalina Gomez-Puerto, M.; Calum, H.; et al. The Environmental Occurrence of Pseudomonas aeruginosa. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2020, 128, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Elias, S.; Banin, E. Multi-Species Biofilms: Living with Friendly Neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef]
- Evseev, P.; Lukianova, A.; Sykilinda, N.; Gorshkova, A.; Bondar, A.; Shneider, M.; Kabilov, M.; Drucker, V.; Miroshnikov, K. Pseudomonas Phage MD8: Genetic Mosaicism and Challenges of Taxonomic Classification of Lambdoid Bacteriophages. Int. J. Mol. Sci. 2021, 22, 10350. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaplin, A.V.; Sykilinda, N.N.; Skvortsov, G.A.; Troshin, K.S.; Vasilyeva, A.A.; Shuraleva, S.A.; Malkov, A.A.; Simonov, V.S.; Efimov, B.A.; Kafarskaia, L.I.; et al. Pseudomonas Phage Banzai: Genomic and Functional Analysis of Novel Pbunavirus with Lytic Activity Against Pseudomonas aeruginosa. Viruses 2025, 17, 1088. https://doi.org/10.3390/v17081088
Chaplin AV, Sykilinda NN, Skvortsov GA, Troshin KS, Vasilyeva AA, Shuraleva SA, Malkov AA, Simonov VS, Efimov BA, Kafarskaia LI, et al. Pseudomonas Phage Banzai: Genomic and Functional Analysis of Novel Pbunavirus with Lytic Activity Against Pseudomonas aeruginosa. Viruses. 2025; 17(8):1088. https://doi.org/10.3390/v17081088
Chicago/Turabian StyleChaplin, Andrei V., Nina N. Sykilinda, George A. Skvortsov, Konstantin S. Troshin, Anna A. Vasilyeva, Sofia A. Shuraleva, Artem A. Malkov, Vladislav S. Simonov, Boris A. Efimov, Lyudmila I. Kafarskaia, and et al. 2025. "Pseudomonas Phage Banzai: Genomic and Functional Analysis of Novel Pbunavirus with Lytic Activity Against Pseudomonas aeruginosa" Viruses 17, no. 8: 1088. https://doi.org/10.3390/v17081088
APA StyleChaplin, A. V., Sykilinda, N. N., Skvortsov, G. A., Troshin, K. S., Vasilyeva, A. A., Shuraleva, S. A., Malkov, A. A., Simonov, V. S., Efimov, B. A., Kafarskaia, L. I., Miroshnikov, K. A., Kuznetsova, A. A., & Evseev, P. V. (2025). Pseudomonas Phage Banzai: Genomic and Functional Analysis of Novel Pbunavirus with Lytic Activity Against Pseudomonas aeruginosa. Viruses, 17(8), 1088. https://doi.org/10.3390/v17081088






