Comparative Endosymbiont Community Structures of Nonviruliferous and Rice Stripe Virus-Viruliferous Laodelphax striatellus (Hemiptera: Delphacidae) in Korea
Abstract
1. Introduction
2. Results
2.1. Differences in the Diversity of SBPH with or Without RSV Infection
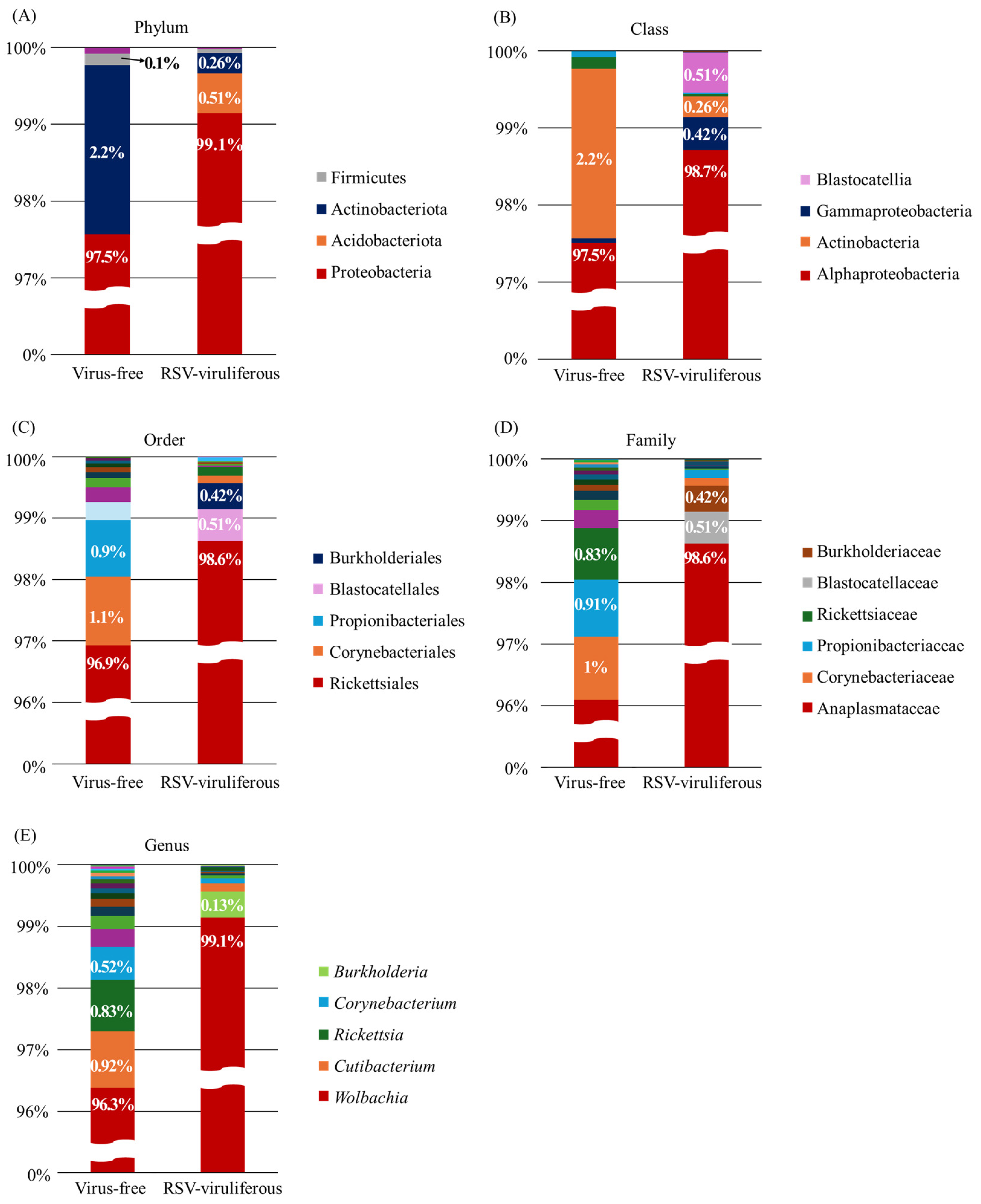
2.2. Bacterial Community Composition
2.3. Validation of Sequencing Data
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. RNA Extraction and RSV Diagnosis
4.3. DNA Extraction, Library Construction and Sequencing
4.4. 16S Taxonomy Analysis
4.4.1. OTU Analysis
4.4.2. ASV Analysis
4.5. PCR for Endosymbiont Detection
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hibino, H. Biology and epidemiology of rice viruses. Annu. Rev. Phytopathol. 1996, 34, 249–274. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, Y.; Li, J. Towards molecular breeding and improvement of rice in China. Trends Plant Sci. 2005, 10, 610–614. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2017, 46, D708–D717. [Google Scholar] [CrossRef]
- Ishikawa, K.; Omura, T.; Hibino, H. Morphological characteristics of rice stripe virus. J. Gen. Virol. 1989, 70, 3465–3468. [Google Scholar] [CrossRef]
- Falk, B.W.; Tsai, J.H. Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 1998, 36, 139–163. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.-Y.; Yang, J.-G.; Liao, F.-L.; Gao, F.-L.; Lu, L.-M.; Zhang, X.-T.; Li, F.; Wu, Z.-J.; Lin, Q.-Y.; Xie, L.-H. Genetic diversity and population structure of rice stripe virus in China. J. Gen. Virol. 2009, 90, 1025–1034. [Google Scholar] [CrossRef]
- Jia, D.; Chen, Q.; Mao, Q.; Zhang, X.; Wu, W.; Chen, H.; Yu, X.; Wang, Z.; Wei, T. Vector mediated transmission of persistently transmitted plant viruses. Curr. Opin. Virol. 2018, 28, 127–132. [Google Scholar] [CrossRef]
- Zhu, J.; Jiang, F.; Wang, X.; Yang, P.; Bao, Y.; Zhao, W.; Wang, W.; Lu, H.; Wang, Q.; Cui, N. Genome sequence of the small brown planthopper, Laodelphax striatellus. Gigascience 2017, 6, gix109. [Google Scholar] [CrossRef]
- Lederberg, J.; McCray, A.T. Ome SweetOmics—A genealogical treasury of words. Scientist 2001, 15, 8. [Google Scholar]
- Coon, K.L.; Vogel, K.J.; Brown, M.R.; Strand, M.R. Mosquitoes rely on their gut microbiota for development. Mol. Ecol. 2014, 23, 2727–2739. [Google Scholar] [CrossRef]
- Minard, G.; Mavingui, P.; Moro, C.V. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasites Vectors 2013, 6, 146. [Google Scholar] [CrossRef]
- Dillon, R.J.; Dillon, V. The gut bacteria of insects: Nonpathogenic interactions. Annu. Rev. Entomol. 2004, 49, 71–92. [Google Scholar] [CrossRef]
