DNAJ Homolog Subfamily C Member 11 Stabilizes SARS-CoV-2 NSP3 to Promote Double-Membrane Vesicle Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Viral Infection and Amplification Test
2.3. Antibodies and Western Blot Analysis
2.4. Plasmid Construction and Cell Transfection
2.5. Immunofluorescence and Confocal Immunofluorescence Microscopy
2.6. Co-Immunoprecipitation (Co-IP) Analysis
2.7. Protein Expression and Purification
2.8. Pull-Down Assay
2.9. Quantitative Real-Time PCR (qRT-PCR)
2.10. Lentivirus-Mediated Stable Cell Line Construction
2.11. Transmission Electron Microscopy (TEM)
2.12. Cell Viability Assay
2.13. Quantification and Statistical Analysis
3. Results
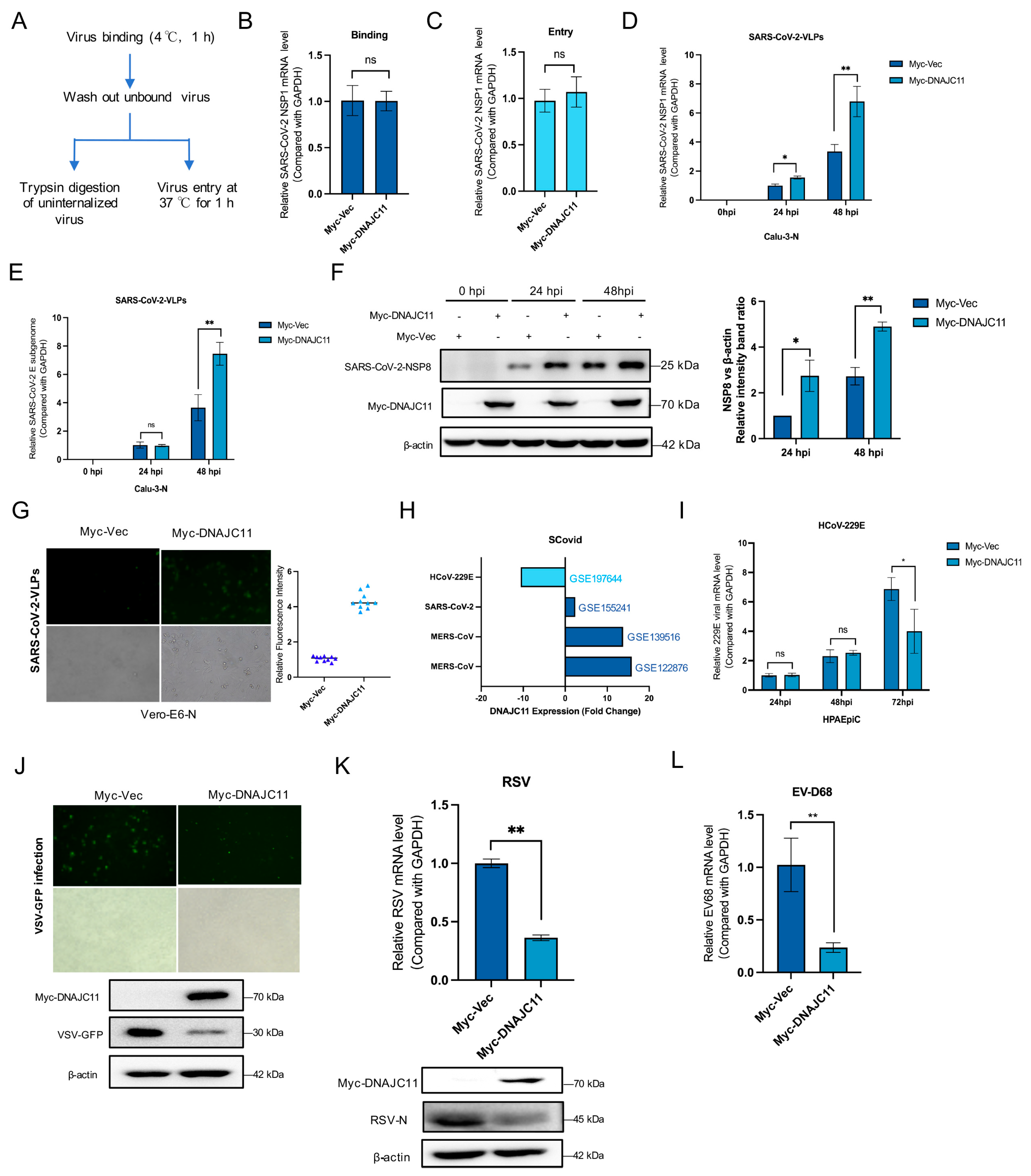
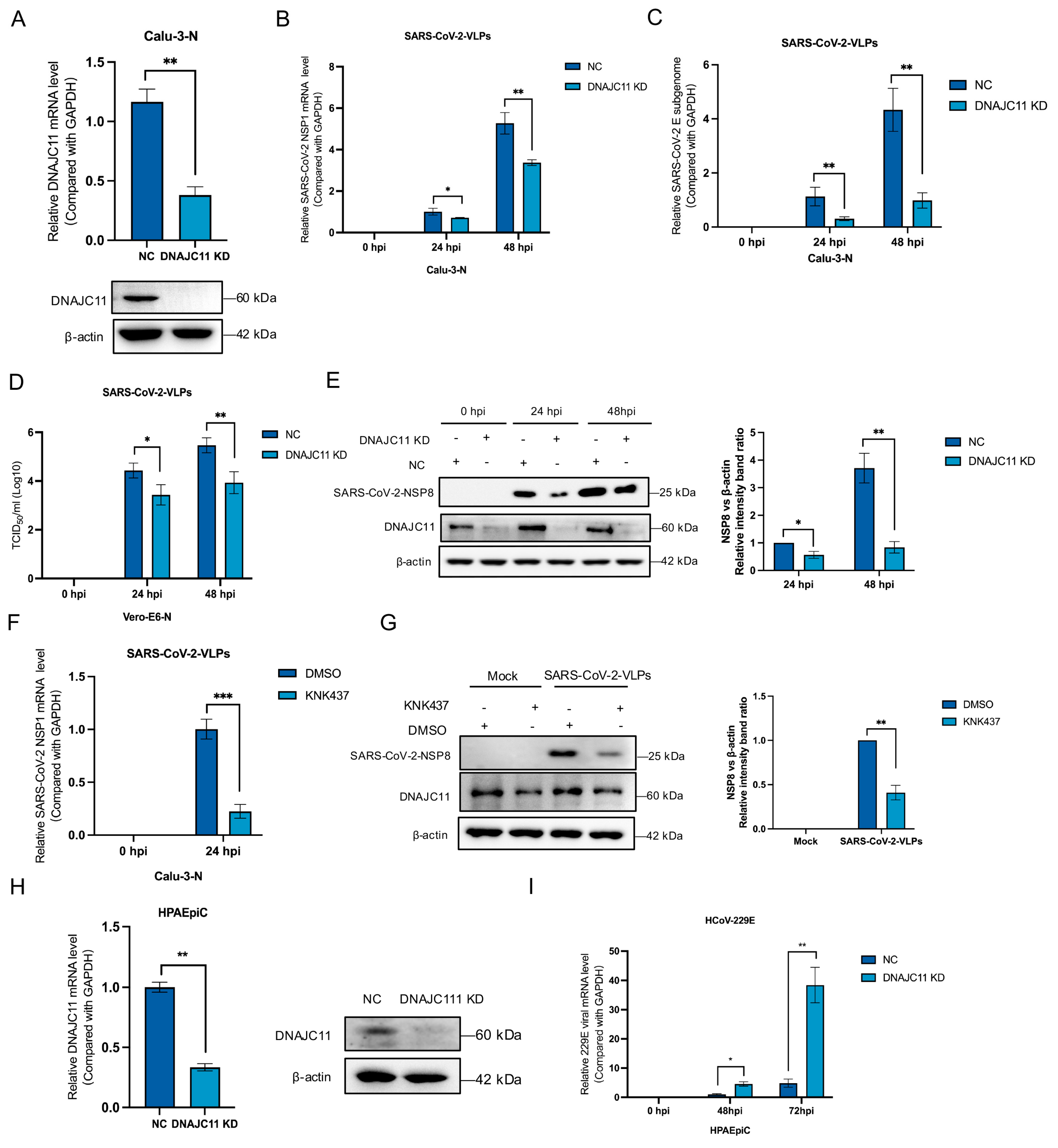
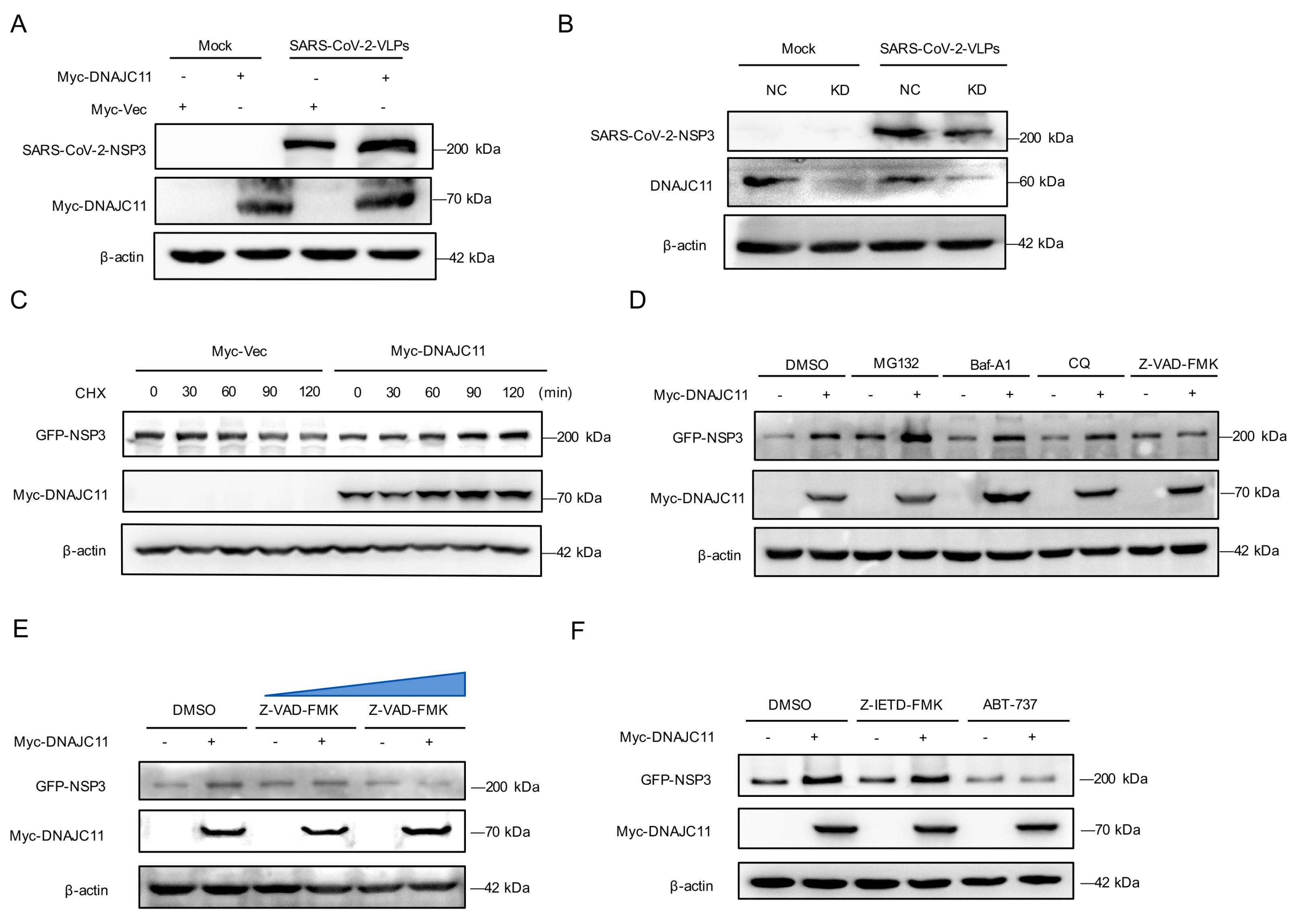
3.1. DNAJC11 Facilitates SARS-CoV-2 Replication
3.2. DNAJC11 Does Not Promote SARS-CoV-2 Infection Through Type I IFN or IFN-Stimulated Gene (ISG) Pathways
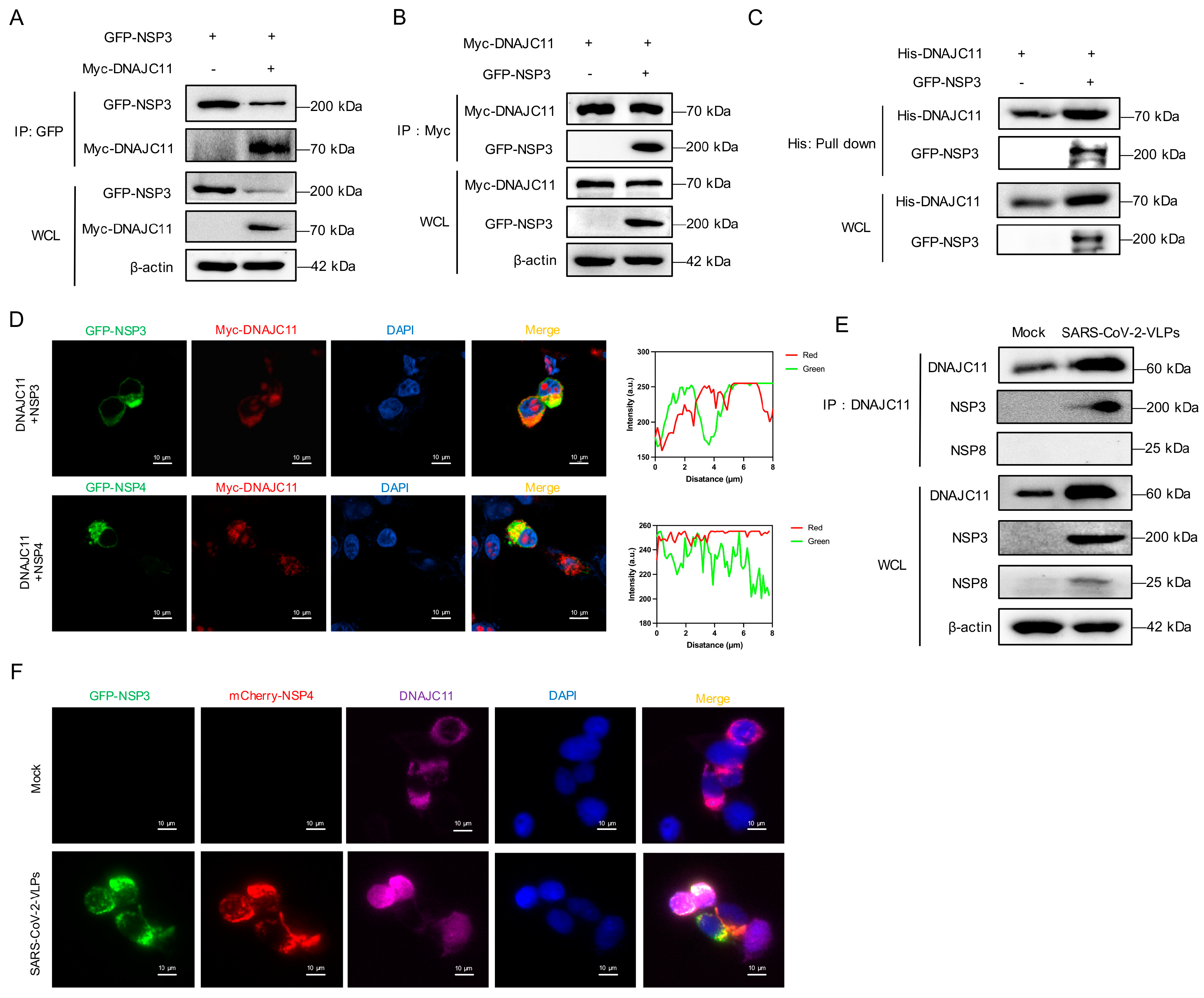
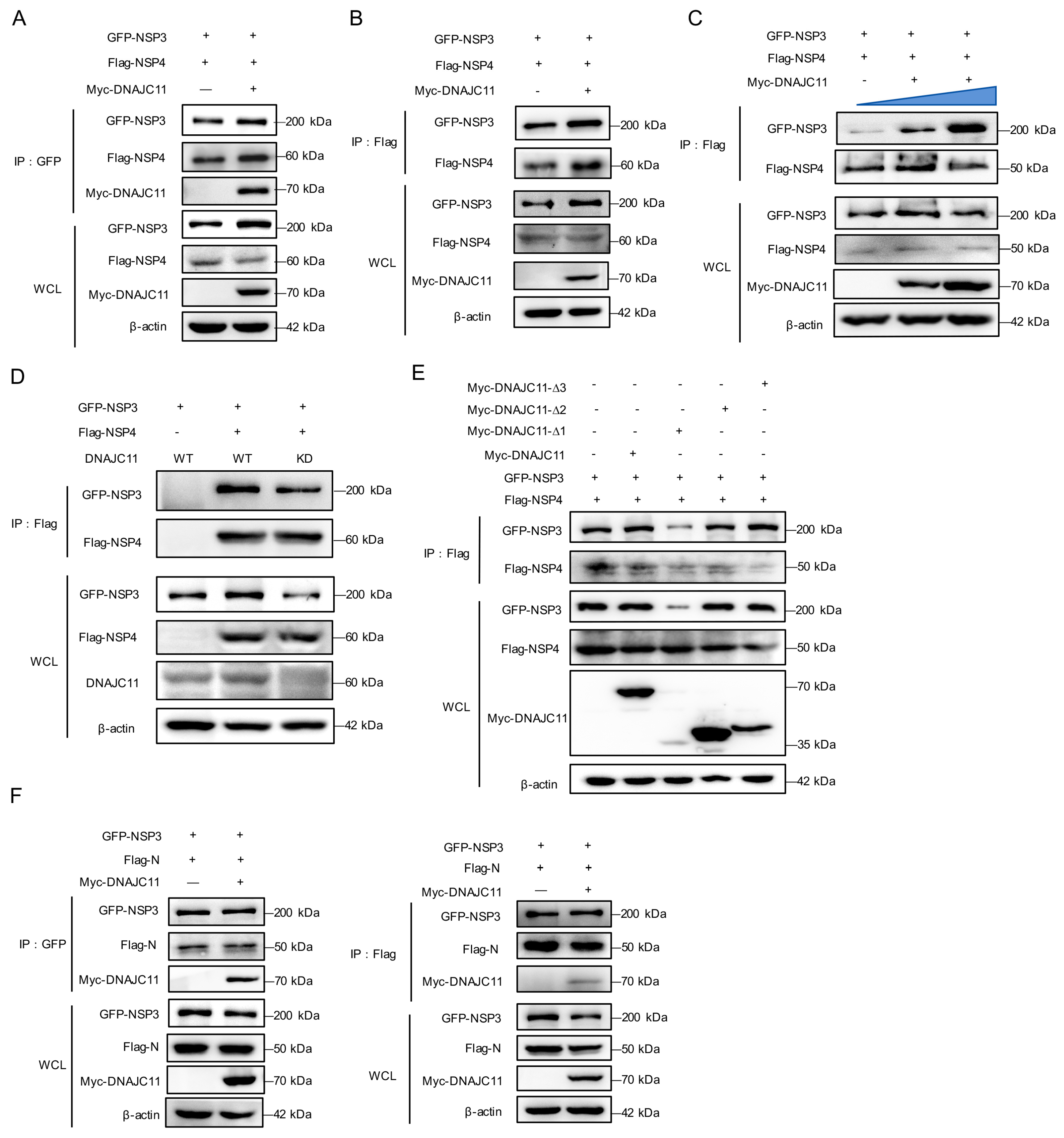
3.3. SARS-CoV-2 NSP3 Interacts with DNAJC11
3.4. DNAJC11 J Domain Interacts with SARS-CoV-2 NSP3
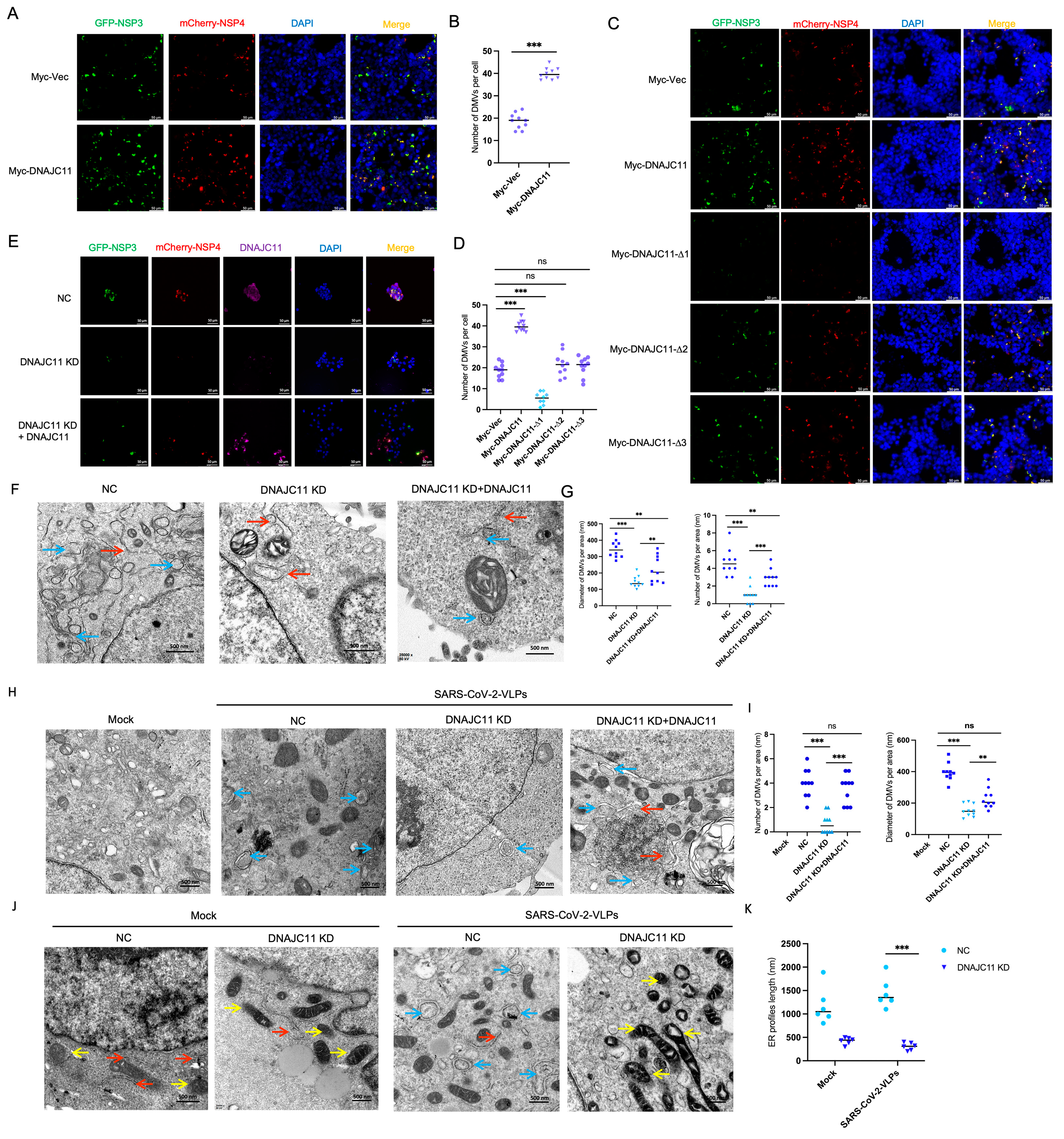
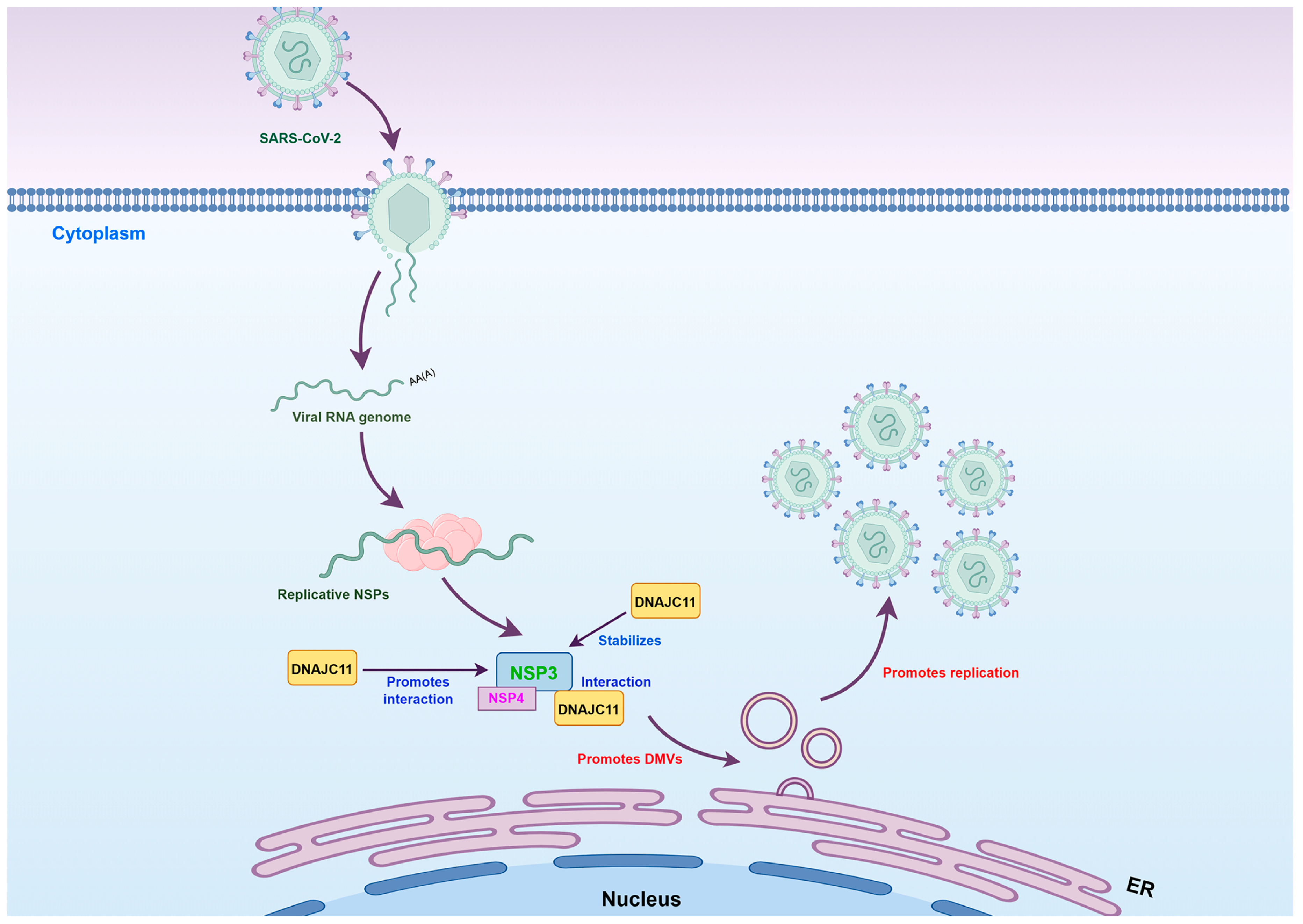
3.5. DNAJC11 Facilitates DMV Formation
3.6. DNAJC11 Depletion Impairs SARS-CoV-2-Induced DMV Formation and Disrupts ER and Mitochondrial Morphology
