Virome Survey of Banana Plantations and Surrounding Plants in Malawi
Abstract
1. Introduction
2. Materials and Methods
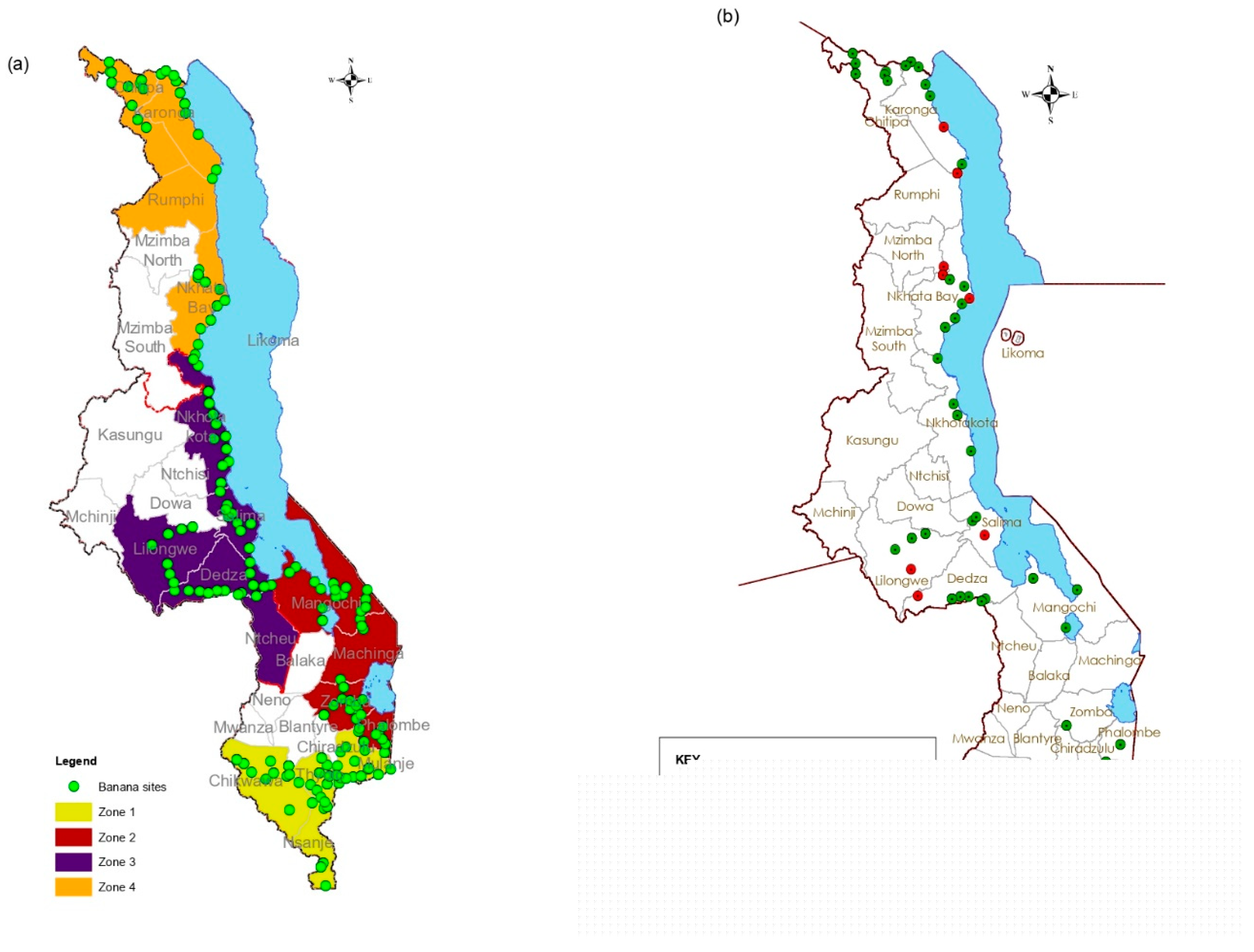
2.1. Plant Sample Collection and Species Identification
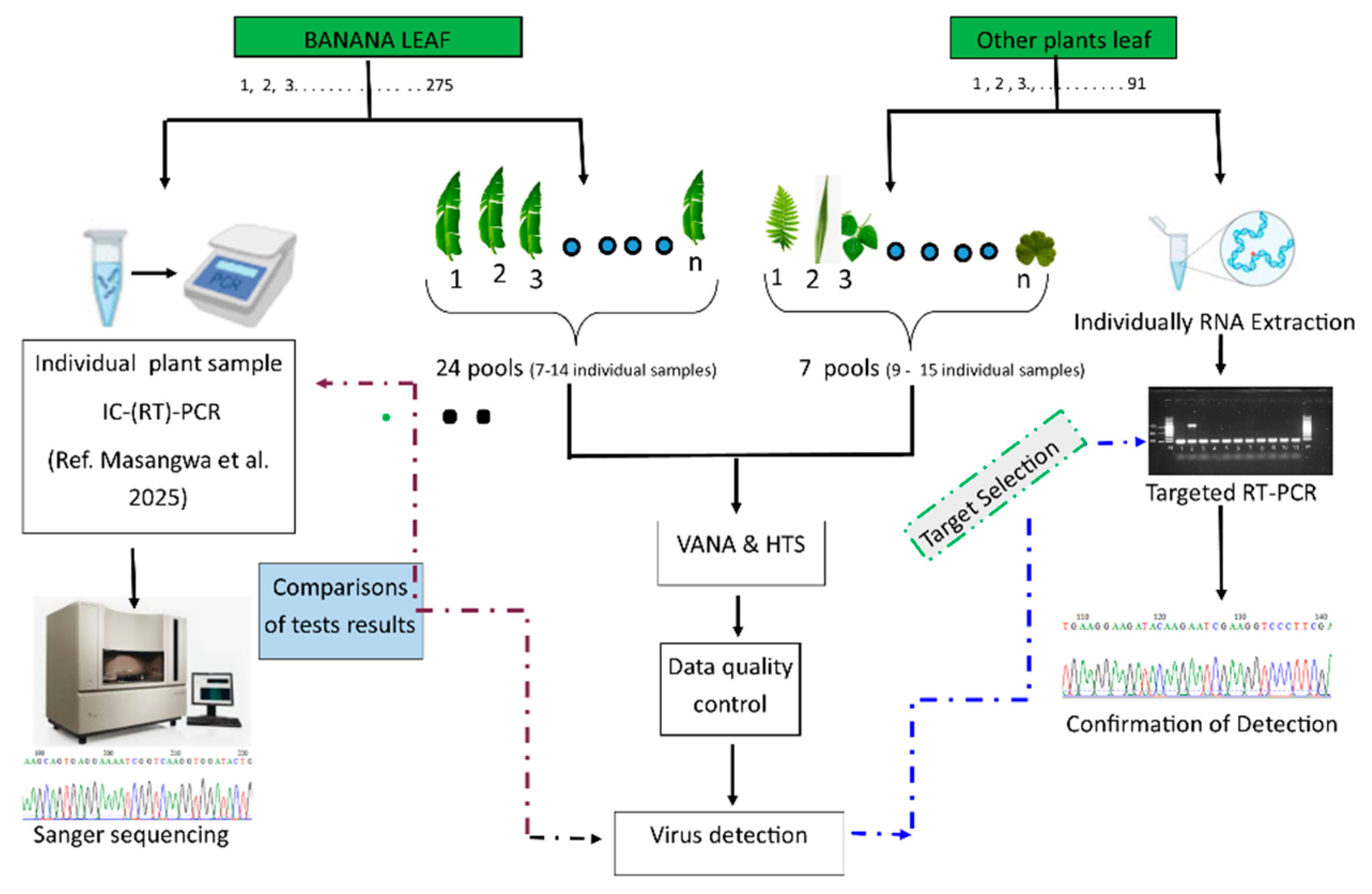
2.2. Sample Poolings Viral Enrichment and High Throughput Sequencing
2.3. Bioinformatics Analyses
2.3.1. HTS Data and Genomic Analyses
2.3.2. Pairwise Genome Comparison and Phylogenetic Analyses
2.4. Confirmation of HTS-Based Virus Detection by (RT)-PCR
2.4.1. Total Nucleic Acid Extraction
2.4.2. Reverse Transcription and PCR
3. Results
3.1. Plant Sample Collection and Identification
3.2. Virome Characterization by VANA-HTS
3.3. Comparison of Banana Virus Detection Between HTS Test on Pooled Samples and PCR-Based Tests on Individual Samples
3.4. New Host Detection
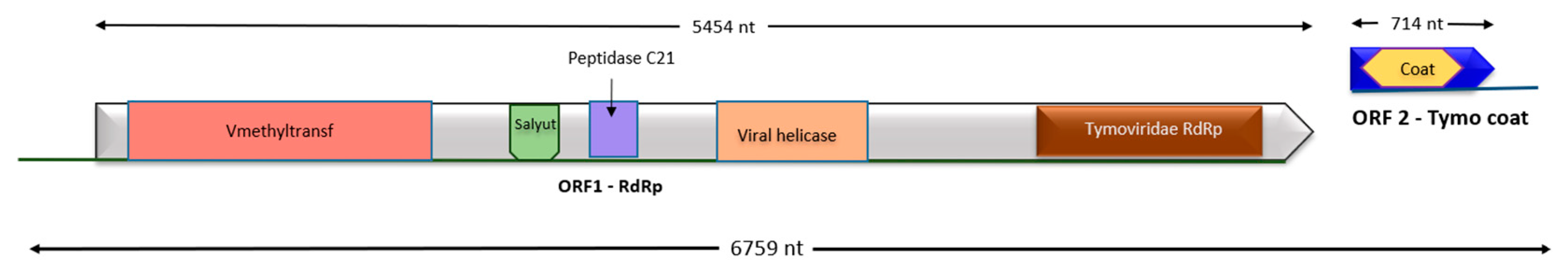
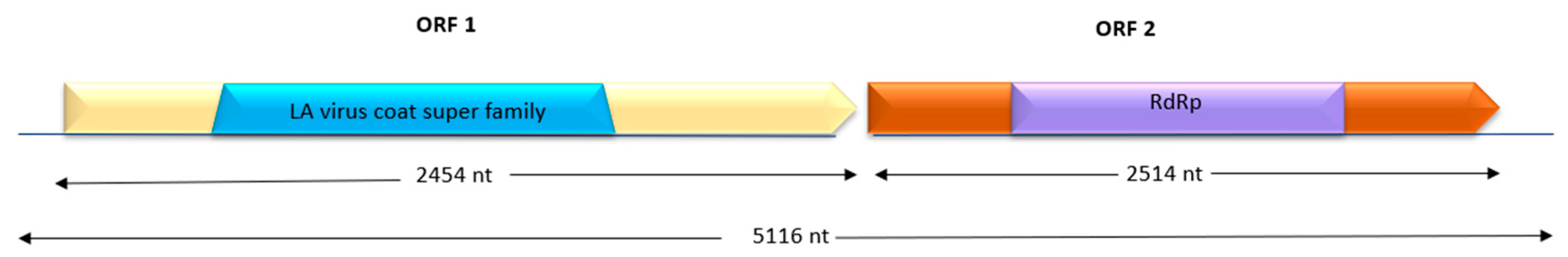
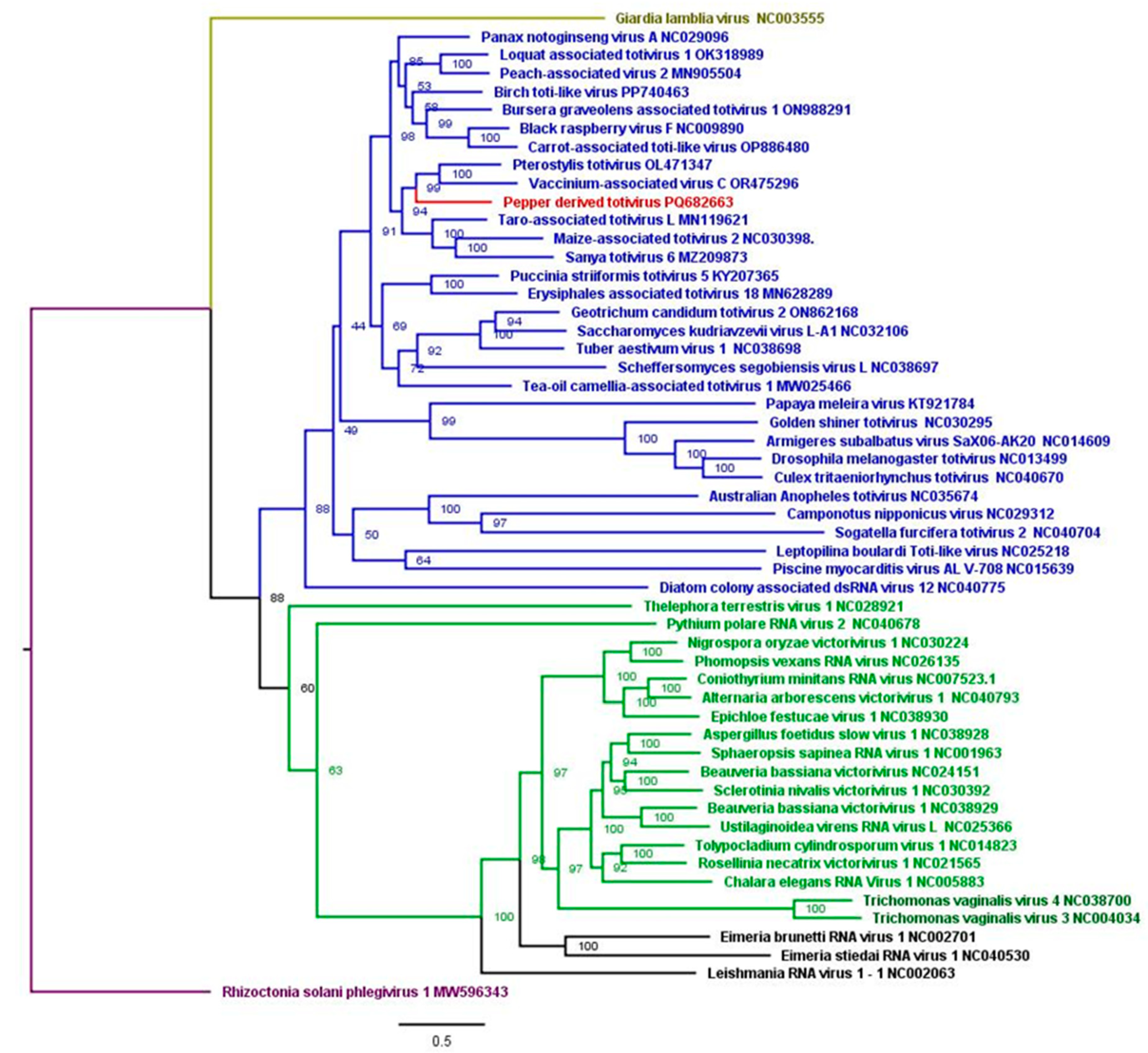
3.5. Identification and Characterization of Two New Viruses
Genome Sequence Reconstruction and Annotation
3.6. Phylogenetic Analyses
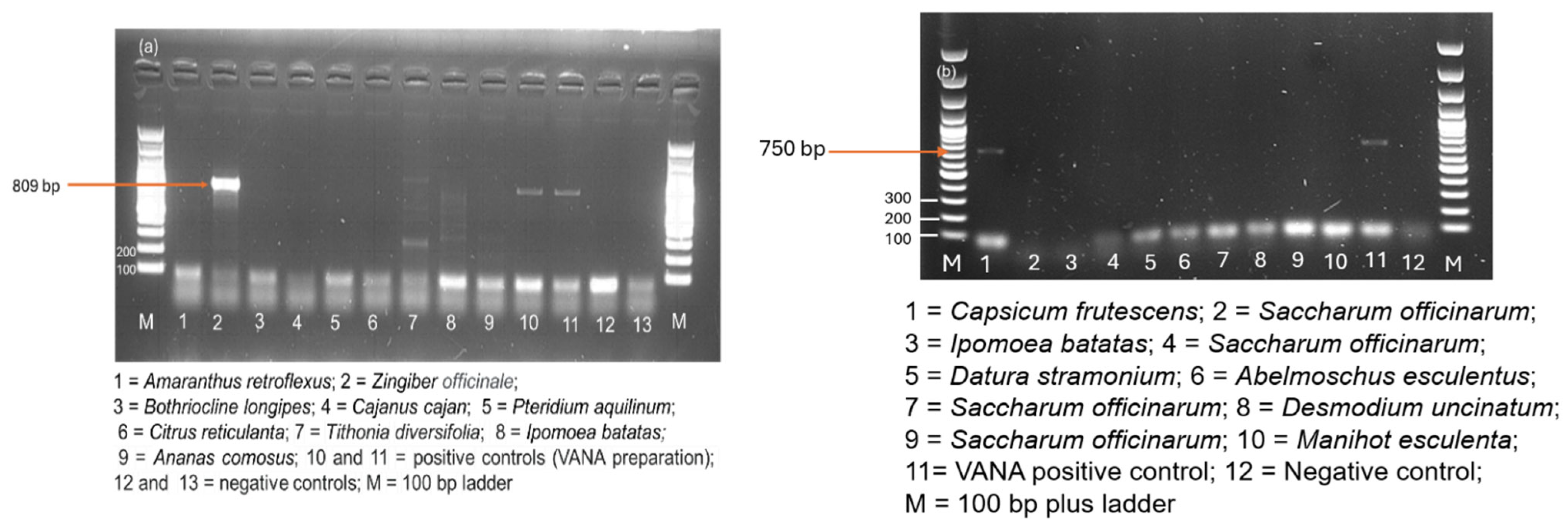
3.7. Confirmation by RT-PCR on Individual Samples
3.7.1. Novel Viruses
3.7.2. New Hosts for Viruses
3.7.3. First Report of Nine Known Viruses in Malawi
4. Discussion
4.1. Plant Sample Collection and Identification
4.2. Sensitivity of HTS Tests on Pooled Samples
4.3. The Results Comparison Between the HTS Test on Pooled Samples and PCR-Based Tests on Individual Samples
4.4. The Absence of Virus Exchange Between Banana and Surrounding Plants Deserve Complementary Surveys
4.5. Novel Hosts for Known Viruses
4.6. A Novel Tymoviridae Species
4.7. A Novel Totiviridae Virus Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soko, M.M. Evaluation of Transgenic RNAi Banana and Plantain Lines for Resistance to Banana Bunchy Top Disease. Ph.D. Thesis, Queensland University of Technology, Brisbane, Australia, 2022. [Google Scholar]
- Omondi, B.A.; Soko, M.M.; Nduwimana, I.; Delano, R.T.; Niyongere, C.; Simbare, A.; Kachigamba, D.; Staver, C. The effectiveness of consistent roguing in managing banana bunchy top disease in smallholder production in Africa. Plant Pathol. 2020, 69, 1754–1766. [Google Scholar] [CrossRef]