- Charroux, B.; Royet, J. Drosophila immune response: From systemic antimicrobial peptide production in fat body cells to local defense in the intestinal tract. Fly 2010, 4, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.-H.; Kim, S.-H.; Lee, H.-Y.; Bai, J.Y.; Nam, Y.-D.; Bae, J.-W.; Lee, D.G.; Shin, S.C.; Ha, E.-M.; Lee, W.-J. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 2008, 319, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Moran, N.A. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 2005, 59, 155–189. [Google Scholar] [CrossRef]
- Engelstädter, J.; Hurst, G.D. The ecology and evolution of microbes that manipulate host reproduction. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 127–149. [Google Scholar] [CrossRef]
- Feldhaar, H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 2011, 36, 533–543. [Google Scholar] [CrossRef]
- Moran, N.A.; McCutcheon, J.P.; Nakabachi, A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008, 42, 165–190. [Google Scholar] [CrossRef]
- Banerjee, S.; Hess, D.; Majumder, P.; Roy, D.; Das, S. The interactions of Allium sativum leaf agglutinin with a chaperonin group of unique receptor protein isolated from a bacterial endosymbiont of the mustard aphid. J. Biol. Chem. 2004, 279, 23782–23789. [Google Scholar] [CrossRef]
- Hancock, P.A.; Sinkins, S.P.; Godfray, H.C.J. Strategies for introducing Wolbachia to reduce transmission of mosquito-borne diseases. PLoS Neglected Trop. Dis. 2011, 5, e1024. [Google Scholar] [CrossRef]
- Kambris, Z.; Cook, P.E.; Phuc, H.K.; Sinkins, S.P. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 2009, 326, 134–136. [Google Scholar] [CrossRef]
- Morin, S.; Ghanim, M.; Zeidan, M.; Czosnek, H.; Verbeek, M.; van den Heuvel, J.F. A GroEL homologue from endosymbiotic bacteria of the whiteflyBemisia tabaciis implicated in the circulative transmission of tomato yellow leaf curl virus. Virology 1999, 256, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, Y.; Zchori-Fein, E.; Mozes-Daube, N.; Kontsedalov, S.; Skaljac, M.; Brumin, M.; Sobol, I.; Czosnek, H.; Vavre, F.; Fleury, F. The transmission efficiency of tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J. Virol. 2010, 84, 9310–9317. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.M.; Banerjee, N. Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 1999, 63, 128–148. [Google Scholar] [CrossRef]
- Van den Heuvel, J.; Bruyere, A.; Hogenhout, S.A.; Ziegler-Graff, V.; Brault, V.; Verbeek, M.; Van Der Wilk, F.; Richards, K. The N-terminal region of the luteovirus readthrough domain determines virus binding to Buchnera GroEL and is essential for virus persistence in the aphid. J. Virol. 1997, 71, 7258–7265. [Google Scholar] [CrossRef] [PubMed]
- Petti, C.; Polage, C.; Schreckenberger, P. The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J. Clin. Microbiol. 2005, 43, 6123–6125. [Google Scholar] [CrossRef]
- Ranjith, M.; Harish, E.; Girija, D.; Nazeem, P. Bacterial communities associated with the gut of tomato fruit borer, Helicoverpa armigera (Hübner)(Lepidoptera: Noctuidae) based on Illumina Next-Generation Sequencing. J. Asia-Pac. Entomol. 2016, 19, 333–340. [Google Scholar] [CrossRef]
