Deciphering Cowpea Resistance to Potyvirus: Assessment of eIF4E Gene Mutations and Their Impact on the eIF4E-VPg Protein Interaction
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of the Coding Sequence of eIF4E Genes and VPg Coding Sequence
2.2. Confirmation of Mutations in the eIF4E Gene in Cowpea
2.3. Analysis of eIF4E Gene Mutations and Assessment of Susceptibility/Resistance to CABMV in Cowpea Cultivars
2.4. Primary Sequences, Alignments, and Conserved Domain of eIF4E Coding Region
2.5. Molecular Modeling, Model Validation, and Molecular Dynamics Simulations
2.6. Molecular Docking and Binding Energy Between eIF4E-VPg Complexes
3. Results
3.1. Mutations in the eIF4E Gene of Vigna Unguiculata
3.2. Susceptibility/Resistance to CABMV Based on Mutations in the eIF4E Gene
3.3. Bioassay of Cowpea Cultivars Inoculated with Potyvirus CABMV
3.4. Genetic Variation in the eIF4E CDS Among 27 Cowpea Cultivars
3.5. Alignment and Conserved Domain of eIF4E and VPg Proteins
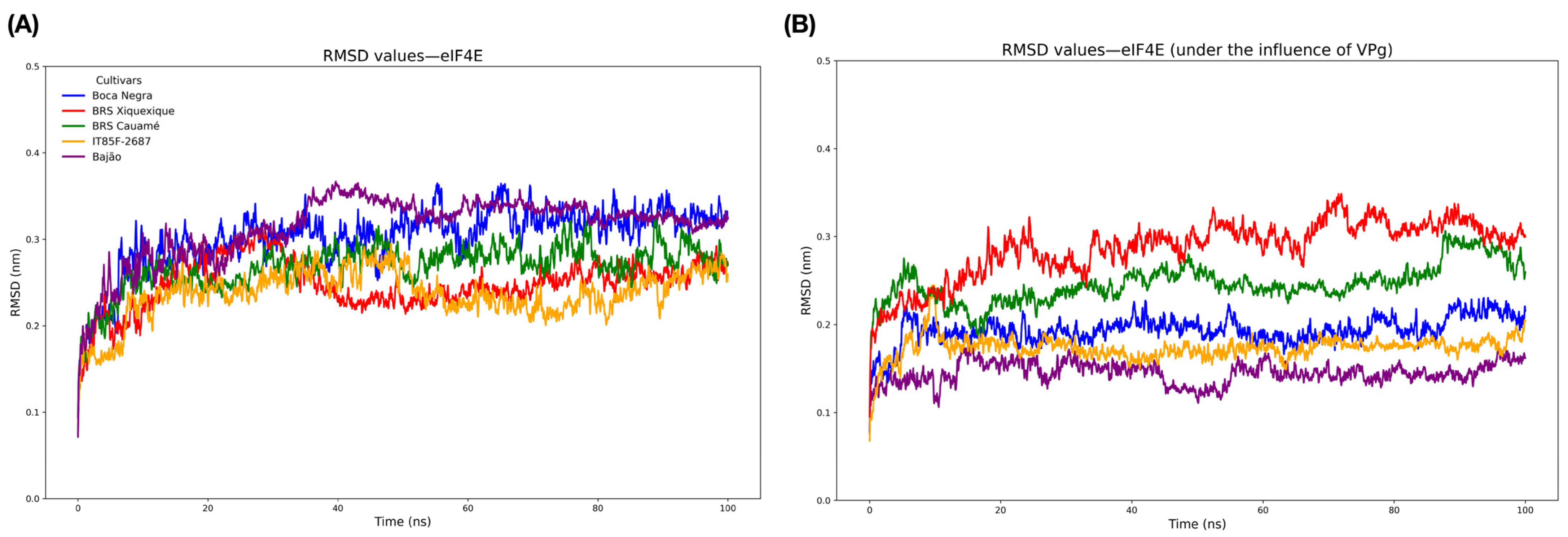
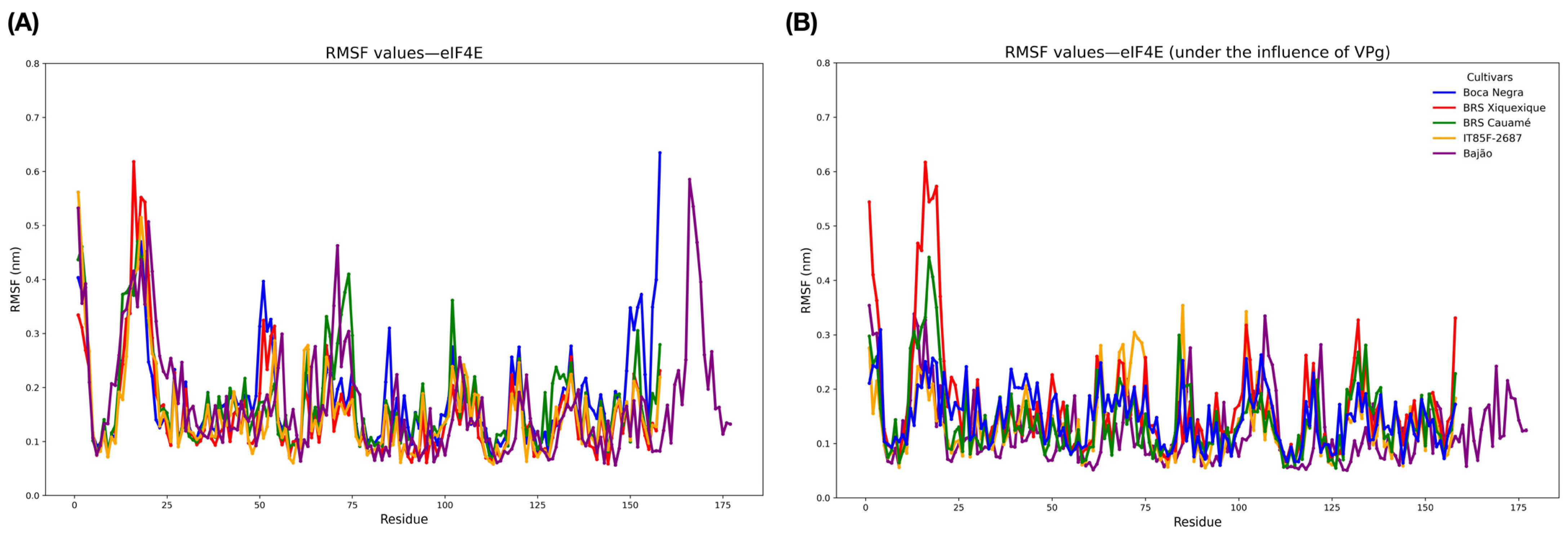
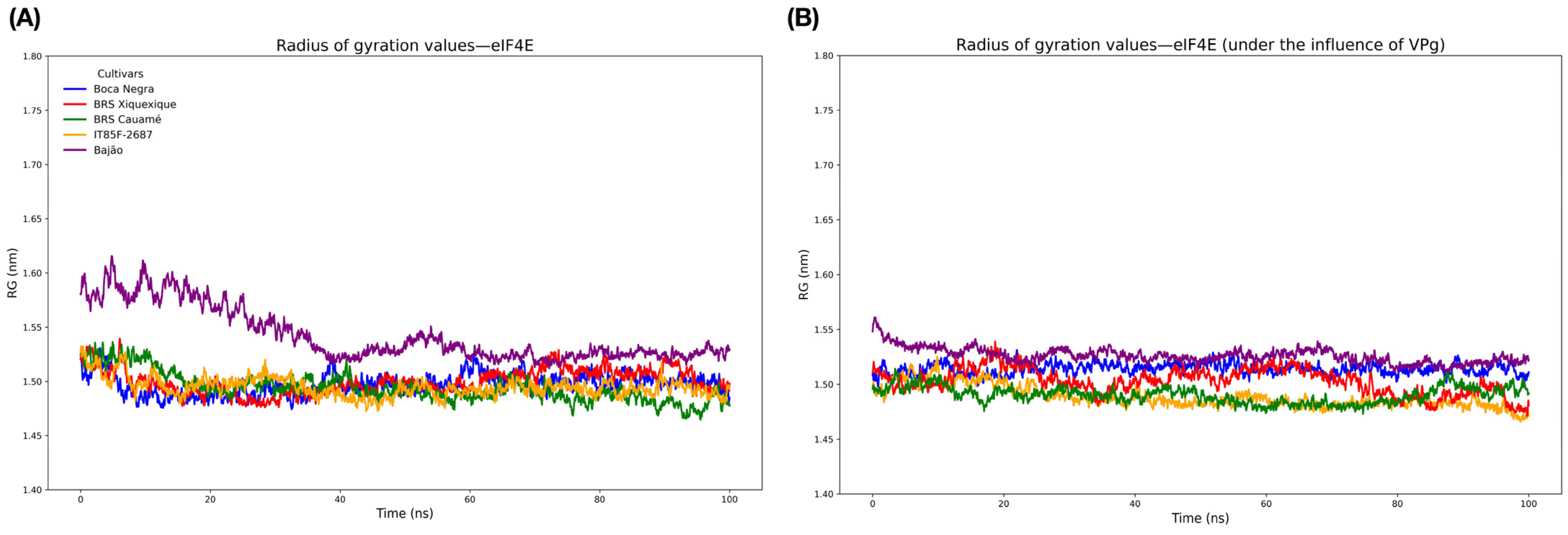
3.6. Molecular Modeling and Structural Validation of eIF4E and VPg Models
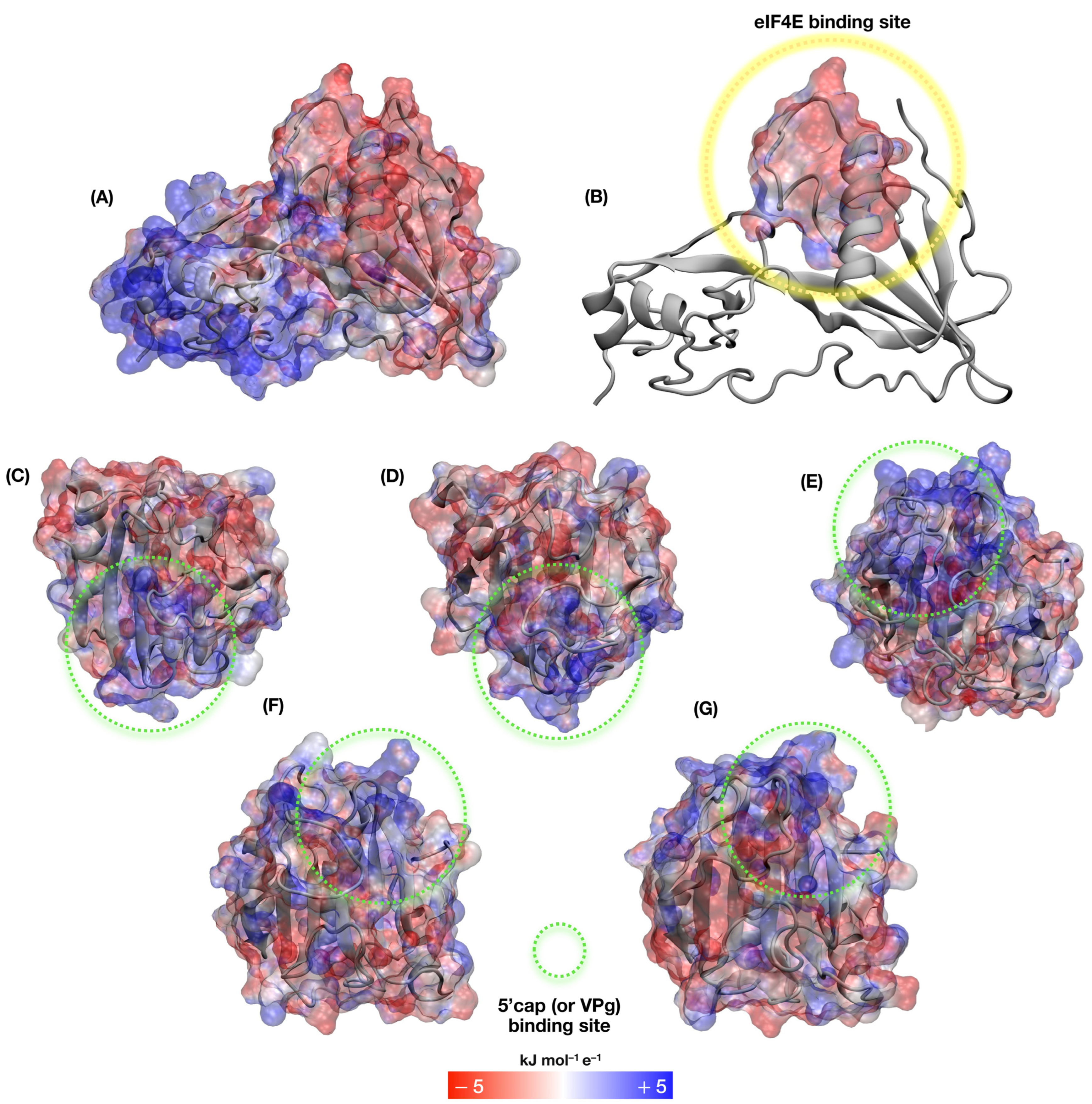
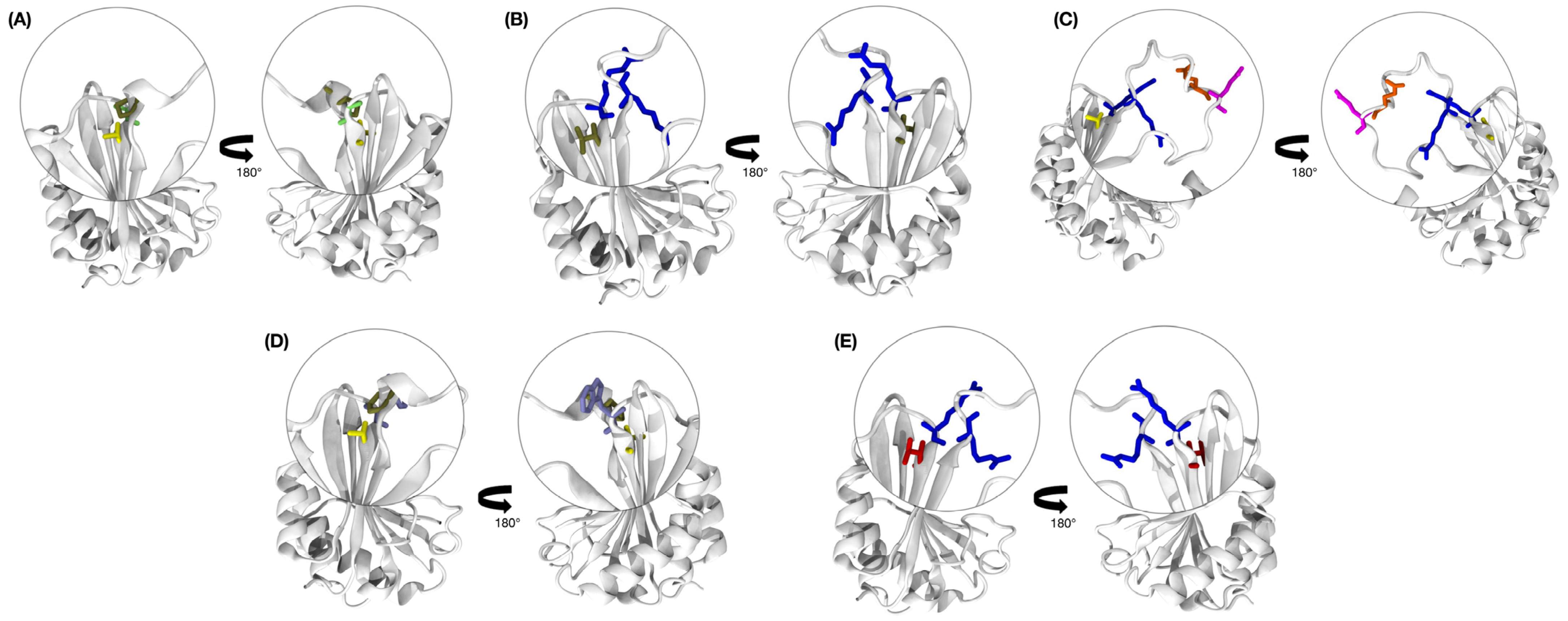
3.7. Molecular Docking, Interface Analysis, and Binding Energy
4. Discussion
5. Conclusions
6. Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| +ssRNA | Positive-sense single-stranded RNA |
| A | Adenine |
| Ala | Alanine |
| Asn | Asparagine |
| Asp | Aspartate |
| C | Cytosine |
| CABMV | Cowpea aphid-borne mosaic virus |
| cDNA | Complementary DNA |
| CDS | Coding sequence |
| CPSMV | Cowpea severe mosaic virus |
| DAIs | Days after inoculation |
| DDG | Delta Delta G |
| eIF4E | Eukaryotic translation initiation factor 4E |
| F | Forward |
| G | Guanine |
| Gln | Glutamine |
| Glu | Glutamate |
| Gly | Glycine |
| HBs | Hydrogen bonds |
| m7GpppN | 7-Methylguanosine |
| MD | Molecular dynamics |
| mRNA | Messenger RNA |
| NMR | Nuclear magnetic resonance |
| PAE | Predicted Aligned Error |
| PDB | Protein Data Bank |
| pLDDT | Predicted Local Distance Difference Test |
| Poly-A | Polyadenylated tail |
| Pro | Proline |
| PSE | Electrostatic Surface Potential |
| PVMV | Pepper veinal mottle virus |
| PVY | Potato virus Y |
| R | Reverse |
| R genes | Resistance genes |
| REU | Rosetta Energy Unit |
| RG | Radius of gyration |
| RMSD | Root Mean Square Deviation |
| RMSF | Root Mean Square Fluctuation |
| S genes | Susceptibility genes |
| SNPs | Single-nucleotide polymorphisms |
| T | Thymine |
| Trp | Tryptophan |
| Tyr | Tyrosine |
| UMP | Uridine monophosphate |
| Val | Valine |
| VPg | Viral protein genome-linked |
References
- Angira, B.; Zhang, Y.; Scheuring, C.F.; Zhang, Y.; Masor, L.; Coleman, J.R.; Liu, Y.-H.; Singh, B.B.; Zhang, H.-B.; Hays, D.B.; et al. Construction of a Single Nucleotide Polymorphism Linkage Map and Identification of Quantitative Trait Loci Controlling Heat Tolerance in Cowpea, Vigna unguiculata (L.) Walp. Mol. Genet. Genom. 2022, 297, 1481–1493. [Google Scholar] [CrossRef]
- Alemu, M.; Asfaw, Z.; Woldu, Z.; Fenta, B.A.; Medvecky, B. Cowpea (Vigna unguiculata (L.) Walp.) (Fabaceae) Landrace Diversity in Northern Ethiopia. Int. J. Biodivers. Conserv. 2016, 8, 297–309. [Google Scholar] [CrossRef]