- Masangwa, J.I.G.; Pareta, N.F.; Moses, P.; Hřibová, E.; Doležel, J.; Fandika, I.; Massart, S. Surveillance and Molecular Characterization of Banana Viruses and Their Association with Musa Germplasm in Malawi. bioRxiv 2024. [Google Scholar] [CrossRef]
- Ndunguru, J.; Sseruwagi, P.; Tairo, F.; Stomeo, F.; Maina, S.; Djinkeng, A.; Kehoe, M.; Boykin, L.M.; Melcher, U. Analyses of Twelve New Whole Genome Sequences of Cassava Brown Streak Viruses and Ugandan Cassava Brown Streak Viruses from East Africa: Diversity, Supercomputing and Evidence for Further Speciation. PLoS ONE 2015, 10, e0139321. [Google Scholar] [CrossRef]
- Fuentes, S.; Gibbs, A.J.; Adams, I.P.; Hajizadeh, M.; Kreuze, J.; Fox, A.; Blouin, A.G.; Jones, R.A.C. Phylogenetics and Evolution of Potato Virus V: Another Potyvirus that Originated in the Andes. Plant Dis. 2022, 106, 691–700. [Google Scholar] [CrossRef]
- Nyirakanani, C.; Tamisier, L.; Bizimana, J.P.; Rollin, J.; Nduwumuremyi, A.; Bigirimana, V.d.P.; Selmi, I.; Lasois, L.; Vanderschuren, H.; Massart, S. Going beyond consensus genome sequences: An innovative SNP-based methodology reconstructs different Ugandan cassava brown streak virus haplotypes at a nationwide scale in Rwanda. Virus Evol. 2023, 9, vead053. [Google Scholar] [CrossRef]
- Kutnjak, D.; Tamisier, L.; Adams, I.; Boonham, N.; Candresse, T.; Chiumenti, M.; De Jonghe, K.; Kreuze, J.F.; Lefebvre, M.; Silva, G.; et al. A Primer on the Analysis of High-Throughput Sequencing Data for Detection of Plant Viruses. Microorganisms 2021, 9, 841. [Google Scholar] [CrossRef]
- Lebas, B.; Adams, I.; Al Rwahnih, M.; Baeyen, S.; Bilodeau, G.J.; Blouin, A.G.; Boonham, N.; Candresse, T.; Chandelier, A.; De Jonghe, K.; et al. Facilitating the adoption of high-throughput sequencing technologies as a plant pest diagnostic test in laboratories: A step-by-step description. EPPO Bull. 2022, 52, 394–418. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Saha, P.; Wiley, G.B.; Quan, J.; White, J.D.; Lai, H.; Chavarría, F.; Shen, G.; Roe, B.A. Ecogenomics: Using massively parallel pyrosequencing to understand virus ecology. Mol. Ecol. 2010, 19, 81–88. [Google Scholar] [CrossRef]