- Patel, J.B. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol. Diagn. 2001, 6, 313–321. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Drancourt, M.; Bollet, C.; Carlioz, A.; Martelin, R.; Gayral, J.-P.; Raoult, D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 2000, 38, 3623–3630. [Google Scholar] [CrossRef]
- Morozova, O.; Marra, M.A. Applications of next-generation sequencing technologies in functional genomics. Genomics 2008, 92, 255–264. [Google Scholar] [CrossRef]
- Mardis, E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar] [CrossRef]
- Forde, B.M.; O’Toole, P.W. Next-generation sequencing technologies and their impact on microbial genomics. Brief. Funct. Genom. 2013, 12, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.E.; Hopkins, K.; Inward, D.J.; Vogler, A.P. Metabarcoding of fungal communities associated with bark beetles. Ecol. Evol. 2016, 6, 1590–1600. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.-O.; Song, S.-L.; Yong, H.-S.; See-Too, W.-S.; Yin, W.-F.; Chan, K.-G. Microbial community composition reveals spatial variation and distinctive core microbiome of the weaver ant Oecophylla smaragdina in Malaysia. Sci. Rep. 2018, 8, 10777. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarczyk-Ziemba, A.; Wagner, G.K.; Grzywnowicz, K.; Kucharczyk, M.; Zielińska, S. The microbiome profiling of fungivorous black tinder fungus beetle Bolitophagus reticulatus reveals the insight into bacterial communities associated with larvae and adults. PeerJ 2019, 7, e6852. [Google Scholar] [CrossRef]
- Bing, X.L.; Zhao, D.S.; Peng, C.W.; Huang, H.J.; Hong, X.Y. Similarities and spatial variations of bacterial and fungal communities in field rice planthopper (Hemiptera: Delphacidae) populations. Insect Sci. 2020, 27, 947–963. [Google Scholar] [CrossRef]
- Shentu, X.; Xiao, Y.; Song, Y.; Cao, Z.; Fan, J.; Yu, X. Comparative analysis of the diversity of the microbial communities between non-fertilized and fertilized eggs of brown planthopper, Nilaparvata lugens Stål. Insects 2020, 11, 49. Insects 2020, 11, 49. [Google Scholar] [CrossRef]
- Nguyen, N.-P.; Warnow, T.; Pop, M.; White, B. A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. NPJ Biofilms Microbiomes 2016, 2, 16004. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv 2016, 081257. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.; Kopylova, E.; Morton, J.; Zech Xu, Z.; Kightley, E.; Thompson, L.; Hyde, E.; Gonzalez, A. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2, R79. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Nearing, J.T.; Douglas, G.M.; Comeau, A.M.; Langille, M.G. Denoising the Denoisers: An independent evaluation of microbiome sequence error-correction approaches. PeerJ 2018, 6, e5364. [Google Scholar] [CrossRef]