3.7. DNAJC11 Stabilizes NSP3 Expression to Promote NSP3-NSP4 Interactions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baggen, J.; Vanstreels, E.; Jansen, S.; Daelemans, D. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 2021, 6, 1219–1232. [Google Scholar] [CrossRef] [PubMed]
- Kirtipal, N.; Bharadwaj, S.; Kang, S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. 2020, 85, 104502. [Google Scholar] [CrossRef] [PubMed]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [PubMed]
- de Wilde, A.H.; Raj, V.S.; Oudshoorn, D.; Bestebroer, T.M.; van Nieuwkoop, S.; Limpens, R.; Posthuma, C.C.; van der Meer, Y.; Barcena, M.; Haagmans, B.L.; et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J. Gen. Virol. 2013, 94 Pt 8, 1749–1760. [Google Scholar] [CrossRef] [PubMed]
- Limpens, R.W.; van der Schaar, H.M.; Kumar, D.; Koster, A.J.; Snijder, E.J.; van Kuppeveld, F.J.; Barcena, M. The transformation of enterovirus replication structures: A three-dimensional study of single- and double-membrane compartments. mBio 2011, 2, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Knoops, K.; Barcena, M.; Limpens, R.W.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. Ultrastructural characterization of arterivirus replication structures: Reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J. Virol. 2012, 86, 2474–2487. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Limpens, R.; de Wilde, A.H.; de Jong, A.W.M.; Zevenhoven-Dobbe, J.C.; Maier, H.J.; Faas, F.; Koster, A.J.; Barcena, M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020, 18, e3000715. [Google Scholar] [CrossRef] [PubMed]