- Maclot, F.; Debue, V.; Malmstrom, C.M.; Filloux, D.; Roumagnac, P.; Eck, M.; Tamisier, L.; Blouin, A.G.; Candresse, T.; Massart, S.; et al. Long-Term Anthropogenic Management and Associated Loss of Plant Diversity Deeply Impact Virome Richness and Composition of Poaceae Communities. Microbiol. Spectr. 2023, 11, e0485022. [Google Scholar] [CrossRef]
- Massart, S.; Olmos, A.; Jijakli, H.; Candresse, T. Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 2014, 188, 90–96. [Google Scholar] [CrossRef]
- Massart, S.; Chiumenti, M.; De Jonghe, K.; Glover, R.; Haegeman, A.; Koloniuk, I.; Komínek, P.; Kreuze, J.; Kutnjak, D.; Lotos, L.; et al. Virus Detection by High-Throughput Sequencing of Small RNAs: Large-Scale Performance Testing of Sequence Analysis Strategies. Phytopathology 2019, 109, 488–497. [Google Scholar] [CrossRef]
- Moubset, O.; François, S.; Maclot, F.; Palanga, E.; Julian, C.; Claude, L.; Fernandez, E.; Rott, P.C.E.; Daugrois, J.-H.; Antoine-Lorquin, A.; et al. Virion-Associated Nucleic Acid-Based Metagenomics: A Decade of Advances in Molecular Characterization of Plant Viruses. Phytopathology 2022, 112, 2253–2272. [Google Scholar] [CrossRef]
- Schönegger, D.; Moubset, O.; Margaria, P.; Menzel, W.; Winter, S.; Roumagnac, P.; Marais, A.; Candresse, T.; Miller, W.A. Benchmarking of virome metagenomic analysis approaches using a large, 60+ members, viral synthetic community. J. Virol. 2023, 97, e0130023. [Google Scholar] [CrossRef]
- Kil, E.-J.; Byun, H.-S.; Kim, S.; Cho, S.; Cho, S.; Roh, K.; Lee, K.-Y.; Choi, H.-S.; Kim, C.-S.; Lee, S. Tomato yellow leaf curl virus Can Overwinter in Stellaria aquatica, a Winter-Hardy TYLCV-Reservoir Weed. Plant Dis. 2015, 99, 588–592. [Google Scholar] [CrossRef]
- Ma, Y.; Marais, A.; Lefebvre, M.; Faure, C.; Candresse, T. Metagenomic analysis of virome cross-talk between cultivated Solanum lycopersicum and wild Solanum nigrum. Virology 2020, 540, 38–44. [Google Scholar] [CrossRef]
- Shi, X.-B.; Zhang, Z.; Li, F.; Preisser, E.L.; Huang, L.-P.; Zhang, D.-Y.; Zhang, Z.-H.; Zhang, S.-B.; Zhou, X.-G.; Zhang, A.-S.; et al. Vector-mediated viral exchange between crops and weedy plants. J. Pest Sci. 2023, 97, 155–171. [Google Scholar] [CrossRef]