- Boukar, O.; Togola, A.; Chamarthi, S.; Belko, N.; Ishikawa, H.; Suzuki, K.; Fatokun, C. Cowpea [Vigna unguiculata (L.) Walp.] Breeding. In Advances in Plant Breeding Strategies: Legumes; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 201–243. ISBN 978-3-030-23399-0. [Google Scholar]
- Kapravelou, G.; Martínez, R.; Martino, J.; Porres, J.M.; Fernández-Fígares, I. Natural Fermentation of Cowpea (Vigna unguiculata) Flour Improves the Nutritive Utilization of Indispensable Amino Acids and Phosphorus by Growing Rats. Nutrients 2020, 12, 2186. [Google Scholar] [CrossRef]
- Herniter, I.A.; Muñoz-Amatriaín, M.; Close, T.J. Genetic, Textual, and Archeological Evidence of the Historical Global Spread of Cowpea ([L.] Walp.). Legume Sci. 2020, 2, e57. [Google Scholar] [CrossRef]
- Amorim, L.L.B.; Ferreira-Neto, J.R.C.; Bezerra-Neto, J.P.; Pandolfi, V.; de Araújo, F.T.; da Silva Matos, M.K.; Santos, M.G.; Kido, E.A.; Benko-Iseppon, A.M. Cowpea and Abiotic Stresses: Identification of Reference Genes for Transcriptional Profiling by qPCR. Plant Methods 2018, 14, 88. [Google Scholar] [CrossRef]
- Borges-Martins, A.N.C.; Ferreira-Neto, J.R.C.; Silva, M.D.d.; Morais, D.A.d.L.; Pandolfi, V.; Silva, R.L.d.O.; Melo, A.L.T.M.d.; da Costa, A.F.; Benko-Iseppon, A.M. Unlocking Cowpea’s Defense Responses: Conserved Transcriptional Signatures in the Battle against CABMV and CPSMV Viruses. Life 2023, 13, 1747. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.R.R.; Aragão, F.J.L. RNAi-Based Enhanced Resistance to Cowpea severe mosaic virus and Cowpea aphid-borne mosaic virus in transgenic cowpea. Plant Pathol. 2014, 63, 831–837. [Google Scholar] [CrossRef]
- Tavert-Roudet, G.; Anne, A.; Barra, A.; Chovin, A.; Demaille, C.; Michon, T. The Potyvirus Particle Recruits the Plant Translation Initiation Factor eIF4E by Means of the VPg Covalently Linked to the Viral RNA. Mol. Plant-Microbe Interact. 2017, 30, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Mäkinen, K. Plant Susceptibility Genes as a Source for Potyvirus Resistance. Ann. Appl. Biol. 2020, 176, 122–129. [Google Scholar] [CrossRef]
- Walter, J.; Charon, J.; Hu, Y.; Lachat, J.; Leger, T.; Lafforgue, G.; Barra, A.; Michon, T. Comparative analysis of mutational robustness of the intrinsically disordered viral protein VPg and of its interactor eIF4E. PLoS ONE 2019, 14, e0211725. [Google Scholar] [CrossRef]
- Wang, A.; Krishnaswamy, S. Eukaryotic Translation Initiation Factor 4E-mediated Recessive Resistance to Plant Viruses and Its Utility in Crop Improvement. Mol. Plant Pathol. 2012, 13, 795–803. [Google Scholar] [CrossRef]
- Coutinho De Oliveira, L.; Volpon, L.; Rahardjo, A.K.; Osborne, M.J.; Culjkovic-Kraljacic, B.; Trahan, C.; Oeffinger, M.; Kwok, B.H.; Borden, K.L.B. Structural Studies of the eIF4E–VPg Complex Reveal a Direct Competition for Capped RNA: Implications for Translation. Proc. Natl. Acad. Sci. USA 2019, 116, 24056–24065. [Google Scholar] [CrossRef]
- Sun, Y.; Guo, Y.; Lou, Z. Formation and Working Mechanism of the Picornavirus VPg Uridylylation Complex. Curr. Opin. Virol. 2014, 9, 24–30. [Google Scholar] [CrossRef]