- Santerre, M.; Arjona, S.P.; Allen, C.N.; Shcherbik, N.; Sawaya, B.E. Why do SARS-CoV-2 NSPs rush to the ER? J. Neurol. 2021, 268, 2013–2022. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, S.; Guarino, A.M.; Giaquinto, L.; Polishchuk, E.V.; Santoro, M.; Di Tullio, G.; Wilson, C.; Panariello, F.; Soares, V.C.; Dias, S.S.G.; et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature 2022, 606, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, L.; Zhao, X.; Makroczyova, J.; Wachsmuth-Melm, M.; Prasad, V.; Hensel, Z.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 nsp3 and nsp4 are minimal constituents of a pore spanning replication organelle. Nat. Commun. 2023, 14, 7894. [Google Scholar] [CrossRef] [PubMed]
- Violitzi, F.; Perivolidi, V.I.; Thireou, T.; Grivas, I.; Haralambous, S.; Samiotaki, M.; Panayotou, G.; Douni, E. Mapping Interactome Networks of DNAJC11, a Novel Mitochondrial Protein Causing Neuromuscular Pathology in Mice. J. Proteome Res. 2019, 18, 3896–3912. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Gierasch, L.M. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 2019, 294, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Zarouchlioti, C.; Parfitt, D.A.; Li, W.; Gittings, L.M.; Cheetham, M.E. DNAJ Proteins in neurodegeneration: Essential and protective factors. Philos. Trans. R Soc. Lond. B Biol. Sci. 2018, 373, 20160534. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ju, X.; Ding, Q. A Nucleocapsid-based Transcomplementation Cell Culture System of SARS-CoV-2 to Recapitulate the Complete Viral Life Cycle. Bio. Protoc. 2021, 11, e4257. [Google Scholar] [CrossRef] [PubMed]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gui, Q.; Liang, F.X.; Sall, J.; Zhang, Q.; Duan, Y.; Dhabaria, A.; Askenazi, M.; Ueberheide, B.; Stapleford, K.A.; et al. The REEP5/TRAM1 complex binds SARS-CoV-2 NSP3 and promotes virus replication. J. Virol. 2023, 97, e0050723. [Google Scholar] [CrossRef] [PubMed]
- Mantlo, E.; Bukreyeva, N.; Maruyama, J.; Paessler, S.; Huang, C. Potent Antiviral Activities of Type I Interferons to SARS-CoV-2 Infection. bioRxiv 2020, 179, 104811. [Google Scholar]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharmacol. 2021, 183, 114316. [Google Scholar] [CrossRef] [PubMed]
- Ioakeimidis, F.; Ott, C.; Kozjak-Pavlovic, V.; Violitzi, F.; Rinotas, V.; Makrinou, E.; Eliopoulos, E.; Fasseas, C.; Kollias, G.; Douni, E. A splicing mutation in the novel mitochondrial protein DNAJC11 causes motor neuron pathology associated with cristae disorganization, and lymphoid abnormalities in mice. PLoS ONE 2014, 9, e104237. [Google Scholar] [CrossRef] [PubMed]
- Fillmore, N.R.; Szalat, R.E.; La, J.; Branch-Elliman, W.; Monach, P.A.; Nguyen, V.; Samur, M.K.; Brophy, M.T.; Do, N.V.; Munshi, N.C. Recent common human coronavirus infection protects against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: A Veterans Affairs cohort study. Proc. Natl. Acad. Sci. USA 2022, 119, e2213783119. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.L.; Guo, D.; Rottier, P.J. Coronavirus: Epidemiology, genome replication and the interactions with their hosts. Virol. Sin. 2016, 31, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Frieman, M. The art of war: Battles between virus and host. Curr. Opin. Virol. 2014, 6, 76–77. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hou, Y.; Zhou, Y.; Yang, Z.; Zhao, H.; Jian, T.; Yu, Q.; Zeng, F.; Liu, X.; Zhang, Z.; et al. LLPS of FXR proteins drives replication organelle clustering for beta-coronaviral proliferation. J. Cell Biol. 2024, 223, e202309140. [Google Scholar] [CrossRef] [PubMed]
- Garvanska, D.H.; Alvarado, R.E.; Mundt, F.O.; Lindqvist, R.; Duel, J.K.; Coscia, F.; Nilsson, E.; Lokugamage, K.; Johnson, B.A.; Plante, J.A.; et al. The NSP3 protein of SARS-CoV-2 binds fragile X mental retardation proteins to disrupt UBAP2L interactions. EMBO Rep. 2024, 25, 902–926. [Google Scholar] [CrossRef] [PubMed]
- Kratzel, A.; Thiel, V. RTN3 and RTN4: Architects of SARS-CoV-2 replication organelles. J. Cell Biol. 2023, 222, e202306020. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Marusich, M.F.; Souda, P.; Whitelegge, J.; Capaldi, R.A. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007, 581, 3545–3549. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liang, C.; Zhou, L. Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 2020, 29, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Knox, C.; Luke, G.A.; Blatch, G.L.; Pesce, E.R. Heat shock protein 40 (Hsp40) plays a key role in the virus life cycle. Virus Res. 2011, 160, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.Q.; Yuan, L.; Zhao, Q.; Yuan, J.L.; Miao, C.; Chang, Y.F.; Wen, X.T.; Wu, R.; Huang, X.B.; Wen, Y.P.; et al. Hsp40 Protein DNAJB6 Interacts with Viral NS3 and Inhibits the Replication of the Japanese Encephalitis Virus. Int. J. Mol. Sci. 2019, 20, 5719. [Google Scholar] [CrossRef] [PubMed]
- Batra, J.; Tripathi, S.; Kumar, A.; Katz, J.M.; Cox, N.J.; Lal, R.B.; Sambhara, S.; Lal, S.K. Human Heat shock protein 40 (Hsp40/DnaJB1) promotes influenza A virus replication by assisting nuclear import of viral ribonucleoproteins. Sci. Rep. 2016, 6, 19063. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Liu, S.; Liu, M.; Wang, S.; Bi, Z.; Fan, W.; Shi, Z.; Song, S.; Yan, L. Hsp70 Inhibits the Replication of Fowl Adenovirus Serotype 4 by Suppressing Viral Hexon with the Assistance of DnaJC7. J. Virol. 2022, 96, e0080722. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Zhong, Q.; Gao, G.F. Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 2022, 65, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Kumari, S.; Pandey, B.; Mistry, H.; Bihani, S.C.; Das, A.; Prashar, V.; Gupta, G.D.; Panicker, L.; Kumar, M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021, 433, 166725. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Li, M.; Sun, L.; Deng, H.; Zhao, Y.G. DMV biogenesis during beta-coronavirus infection requires autophagy proteins VMP1 and TMEM41B. Autophagy 2023, 19, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, T.; Zhong, L.; Zhang, W.; Zhang, Y.; Yu, X.; Yuan, S.; Ni, T. Molecular architecture of coronavirus double-membrane vesicle pore complex. Nature 2024, 633, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, E.; Roingeard, P. Virus-induced double-membrane vesicles. Cell Microbiol. 2015, 17, 45–50. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Yu, H.; Liu, S.; Wan, H.; Fu, S.; Liu, S.; Yang, J.; Zhang, Z.; Huang, H.; Li, Q.; et al. Mitochondrial cristae architecture protects against mtDNA release and inflammation. Cell Rep. 2022, 41, 111774. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.K.; Tikoo, S.K. Viruses as modulators of mitochondrial functions. Adv. Virol. 2013, 2013, 738794. [Google Scholar] [CrossRef] [PubMed]
- Twu, W.I.; Lee, J.Y.; Kim, H.; Prasad, V.; Cerikan, B.; Haselmann, U.; Tabata, K.; Bartenschlager, R. Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation. Cell Rep. 2021, 37, 110049. [Google Scholar] [CrossRef] [PubMed]
- Roingeard, P.; Eymieux, S.; Burlaud-Gaillard, J.; Hourioux, C.; Patient, R.; Blanchard, E. The double-membrane vesicle (DMV): A virus-induced organelle dedicated to the replication of SARS-CoV-2 and other positive-sense single-stranded RNA viruses. Cell. Mol. Life. Sci. 2022, 79, 425. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.T.; Zeb, M.T.; Ahsan, H.; Ahmed, A.; Ali, A.; Akhtar, K.; Malik, S.I.; Cui, Z.; Ali, S.; Khan, A.S.; et al. SARS-CoV-2 nucleocapsid and Nsp3 binding: An in silico study. Arch. Microbiol. 2021, 203, 59–66. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Yang, S.; Li, X.; Xiang, J.; Cai, J.; Wang, Y.; Li, Q.; Zang, N.; Wang, J.; Shang, J.; et al. DNAJ Homolog Subfamily C Member 11 Stabilizes SARS-CoV-2 NSP3 to Promote Double-Membrane Vesicle Formation. Viruses 2025, 17, 1025. https://doi.org/10.3390/v17081025
Chen S, Yang S, Li X, Xiang J, Cai J, Wang Y, Li Q, Zang N, Wang J, Shang J, et al. DNAJ Homolog Subfamily C Member 11 Stabilizes SARS-CoV-2 NSP3 to Promote Double-Membrane Vesicle Formation. Viruses. 2025; 17(8):1025. https://doi.org/10.3390/v17081025
Chicago/Turabian StyleChen, Shuying, Shanrong Yang, Xiaoning Li, Junqi Xiang, Jiangyu Cai, Yaokai Wang, Qingqing Li, Na Zang, Jiaxu Wang, Jian Shang, and et al. 2025. "DNAJ Homolog Subfamily C Member 11 Stabilizes SARS-CoV-2 NSP3 to Promote Double-Membrane Vesicle Formation" Viruses 17, no. 8: 1025. https://doi.org/10.3390/v17081025
APA StyleChen, S., Yang, S., Li, X., Xiang, J., Cai, J., Wang, Y., Li, Q., Zang, N., Wang, J., Shang, J., & Wan, Y. (2025). DNAJ Homolog Subfamily C Member 11 Stabilizes SARS-CoV-2 NSP3 to Promote Double-Membrane Vesicle Formation. Viruses, 17(8), 1025. https://doi.org/10.3390/v17081025