- Maachi, A.; Donaire, L.; Hernando, Y.; Aranda, M.A.; Simon, A.E. Genetic Differentiation and Migration Fluxes of Viruses from Melon Crops and Crop Edge Weeds. J. Virol. 2022, 96, e0042122. [Google Scholar] [CrossRef]
- Orfanidou, C.; Dimitriou, C.; Papayiannis, L.; Maliogka, V.; Katis, N. Epidemiology and genetic diversity of criniviruses associated with tomato yellows disease in Greece. Virus Res. 2013, 186, 120–129. [Google Scholar] [CrossRef]
- Guyot, V.; Ly, N.-S.; Trieu, T.-D.; Insisiengmay, O.; Zhang, T.; Iskra-Caruana, M.-L.; BforBB Consortium; Pooggin, M.M. Evidence for Dicot Plants as Alternative Hosts of Banana Bunchy Top Virus and Its Alphasatellites in South-East Asia. Pathogens 2023, 12, 1289. [Google Scholar] [CrossRef]
- Hamim, I.; Green, J.C.; Borth, W.B.; Melzer, M.J.; Wang, Y.N.; Hu, J.S. First Report of Banana bunchy top virus in Heliconia spp. on Hawaii. Plant Dis. 2017, 101, 2153. [Google Scholar] [CrossRef]
- Siljo, A.; Bhat, A.I.; Biju, C.N. Detection of Cardamom mosaic virus and Banana bract mosaic virus in cardamom using SYBR Green based reverse transcription-quantitative PCR. VirusDisease 2013, 25, 137–141. [Google Scholar] [CrossRef]
- Diouf, M.B.; Guyader, S.; Nopoly, M.-M.; Gaspard, O.; Filloux, D.; Candresse, T.; Marais, A.; Teycheney, P.-Y.; Umber, M. Molecular diversity of yam virus Y and identification of banana mild mosaic virus isolates infecting yam (Dioscorea spp.). Arch. Virol. 2023, 168, 180. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.; Lee, H.-J.; Jeong, R.-D.; Park, M.-R. First report of cactus virus X infecting banana (Musa spp.) in Korea. J. Plant Pathol. 2022, 104, 1137. [Google Scholar] [CrossRef]
- Maclot, F.J.; Debue, V.; Blouin, A.G.; Pareta, N.F.; Tamisier, L.; Filloux, D.; Massart, S. Identification, molecular and biological characterization of two novel secovirids in wild grass species in Belgium. Virus Res. 2021, 298, 198397. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Marais, A.; Lefebvre, M.; Theil, S.; Svanella-Dumas, L.; Faure, C.; Candresse, T.; Simon, A.E. Phytovirome Analysis of Wild Plant Populations: Comparison of Double-Stranded RNA and Virion-Associated Nucleic Acid Metagenomic Approaches. J. Virol. 2019, 94, e01462-19. [Google Scholar] [CrossRef] [PubMed]