- Rantalainen, K.I.; Eskelin, K.; Tompa, P.; Mäkinen, K. Structural Flexibility Allows the Functional Diversity of Potyvirus Genome-Linked Protein VPg. J. Virol. 2011, 85, 2449–2457. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Shi, Y.; Dai, Z.; Wang, A. The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection. Viruses 2020, 12, 77. [Google Scholar] [CrossRef]
- Zlobin, N.; Taranov, V. Plant eIF4E Isoforms as Factors of Susceptibility and Resistance to Potyviruses. Front. Plant Sci. 2023, 14, 1041868. [Google Scholar] [CrossRef]
- Garcia-Ruiz, H.; Szurek, B.; Van den Ackerveken, G. Stop Helping Pathogens: Engineering Plant Susceptibility Genes for Durable Resistance. Curr. Opin. Biotechnol. 2021, 70, 187–195. [Google Scholar] [CrossRef]
- Van Schie, C.C.N.; Takken, F.L.W. Susceptibility Genes 101: How to Be a Good Host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef]
- Gallois, J.-L.; Moury, B.; German-Retana, S. Role of the Genetic Background in Resistance to Plant Viruses. Int. J. Mol. Sci. 2018, 19, 2856. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Aranda, M.A. Recessive Resistance to Plant Viruses. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2009; Volume 75, pp. 119–231. ISBN 978-0-12-381397-8. [Google Scholar]
- Sotomayor-Vivas, C.; Hernández-Lemus, E.; Dorantes-Gilardi, R. Linking Protein Structural and Functional Change to Mutation Using Amino Acid Networks. PLoS ONE 2022, 17, e0261829. [Google Scholar] [CrossRef] [PubMed]
- Teng, S.; Wang, L.; Srivastava, A.K.; Schwartz, C.E.; Alexov, E. Structural Assessment of the Effects of Amino Acid Substitutions on Protein Stability and Protein-Protein Interaction. Int. J. Comput. Biol. Drug Des. 2010, 3, 334–349. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, M.; Nikonova, E.; Babakov, A.; Kolesnikova, V.; Razhina, O.; Zlobin, N.; Taranov, V.; Nikonov, O. Interaction of Solanum Tuberosum L. Translation Initiation Factors eIF4E with Potato virus Y VPg: Apprehend and Avoid. Biochimie 2024, 219, 1–11. [Google Scholar] [CrossRef]
- Urquidi Camacho, R.A.; Lokdarshi, A.; Von Arnim, A.G. Translational gene regulation in plants: A green new deal. WIREs RNA 2020, 11, e1597. [Google Scholar] [CrossRef]
- Joshi, B.; Lee, K.; Maeder, D.L.; Jagus, R. Phylogenetic Analysis of eIF4E-Family Members. BMC Evol. Biol. 2005, 5, 48. [Google Scholar] [CrossRef]
- Patrick, R.M.; Browning, K.S. The eIF4F and eIFiso4F Complexes of Plants: An Evolutionary Perspective. Int. J. Genom. 2012, 2012, 287814. [Google Scholar] [CrossRef]
- Duprat, A.; Caranta, C.; Revers, F.; Menand, B.; Browning, K.S.; Robaglia, C. The Arabidopsis Eukaryotic Initiation Factor (iso)4E Is Dispensable for Plant Growth but Required for Susceptibility to Potyviruses. Plant J. 2002, 32, 927–934. [Google Scholar] [CrossRef]
- Estevan, J.; Maréna, A.; Callot, C.; Lacombe, S.; Moretti, A.; Caranta, C.; Gallois, J.-L. Specific Requirement for Translation Initiation Factor 4E or Its Isoform Drives Plant Host Susceptibility to Tobacco Etch virus. BMC Plant Biol. 2014, 14, 67. [Google Scholar] [CrossRef]