- Prodan, A.; Tremaroli, V.; Brolin, H.; Zwinderman, A.H.; Nieuwdorp, M.; Levin, E. Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing. PLoS ONE 2020, 15, e0227434. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhou, C.; Chen, G.; Zhou, Y. Bacterial microbiota in small brown planthopper populations with different rice viruses. J. Basic Microbiol. 2017, 57, 590–596. [Google Scholar] [CrossRef]
- Werren, J.H. Biology of wolbachia. Annu. Rev. Entomol. 1997, 42, 587–609. [Google Scholar] [CrossRef]
- Zug, R.; Hammerstein, P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE 2012, 7, e38544. [Google Scholar] [CrossRef]
- Zhang, K.J.; Han, X.; Hong, X.Y. Various infection status and molecular evidence for horizontal transmission and recombination of Wolbachia and Cardinium among rice planthoppers and related species. Insect Sci. 2013, 20, 329–344. [Google Scholar] [CrossRef]
- Turelli, M.; Cooper, B.S.; Richardson, K.M.; Ginsberg, P.S.; Peckenpaugh, B.; Antelope, C.X.; Kim, K.J.; May, M.R.; Abrieux, A.; Wilson, D.A. Rapid global spread of wRi-like Wolbachia across multiple Drosophila. Curr. Biol. 2018, 28, 963–971.e8. [Google Scholar] [CrossRef]
- Weeks, A.; Breeuwer, J. Wolbachia–induced parthenogenesis in a genus of phytophagous mites. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2001, 268, 2245–2251. [Google Scholar] [CrossRef]
- Zheng, Y.; Ren, P.-P.; Wang, J.-L.; Wang, Y.-F. Wolbachia-induced cytoplasmic incompatibility is associated with decreased Hira expression in male Drosophila. PLoS ONE 2011, 6, e19512. [Google Scholar] [CrossRef][Green Version]
- Beckmann, J.F.; Ronau, J.A.; Hochstrasser, M. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2017, 2, 17007. [Google Scholar] [CrossRef] [PubMed]
- LePage, D.P.; Metcalf, J.A.; Bordenstein, S.R.; On, J.; Perlmutter, J.I.; Shropshire, J.D.; Layton, E.M.; Funkhouser-Jones, L.J.; Beckmann, J.F.; Bordenstein, S.R. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 2017, 543, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Miyata, M.; Konagaya, T.; Yukuhiro, K.; Nomura, M.; Kageyama, D. Wolbachia-induced meiotic drive and feminization is associated with an independent occurrence of selective mitochondrial sweep in a butterfly. Biol. Lett. 2017, 13, 20170153. [Google Scholar] [CrossRef] [PubMed]
- Harumoto, T.; Fukatsu, T.; Lemaitre, B. Common and unique strategies of male killing evolved in two distinct Drosophila symbionts. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172167. [Google Scholar]
- Teixeira, L.; Ferreira, Á.; Ashburner, M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008, 6, e1000002. [Google Scholar] [CrossRef]
- Bian, G.; Xu, Y.; Lu, P.; Xie, Y.; Xi, Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 2010, 6, e1000833. [Google Scholar] [CrossRef]