- Candresse, T.; Faure, C.; Marais, A. First Report of Grapevine Red Globe Virus (GRGV) and Grapevine Rupestris Vein Feathering Virus (GRVFV) Infecting Grapevine (Vitis vinifera) in Portugal. Plant Dis. 2023, 107, 974. [Google Scholar] [CrossRef]
- Temple-Boyer-Dury, C. Exploring the Risks Associated with Tomato Viruses in Belgian Diversified Production Systems and Biological Characterization of Physostegia Chlorotic Mottle Virus. Ph.D. Thesis, University of Liege, Liège, Belgium, 2023. [Google Scholar]
- Massart, S.; Adams, I.; Al Rwahnih, M.; Baeyen, S.; Bilodeau, G.J.; Blouin, A.G.; Boonham, N.; Candresse, T.; Chandellier, A.; De Jonghe, K.; et al. Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pests. Peer Community J. 2022, 2, e62. [Google Scholar] [CrossRef]
- Kechin, A.; Boyarskikh, U.; Kel, A.; Filipenko, M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 2017, 24, 1138–1143. [Google Scholar] [CrossRef]
- Sukhorukov, G.; Khalili, M.; Gascuel, O.; Candresse, T.; Marais-Colombel, A.; Nikolski, M. VirHunter: A Deep Learning-Based Method for Detection of Novel RNA Viruses in Plant Sequencing Data. Front. Bioinform. 2022, 2, 867111. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Pareta, N.F.; Khalili, M.; Maachi, A.; Rivarez, M.P.S.; Rollin, J.; Salavert, F.; Temple, C.; Aranda, M.A.; Boonham, N.; Botermans, M.; et al. Managing the deluge of newly discovered plant viruses and viroids: An optimized scientific and regulatory framework for their characterization and risk analysis. Front. Microbiol. 2023, 14, 1181562. [Google Scholar] [CrossRef] [PubMed]
- Murigi, M.; Ngui, D.; Ogada, M.J. Impact of smallholder banana contract farming on farm productivity and income in Kenya. Cogent Econ. Financ. 2024, 12, 2364353. [Google Scholar] [CrossRef]
- Feyso, A.; Mensa, A.; Moral, M.T. Smallholder banana—Based farming system dynamics of Arba Minch Zuria District, the case of Gamo Zone, Ethiopia: Qualitative exploration. Cogent Food Agric. 2021, 7, 1930425. [Google Scholar] [CrossRef]