- Nicaise, V.; Gallois, J.-L.; Chafiai, F.; Allen, L.M.; Schurdi-Levraud, V.; Browning, K.S.; Candresse, T.; Caranta, C.; Le Gall, O.; German-Retana, S. Coordinated and Selective Recruitment of eIF4E and eIF4G Factors for Potyvirus Infection in Arab. Thaliana. FEBS Lett. 2007, 581, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Michel, V.; Julio, E.; Candresse, T.; Cotucheau, J.; Decorps, C.; Volpatti, R.; Moury, B.; Glais, L.; Jacquot, E.; de Borne, F.D.; et al. A complex eIF4E Locus Impacts the Durability of va Resistance to Potato virus Y in Tobacco. Mol. Plant Pathol. 2019, 20, 1051–1066. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, M.V.; Nikonova, E.Y.; Terentiev, A.A.; Taranov, V.V.; Babakov, A.V.; Nikonov, O.S. VPg of Potato virus Y and Potato Cap-Binding eIF4E Factors: Selective Interaction and Its Supposed Mechanism. Biochem. Mosc. 2021, 86, 1128–1138. [Google Scholar] [CrossRef]
- Moury, B.; Lebaron, C.; Szadkowski, M.; Ben Khalifa, M.; Girardot, G.; Bolou Bi, B.A.; Koné, D.; Nitiema, L.W.; Fakhfakh, H.; Gallois, J.-L. Knock-out Mutation of Eukaryotic Initiation Factor 4E2 (eIF4E2) Confers Resistance to Pepper veinal mottle virus in Tomato. Virology 2020, 539, 11–17. [Google Scholar] [CrossRef]
- Ruffel, S.; Gallois, J.-L.; Moury, B.; Robaglia, C.; Palloix, A.; Caranta, C. Simultaneous Mutations in Translation Initiation Factors eIF4E and eIF(Iso)4E Are Required to Prevent Pepper veinal mottle virus Infection of Pepper. J. Gen. Virol. 2006, 87, 2089–2098. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Oliveira, C.R.R.D.; Freire Filho, F.R.; Nogueira, M.D.S.D.R.; Barros, G.B.; Eiras, M.; Ribeiro, V.Q.; Lopes, Â.C.D.A. Reação de genótipos de feijão-caupi revela resistência às coinfecções pelo Cucumber mosaic virus, Cowpea aphid-borne mosaic virus e Cowpea severe mosaic virus. Bragantia 2012, 71, 59–66. [Google Scholar] [CrossRef]
- Lima, J.A.A.; Silva, A.K.F.D.; Aragão, M.D.L.; Ferreira, N.R.D.A.; Teófilo, E.M. Simple and Multiple Resistances to Viruses in Cowpea Genotypes. Pesqui. Agropecuária Bras. 2011, 46, 1432–1438. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinforma. Oxf. Engl. 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; Van Gunsteren, W.F. A Biomolecular Force Field Based on the Free Enthalpy of Hydration and Solvation: The GROMOS Force-Field Parameter Sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces: Proceedings of the Fourteenth Jerusalem Symposium on Quantum Chemistry and Biochemistry Held in Jerusalem, Israel, April 13–16, 1981; Pullman, B., Ed.; Springer: Dordrecht, The Netherlands, 1981; pp. 331–342. ISBN 978-94-015-7658-1. [Google Scholar]
- Hess, B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 116–122. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Berendsen, H.J.C. A Leap-Frog Algorithm for Stochastic Dynamics. Mol. Simul. 1988, 1, 173–185. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. Publ. Protein Soc. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, L.; Weng, G.; Shen, C.; Zhang, O.; Liu, M.; Zhang, C.; Gu, S.; Wang, J.; Wang, X.; et al. HawkDock Version 2: An Updated Web Server to Predict and Analyze the Structures of Protein-Protein Complexes. Nucleic Acids Res. 2025, gkaf379. [Google Scholar] [CrossRef]
- Oliveira, C.R.R. Reação de Genótipos de Feijão-Caupi às Coinfecções pelo Cucumber mosaic virus, Cowpea aphid-borne mosaic virus e Cowpea severe mosaic virus. Master’s Thesis, Universidade Federal do Piauí, Teresina, Brazil, 2011. [Google Scholar]