- Pimentel, A.C.; Cesar, C.S.; Martins, M.; Cogni, R. The antiviral effects of the symbiont bacteria Wolbachia in insects. Front. Immunol. 2021, 11, 626329. [Google Scholar] [CrossRef]
- Kang, L.; Ma, X.; Cai, L.; Liao, S.; Sun, L.; Zhu, H.; Chen, X.; Shen, D.; Zhao, S.; Li, C. Superinfection of Laodelphax striatellus with Wolbachia from Drosophila simulans. Heredity 2003, 90, 71–76. [Google Scholar] [CrossRef]
- Noda, H.; Koizumi, Y.; Zhang, Q.; Deng, K. Infection density of Wolbachia and incompatibility level in two planthopper species, Laodelphax striatellus and Sogatella furcifera. Insect Biochem. Mol. Biol. 2001, 31, 727–737. [Google Scholar] [CrossRef]
- Kawai, S.; Matsumoto, Y.; Gotoh, T.; Noda, H. Transinfection of Wolbachia in planthoppers: Nymphal injection of cultured Wolbachia and infection dynamics. Environ. Entomol. 2009, 38, 1626–1633. [Google Scholar] [CrossRef]
- Breeuwer, J.A. Wolbachia and cytoplasmic incompatibility in the spider mites Tetranychus urticae and T. turkestani. Heredity 1997, 79, 41–47. [Google Scholar] [CrossRef]
- Bourtzis, K.; Dobson, S.L.; Braig, H.R.; O’Neill, S.L. Rescuing Wolbachia have been overlooked. Nature 1998, 391, 852–853. [Google Scholar] [CrossRef]
- Zhang, F.; Guo, H.; Zheng, H.; Zhou, T.; Zhou, Y.; Wang, S.; Fang, R.; Qian, W.; Chen, X. Research article Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV). BMC Genom. 2010, 11, 303. [Google Scholar] [CrossRef]
- Gill, A.C.; Darby, A.C.; Makepeace, B.L. Iron necessity: The secret of Wolbachia’s success? PLoS Neglected Trop. Dis. 2014, 8, e3224. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Vandamme, P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 2003, 5, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Nowak, J.; Coenye, T.; Clément, C.; Ait Barka, E. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 2008, 32, 607–626. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M.; Flórez, L.V. Versatile and dynamic symbioses between insects and Burkholderia bacteria. Annu. Rev. Entomol. 2020, 65, 145–170. [Google Scholar] [CrossRef]
- Viallard, V.; Poirier, I.; Cournoyer, B.; Haurat, J.; Wiebkin, S.; Ophel-Keller, K.; Balandreau, J. Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of [Pseudomonas] phenazinium, [Pseudomonas] pyrrocinia and [Pseudomonas] glathei as Burkholderia. Int. J. Syst. Evol. Microbiol. 1998, 48, 549–563. [Google Scholar] [CrossRef]
- Michalik, K.; Szklarzewicz, T.; Kalandyk-Kołodziejczyk, M.; Jankowska, W.; Michalik, A. Bacteria belonging to the genus Burkholderia are obligatory symbionts of the eriococcids Acanthococcus aceris Signoret, 1875 and Gossyparia spuria (Modeer, 1778) (Insecta, Hemiptera, Coccoidea). Arthropod Struct. Dev. 2016, 45, 265–272. [Google Scholar] [CrossRef]
- Flórez, L.V.; Kaltenpoth, M. Symbiont dynamics and strain diversity in the defensive mutualism between Lagria beetles and Burkholderia. Environ. Microbiol. 2017, 19, 3674–3688. [Google Scholar] [CrossRef]