- Changadeya, W.; Ambali, A.J.D.; Kambewa, D. Farmers’ adoption potential of improved banana production techniques in Malawi. Int. J. Phys. Soc. Sci. 2012, 2, 32–48. [Google Scholar]
- Ouma, G. Intercropping and its application to banana production in East Africa. J. Plant Breed. Crop Sci. 2009, 1, 13–15. [Google Scholar]
- Blomme, G.; Ntamwira, J.; Kearsley, E.; Bahati, L.; Amini, D.; Safari, N.; Ocimati, W. Sensitivity and Tolerance of Different Annual Crops to Different Levels of Banana Shade and Dry Season Weather. Front. Sustain. Food Syst. 2020, 4, 545926. [Google Scholar] [CrossRef]
- Van Asten, P.J.; Wairegi, L.W.I.; Mukasa, D.; Uringi, N.O. Agronomic and economic benefits of coffee–banana intercropping in Uganda’s smallholder farming systems. Agric. Syst. 2011, 104, 326–334. [Google Scholar] [CrossRef]
- Ntamwira, J.; Pypers, P.; Van Asten, P.J.A.; Vanlauwe, B.; Ruhigwa, B.; Lepoint, P.; Dheda, B.; Monde, T.; Kamira, M.; Blomme, G. Effect of banana leaf pruning on banana and legume yield under intercropping in farmers fields in eastern Democratic Republic of Congo. J. Hortic. For. 2014, 6, 72–80. [Google Scholar] [CrossRef]
- Hanafi, M.; Rong, W.; Tamisier, L.; Berhal, C.; Roux, N.; Massart, S. Detection of Banana Mild Mosaic Virus in Musa In Vitro Plants: High-Throughput Sequencing Presents Higher Diagnostic Sensitivity Than (IC)-RT-PCR and Identifies a New Betaflexiviridae Species. Plants 2022, 11, 226. [Google Scholar] [CrossRef]
- Rong, W.; Rollin, J.; Hanafi, M.; Roux, N.; Massart, S. Validation of High-Throughput Sequencing as Virus Indexing Test for Musa Germplasm: Performance Criteria Evaluation and Contamination Monitoring Using an Alien Control. Phytofrontiers™ 2023, 3, 91–102. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef]
- Haible, D.; Kober, S.; Jeske, H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef]
- Filloux, D.; Dallot, S.; Delaunay, A.; Galzi, S.; Jacquot, E.; Roumagnac, P. Metagenomics Approaches Based on Virion-Associated Nucleic Acids (VANA): An Innovative Tool for Assessing Without A Priori Viral Diversity of Plants. In Plant Pathology; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2015; Volume 1302, pp. 249–257. [Google Scholar]