- Silva, J.A.; Costa, A.F.D.; Benko-Iseppon, A.M.; Guimarães, L.M.P.; Nicoli, A. Resistência de Vigna unguiculata ao Cowpea aphid-borne mosaic virus. Pesqui. Agropecuária Pernambucana 2021, 26, 1–3. [Google Scholar] [CrossRef]
- Barros, G.B.; Nogueira, M.D.S.D.R.; Oliveira, C.R.R.D.; Freire Filho, F.R.; Ribeiro, V.Q.; Veiga, C.F.D.M.; Brioso, P.S.T.; Eiras, M. Obtenção de plantas de feijão-caupi resistentes ao Cowpea severe mosaic virus e ao Cowpea aphid-borne mosaic virus. Summa Phytopathol. 2013, 39, 130–136. [Google Scholar] [CrossRef]
- de Santana, S.R.A.; da Silva Santana, J.T.; da Silva Costa, K.D.; da Costa, R.R.; da Costa, A.F.; de Carvalho Filho, J.L.S. Herança Da Resistência do Feijão-Caupi Ao Cowpea severe mosaic virus e Cowpea aphid-borne mosaic vírus: Inheritance of Cowpea Resistance to Cowpea severe mosaic virus and Cowpea aphid-borne mosaic virus. Braz. J. Dev. 2022, 8, 69367–69383. [Google Scholar] [CrossRef]
- Santos-Silva, C.A.D.; Zupin, L.; Oliveira-Lima, M.; Vilela, L.M.B.; Bezerra-Neto, J.P.; Ferreira-Neto, J.R.; Ferreira, J.D.C.; Oliveira-Silva, R.L.D.; Pires, C.D.J.; Aburjaile, F.F.; et al. Plant Antimicrobial Peptides: State of the Art, In Silico Prediction and Perspectives in the Omics Era. Bioinforma. Biol. Insights 2020, 14, 1177932220952739. [Google Scholar] [CrossRef]
- Santos-Silva, C.A.D.; Vilela, L.M.B.; Oliveira-Silva, R.L.D.; Silva, J.B.D.; Machado, A.R.; Bezerra-Neto, J.P.; Crovella, S.; Benko-Iseppon, A.M. Cassava (Manihot esculenta) Defensins: Prospection, Structural Analysis and Tissue-Specific Expression under Biotic/Abiotic Stresses. Biochimie 2021, 186, 1–12. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—Distance Constraints Applied on Model Quality Estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Barra, A.; Charon, J.; Tavert-Roudet, G.; Michon, T. Spectroscopic Investigation of the Kinetic Mechanism Involved in the Association of Potyviral VPg with the Host Plant Translation Initiation Factor eIF4E. Int. J. Mol. Sci. 2020, 21, 5618. [Google Scholar] [CrossRef]
- Okade, H.; Fujita, Y.; Miyamoto, S.; Tomoo, K.; Muto, S.; Miyoshi, H.; Natsuaki, T.; Rhoads, R.E.; Ishida, T. Turnip Mosaic Virus Genome-Linked Protein VPg Binds C-Terminal Region of Cap-Bound Initiation Factor 4E Orthologue without Exhibiting Host Cellular Specificity. J. Biochem. 2009, 145, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Yeam, I.; Cavatorta, J.R.; Ripoll, D.R.; Kang, B.-C.; Jahn, M.M. Functional Dissection of Naturally Occurring Amino Acid Substitutions in eIF4E That Confers Recessive Potyvirus Resistance in Plants. Plant Cell 2007, 19, 2913–2928. [Google Scholar] [CrossRef] [PubMed]
- Sanfaçon, H. Plant Translation Factors and Virus Resistance. Viruses 2015, 7, 3392–3419. [Google Scholar] [CrossRef]
- Horovitz, O.; Paşca, R.-D. Classification of Amino Acids by Multivariate Data Analysis, Based on Thermodynamic and Structural Characteristics. Stud. Univ. Babes-Bolyai Chemia. 2017, 62, 19–31. [Google Scholar] [CrossRef]
- Holzgräfe, C.; Wallin, S. Local versus Global Fold Switching in Protein Evolution: Insight from a Three-Letter Continuous Model. Phys. Biol. 2015, 12, 026002. [Google Scholar] [CrossRef] [PubMed]