- Takeshita, K.; Kikuchi, Y. Riptortus pedestris and Burkholderia symbiont: An ideal model system for insect–microbe symbiotic associations. Res. Microbiol. 2017, 168, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Wierz, J.C.; Gaube, P.; Klebsch, D.; Kaltenpoth, M.; Flórez, L.V. Transmission of bacterial symbionts with and without genome erosion between a beetle host and the plant environment. Front. Microbiol. 2021, 12, 715601. [Google Scholar] [CrossRef] [PubMed]
- Kato, C.Y.; Chung, I.H.; Robinson, L.K.; Austin, A.L.; Dasch, G.A.; Massung, R.F. Assessment of real-time PCR assay for detection of Rickettsia spp. and Rickettsia rickettsii in banked clinical samples. J. Clin. Microbiol. 2013, 51, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.K.; Cho, M.S.; Kim, M.H.; Choi, H.J.; Kang, M.J.; Shim, H.S.; Ahn, T.Y.; Kim, J.; Park, D.S. Rapid and specific detection of Burkholderia glumae in rice seed by real-time Bio-PCR using species-specific primers based on an rhs family gene. Plant Dis. 2012, 96, 577–580. [Google Scholar] [CrossRef]
- Lee, B.C.; Hong, Y.K.; Kwak, D.Y.; Oh, B.G.; Park, S.T.; Kim, S.C. Detection of rice stripe virus using RT-PCR. Res. Plant Dis. 2004, 10, 30–33. [Google Scholar] [CrossRef]

| Population | Sample | Total Reads After Trimming | OTUs or ASVs | Total OTUs or ASVs of Population | Shannon Entropy | Phylogenetic Diversity | Simpson’s Index |
|---|---|---|---|---|---|---|---|
| Nonviruliferous | VF-1-OTU | 241,586 | 54 | 61 | 0.62 | 1.59 | 0.15 |
| RSV-viruliferous | RSV-1-OTU | 327,278 | 27 | 0.63 | 1.11 | 0.18 | |
| Nonviruliferous | VF-1-ASV | 241,586 | 24 | 34 | 5.78 | 1.46 | 0.98 |
| RSV-viruliferous | RSV-1-ASV | 327,278 | 17 | 5.72 | 1.08 | 0.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, J.; Kwon, M.; Lee, B.C.; Kil, E.-J. Comparative Endosymbiont Community Structures of Nonviruliferous and Rice Stripe Virus-Viruliferous Laodelphax striatellus (Hemiptera: Delphacidae) in Korea. Viruses 2025, 17, 1074. https://doi.org/10.3390/v17081074
Jeon J, Kwon M, Lee BC, Kil E-J. Comparative Endosymbiont Community Structures of Nonviruliferous and Rice Stripe Virus-Viruliferous Laodelphax striatellus (Hemiptera: Delphacidae) in Korea. Viruses. 2025; 17(8):1074. https://doi.org/10.3390/v17081074
Chicago/Turabian StyleJeon, Jiho, Minhyeok Kwon, Bong Choon Lee, and Eui-Joon Kil. 2025. "Comparative Endosymbiont Community Structures of Nonviruliferous and Rice Stripe Virus-Viruliferous Laodelphax striatellus (Hemiptera: Delphacidae) in Korea" Viruses 17, no. 8: 1074. https://doi.org/10.3390/v17081074
APA StyleJeon, J., Kwon, M., Lee, B. C., & Kil, E.-J. (2025). Comparative Endosymbiont Community Structures of Nonviruliferous and Rice Stripe Virus-Viruliferous Laodelphax striatellus (Hemiptera: Delphacidae) in Korea. Viruses, 17(8), 1074. https://doi.org/10.3390/v17081074