- Handayani, R.; Fans, K.; Mastuti, T.S.; Rosa, D. Comparison study of antioxidant activity from three banana leaves extracts. J. Teknol. Dan Ind. Pangan 2021, 32, 92–97. [Google Scholar] [CrossRef]
- Pinili, M.S.; Nagashima, I.; Dizon, T.O.; Natsuaki, K.T. Cross-transmission and new alternate hosts of banana bunchy top virus. Trop. Agric. Dev. 2013, 57, 1–7. [Google Scholar]
- Vikrant; Singh, J.; Majumder, S. First report of banana bunchy top virus infecting Colocasia esculenta (L.) Schott, from India. J. Plant Pathol. 2024, 106, 757. [Google Scholar] [CrossRef]
- Bernardo, P.; Golden, M.; Akram, M.; Naimuddin; Nadarajan, N.; Fernandez, E.; Granier, M.; Rebelo, A.G.; Peterschmitt, M.; Martin, D.P.; et al. Identification and characterisation of a highly divergent geminivirus: Evolutionary and taxonomic implications. Virus Res. 2013, 177, 35–45. [Google Scholar] [CrossRef]
- Rao, X.-Q.; Wu, Z.-L.; Wang, W.; Zhou, L.; Sun, J.; Li, H.-P. Genetic diversity analysis reveals new badnaviruses infecting banana in South China. J. Plant Pathol. 2020, 102, 1065–1075. [Google Scholar] [CrossRef]
- CABI. Potato Virus Y (Potato Mottle). CABI Compend. 2021, 43762. [Google Scholar] [CrossRef]
- Chatzivassiliou, E.K. An Annotated List of Legume-Infecting Viruses in the Light of Metagenomics. Plants 2021, 10, 1413. [Google Scholar] [CrossRef]
- Laiton-Donato, K.; Guzmán, C.; Perdomo-Balaguera, E.; Sarmiento, L.; Torres-Fernandez, O.; Ruiz, H.A.; Rosales-Munar, A.; Peláez-Carvajal, D.; Navas, M.-C.; Wong, M.C.; et al. Novel Putative Tymoviridae-like Virus Isolated from Culex Mosquitoes in Colombia. Viruses 2023, 15, 953. [Google Scholar] [CrossRef] [PubMed]
- Martelli, G.P.; Sabanadzovic, S.; Ghanem-Sabanadzovic, N.A.; Saldarelli, P. Maculavirus, a new genus of plant viruses. Arch. Virol. 2002, 147, 1847–1853. [Google Scholar] [CrossRef]
- Katsuma, S.; Tanaka, S.; Omuro, N.; Takabuchi, L.; Daimon, T.; Imanishi, S.; Yamashita, S.; Iwanaga, M.; Mita, K.; Maeda, S.; et al. Novel Macula-Like Virus Identified in Bombyx mori Cultured Cells. J. Virol. 2005, 79, 5577–5584. [Google Scholar] [CrossRef]