- Charron, C.; Nicolaï, M.; Gallois, J.; Robaglia, C.; Moury, B.; Palloix, A.; Caranta, C. Natural Variation and Functional Analyses Provide Evidence for Co-evolution between Plant eIF4E and Potyviral VPg. Plant J. 2008, 54, 56–68. [Google Scholar] [CrossRef]
- Gao, Z.; Johansen, E.; Eyers, S.; Thomas, C.L.; Noel Ellis, T.H.; Maule, A.J. The Potyvirus Recessive Resistance Gene, sbm1, Identifies a Novel Role for Translation Initiation Factor eIF4E in Cell-to-cell Trafficking. Plant J. 2004, 40, 376–385. [Google Scholar] [CrossRef]
- Zhou, L.; Tian, Y.; Ren, L.; Yan, Z.; Jiang, J.; Shi, Q.; Geng, C.; Li, X. A Natural Substitution of a Conserved Amino Acid in eIF4E Confers Resistance against Multiple Potyviruses. Mol. Plant Pathol. 2024, 25, e13418. [Google Scholar] [CrossRef]
- Duff-Farrier, C.R.A.; Candresse, T.; Bailey, A.M.; Boonham, N.; Foster, G.D. Evidence for Different, Host-Dependent Functioning of Rx against Both Wild-Type and Recombinant Pepino mosaic virus. Mol. Plant Pathol. 2016, 17, 120–126. [Google Scholar] [CrossRef]
- Moury, B.; Janzac, B.; Ruellan, Y.; Simon, V.; Ben Khalifa, M.; Fakhfakh, H.; Fabre, F.; Palloix, A. Interaction Patterns between Potato virus Y and eIF4E-Mediated Recessive Resistance in the Solanaceae. J. Virol. 2014, 88, 9799–9807. [Google Scholar] [CrossRef]
- Bruun-Rasmussen, M.; Møller, I.S.; Tulinius, G.; Hansen, J.K.R.; Lund, O.S.; Johansen, I.E. The Same Allele of Translation Initiation Factor 4E Mediates Resistance Against Two Potyvirus Spp. in Pisum sativum. Mol. Plant-Microbe Interact. 2007, 20, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Lebaron, C.; Rosado, A.; Sauvage, C.; Gauffier, C.; German-Retana, S.; Moury, B.; Gallois, J.-L. A New eIF4E1 Allele Characterized by RNAseq Data Mining is Associated with Resistance to Potato virus Y in Tomato Albeit with a Low Durability. J. Gen. Virol. 2016, 97, 3063–3072. [Google Scholar] [CrossRef]
- Naderpour, M.; Lund, O.S.; Larsen, R.; Johansen, E. Potyviral Resistance Derived from Cultivars of Phaseolus Vulgaris Carrying Bc-3 Is Associated with the Homozygotic Presence of a Mutated eIF4E allele. Mol. Plant Pathol. 2010, 11, 255–263. [Google Scholar] [CrossRef]
- Sikosek, T.; Krobath, H.; Chan, H.S. Theoretical Insights into the Biophysics of Protein Bi-Stability and Evolutionary Switches. PLoS Comput. Biol. 2016, 12, e1004960. [Google Scholar] [CrossRef]
- Trotter, D.; Wallin, S. Effects of Topology and Sequence in Protein Folding Linked via Conformational Fluctuations. Biophys. J. 2020, 118, 1370–1380. [Google Scholar] [CrossRef]
- Schlessinger, A.; Rost, B. Protein Flexibility and Rigidity Predicted from Sequence. Proteins 2005, 61, 115–126. [Google Scholar] [CrossRef]
- Schmid, S.; Hugel, T. Controlling Protein Function by Fine-Tuning Conformational Flexibility. eLife 2020, 9, e57180. [Google Scholar] [CrossRef] [PubMed]
- Amaral, M.; Kokh, D.B.; Bomke, J.; Wegener, A.; Buchstaller, H.P.; Eggenweiler, H.M.; Matias, P.; Sirrenberg, C.; Wade, R.C.; Frech, M. Protein Conformational Flexibility Modulates Kinetics and Thermodynamics of Drug Binding. Nat. Commun. 2017, 8, 2276. [Google Scholar] [CrossRef] [PubMed]
- Grüner, S.; Peter, D.; Weber, R.; Wohlbold, L.; Chung, M.-Y.; Weichenrieder, O.; Valkov, E.; Igreja, C.; Izaurralde, E. The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation. Mol. Cell 2016, 64, 467–479. [Google Scholar] [CrossRef]
- Michon, T.; Estevez, Y.; Walter, J.; German-Retana, S.; Le Gall, O. The Potyviral Virus Genome-Linked Protein VPg Forms a Ternary Complex with the Eukaryotic Initiation Factors eIF4E and eIF4G and Reduces eIF4E Affinity for a mRNA Cap Analogue. FEBS J. 2006, 273, 1312–1322. [Google Scholar] [CrossRef]
- Chung, L.; Bailey, D.; Leen, E.N.; Emmott, E.P.; Chaudhry, Y.; Roberts, L.O.; Curry, S.; Locker, N.; Goodfellow, I.G. Norovirus Translation Requires an Interaction between the C Terminus of the Genome-Linked Viral Protein VPg and Eukaryotic Translation Initiation Factor 4G. J. Biol. Chem. 2014, 289, 21738–21750. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xie, H.; Wu, J.; Xie, L.; Yang, J.; Chi, Y. Translation Initiation Factor eIF4E and eIFiso4E Are Both Required for Peanut Stripe Virus Infection in Peanut (Arachis hypogaea L.). Front. Microbiol. 2017, 8, 338. [Google Scholar] [CrossRef] [PubMed]
- Ala-Poikela, M.; Rajamäki, M.-L.; Valkonen, J.P.T. A Novel Interaction Network Used by Potyviruses in Virus–Host Interactions at the Protein Level. Viruses 2019, 11, 1158. [Google Scholar] [CrossRef] [PubMed]
- Pairas, G.N.; Tsoungas, P.G. H -Bond: Τhe Chemistry-Biology H -Bridge. ChemistrySelect 2016, 1, 4520–4532. [Google Scholar] [CrossRef]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O.V. Radius of Gyration as an Indicator of Protein Structure Compactness. Mol. Biol. 2008, 42, 623–628. [Google Scholar] [CrossRef]





| POSITION | TYPE | CULTIVAR/Condition | ||
|---|---|---|---|---|
| Susceptible | Resistant | |||
| BOCA NEGRA | BR14 MULATO | IT85F-2687 | ||
| 203 | Nucleotide | Cytosine (C) | Cytosine (C) | Guanine (G) * |
| 224 | Nucleotide | Cytosine (C) | Adenine (A) | Adenine (A) |
| 325 | Nucleotide | Guanine (G) | Guanine (G) | Cytosine (C) * |
| 329 | Nucleotide | Cytosine (C) | Cytosine (C) | Thymine (T) * |
| 520 | Nucleotide | Adenine (A) | Thymine (T) | Adenine (A) |
| 68 | Amino acid | Proline (Pro) | Proline (Pro) | Arginine (Arg) * |
| 75 | Amino acid | Alanine (Ala) | Aspartate (Asp) | Aspartate (Asp) |
| 109 | Amino acid | Glycine (Gly) | Glycine (Gly) | Arginine (Arg) * |
| 110 | Amino acid | Alanine (Ala) | Alanine (Ala) | Valine (Val) * |
| 174 | Amino acid | Tyrosine (Tyr) | Asparagine (Asn) | Asparagine (Asn) |
| GENOTYPES | THIS STUDY | LITERATURE DATA | ||
|---|---|---|---|---|
| PCR | Bioassay | Reaction | Reference | |
| 1—Santo Inácio | Susceptible | Susceptible | ** | |
| 2—Pingo de Ouro | Susceptible | Susceptible | Susceptible | [37] |
| 3—BR 14 Mulato | Susceptible | Susceptible | Susceptible | [47,48] |
| 4—BRS Xiquexique | Susceptible | Susceptible | Susceptible | [48] |
| 5—BRS Tumucumaque | Resistant | Susceptible | Susceptible | [48] |
| 6—Inhuma | Susceptible | Susceptible | ** | |
| 7—Boca Negra | Susceptible | Susceptible | ** | |
| 8—João Paulo II | Susceptible | Susceptible | ** | |
| 9—IT85F-2687 | Resistant | Resistant | Resistant | [48] |
| 10—BR 1 Poty | Susceptible | Susceptible | Susceptible | [37] |
| 11—BRS Maratoã | Susceptible | Susceptible | Susceptible/ Resistant | [37,47] |