- Charles, J.; Tangudu, C.S.; Hurt, S.L.; Tumescheit, C.; Firth, A.E.; Garcia-Rejon, J.E.; Machain-Williams, C.; Blitvich, B.J. Discovery of a novel Tymoviridae-like virus in mosquitoes from Mexico. Arch. Virol. 2018, 164, 649–652. [Google Scholar] [CrossRef]
- Gasparro, M.; Caputo, A.R.; Forleo, L.R.; Perniola, R.; Alba, V.; Milella, R.A.; Antonacci, D.; Aurand, J.-M. Study of main grapevine viruses transmission in breeding programs. BIO Web Conf. 2016, 7, 01039. [Google Scholar] [CrossRef]
- Engelbrecht, D.J.; Kasdorf, G.G.F. Field spread of corky bark, fleck, leafroll and Shiraz decline diseases and associated viruses in South African grapevines. Phytophylactica 1990, 22, 347–354. [Google Scholar]
- Woodham, R.C.; Krake, I.R. Investigations on Transmission of Grapevine Leafroll, Yellow Speckle and Fleck Diseases by Dodder. J. Phytopathol. 1983, 106, 193–198. [Google Scholar] [CrossRef]
- Khalifa, M.E.; MacDiarmid, R.M. Molecular Characterization of Two Totiviruses from the Commensal Yeast Geotrichum candidum. Viruses 2023, 15, 2150. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masangwa, J.I.G.; Temple, C.; Rollin, J.; Maclot, F.; Önder, S.; Kamwendo, J.; Mwafongo, E.; Moses, P.; Fandika, I.; Massart, S. Virome Survey of Banana Plantations and Surrounding Plants in Malawi. Viruses 2025, 17, 1068. https://doi.org/10.3390/v17081068
Masangwa JIG, Temple C, Rollin J, Maclot F, Önder S, Kamwendo J, Mwafongo E, Moses P, Fandika I, Massart S. Virome Survey of Banana Plantations and Surrounding Plants in Malawi. Viruses. 2025; 17(8):1068. https://doi.org/10.3390/v17081068
Chicago/Turabian StyleMasangwa, Johnny Isaac Gregorio, Coline Temple, Johan Rollin, François Maclot, Serkan Önder, Jamestone Kamwendo, Elizabeth Mwafongo, Philemon Moses, Isaac Fandika, and Sebastien Massart. 2025. "Virome Survey of Banana Plantations and Surrounding Plants in Malawi" Viruses 17, no. 8: 1068. https://doi.org/10.3390/v17081068
APA StyleMasangwa, J. I. G., Temple, C., Rollin, J., Maclot, F., Önder, S., Kamwendo, J., Mwafongo, E., Moses, P., Fandika, I., & Massart, S. (2025). Virome Survey of Banana Plantations and Surrounding Plants in Malawi. Viruses, 17(8), 1068. https://doi.org/10.3390/v17081068