| 12—BRS Cauamé | Resistant | Susceptible | Susceptible/Resistant | [45,46] |
| 13—BRS Guariba | Resistant | Resistant | Resistant/Susceptible | [47,48,49] |
| 14—BRS Itaim | Susceptible | Susceptible | Resistant | [47] |
| 15—BRS Paraguaçu | Susceptible | Susceptible | ** | |
| 16—L.950.002 | Resistant | Resistant | ** | |
| 17—Miranda IPA 207 | Susceptible | Resistant | ** | |
| 18—IPA 206 | Resistant | Resistant | ** | |
| 19—BRS Juruá | Susceptible | Susceptible | Susceptible/Resistant | [36,47] |
| 20—Canapu | Susceptible | Susceptible | ** | |
| 21—BR10 Piauí | Susceptible | Susceptible | Susceptible | [48] |
| 22—Corujinha | Resistant | Susceptible | ** | |
| 23—TVU-966 | * | Resistant | Resistant | [47,50] |
| 24—Manteguinha Santarém | Susceptible | Resistant | Resistant | [48] |
| 25—IT81D-1053 | Resistant | Resistant | Resistant | [48] |
| 26—Sempre Verde Salgueiro | Susceptible | Susceptible | Susceptible | [50] |
| 27—Bajão | * | Resistant | Resistant | [48] |
| COMPLEXES | HADDOCK | ROSETTA | HAWKDOCK MM-GBSA | ||
|---|---|---|---|---|---|
| Score | Interaction Area (Å2) | Theorical DDG (REUs) | Molecular Surface (Å2) | Predicted Binding Free Energy of Complex (kcal/mol) | |
| eIF4E (Bajão)/VPg | −75.9 | 1195.7 | −35.724 | 632.566 | −84.23 |
| eIF4E (Boca Negra)/VPg | −91.2 | 1424.1 | −41.459 | 684.034 | −98.21 |
| eIF4E (BRS Cauamé)/VPg | −72.3 | 1316.4 | −20.840 | 455.246 | −68.52 |
| eIF4E (BRS Xiquexique)/VPg | −102.9 | 1944.6 | −23.612 | 453.629 | −72.97 |
| eIF4E (IT85F-2687)/VPg | −115.5 | 1820.6 | −30.910 | 518.889 | −82.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrade, F.A.d.; Luna-Aragão, M.A.d.; Ferreira, J.D.C.; Souza, F.F.; Rocha Oliveira, A.C.d.; Costa, A.F.d.; Aragão, F.J.L.; Santos-Silva, C.A.d.; Benko-Iseppon, A.M.; Pandolfi, V. Deciphering Cowpea Resistance to Potyvirus: Assessment of eIF4E Gene Mutations and Their Impact on the eIF4E-VPg Protein Interaction. Viruses 2025, 17, 1050. https://doi.org/10.3390/v17081050
Andrade FAd, Luna-Aragão MAd, Ferreira JDC, Souza FF, Rocha Oliveira ACd, Costa AFd, Aragão FJL, Santos-Silva CAd, Benko-Iseppon AM, Pandolfi V. Deciphering Cowpea Resistance to Potyvirus: Assessment of eIF4E Gene Mutations and Their Impact on the eIF4E-VPg Protein Interaction. Viruses. 2025; 17(8):1050. https://doi.org/10.3390/v17081050
Chicago/Turabian StyleAndrade, Fernanda Alves de, Madson Allan de Luna-Aragão, José Diogo Cavalcanti Ferreira, Fernanda Freitas Souza, Ana Carolina da Rocha Oliveira, Antônio Félix da Costa, Francisco José Lima Aragão, Carlos André dos Santos-Silva, Ana Maria Benko-Iseppon, and Valesca Pandolfi. 2025. "Deciphering Cowpea Resistance to Potyvirus: Assessment of eIF4E Gene Mutations and Their Impact on the eIF4E-VPg Protein Interaction" Viruses 17, no. 8: 1050. https://doi.org/10.3390/v17081050
APA StyleAndrade, F. A. d., Luna-Aragão, M. A. d., Ferreira, J. D. C., Souza, F. F., Rocha Oliveira, A. C. d., Costa, A. F. d., Aragão, F. J. L., Santos-Silva, C. A. d., Benko-Iseppon, A. M., & Pandolfi, V. (2025). Deciphering Cowpea Resistance to Potyvirus: Assessment of eIF4E Gene Mutations and Their Impact on the eIF4E-VPg Protein Interaction. Viruses, 17(8), 1050. https://doi.org/10.3390/v17081050










