The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications
Abstract
1. Introduction
Review Methodology
2. Genomic Landscape
3. Epidemiological Dynamics
3.1. Global Distribution Patterns
3.2. Transmission Efficiency
3.3. Comparative Transmissibility Metrics
4. Clinical and Immunological Insights
4.1. Symptom Profile Variations
4.2. Immune Response Characteristics
4.3. Potential Impacts on Vaccine Effectiveness
5. Future Aspects
5.1. Critical Knowledge Gaps
5.1.1. Variant-Specific Virological Characteristics
5.1.2. Immune Evasion Dynamics
5.1.3. Pathogenesis and Clinical Impact
5.1.4. Transmission and Epidemiologic Fitness
5.1.5. Geographic and Host-Specific Heterogeneity
5.1.6. Long-Term Sequelae and Psychosocial Impact
5.2. Emerging Research Priorities
5.2.1. Enhanced Molecular Surveillance
5.2.2. In Vitro and In Vivo Models
5.2.3. Immune Correlates of Protection
5.2.4. Antiviral and Therapeutic Resistance
5.2.5. Longitudinal Clinical Studies
5.2.6. Integrating Computational Approaches
5.3. Potential Intervention Strategies
5.3.1. Optimized Vaccine Development
5.3.2. Next-Generation Antivirals
5.3.3. Monoclonal Antibody Therapies
5.3.4. Non-Pharmaceutical Interventions (NPIs)
5.3.5. One Health Approach to Understanding the XEC Mutation of SARS-CoV-2
5.3.6. Global Equity Initiatives and Rapid Response Systems
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. World Health Organization Tracking SARS-CoV-2 Variants. 2024. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 13 February 2025).
- World Health Organization. Coronavirus Disease (COVID-19) Weekly Epidemiological Updates and Monthly Operational Updates. 2023. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 13 February 2025).
- World Health Organization. COVID-19 Weekly Epidemiological Update, 9 March 2021. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update---10-march-2021 (accessed on 13 February 2025).
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Holmes, E.C.; Goldstein, S.A.; Rasmussen, A.L.; Robertson, D.L.; Crits-Christoph, A.; Wertheim, J.O.; Anthony, S.J.; Barclay, W.S.; Boni, M.F.; Doherty, P.C. The origins of SARS-CoV-2: A critical review. Cell 2021, 184, 4848–4856. [Google Scholar] [CrossRef] [PubMed]
- Crits-Christoph, A.; Levy, J.I.; Pekar, J.E.; Goldstein, S.A.; Singh, R.; Hensel, Z.; Gangavarapu, K.; Rogers, M.B.; Moshiri, N.; Garry, R.F. Genetic tracing of market wildlife and viruses at the epicenter of the COVID-19 pandemic. Cell 2024, 187, 5468–5482. [Google Scholar] [CrossRef]
- Anderson, R.M.; Fraser, C.; Ghani, A.C.; Donnelly, C.A.; Riley, S.; Ferguson, N.M.; Leung, G.M.; Lam, T.H.; Hedley, A.J. Epidemiology, transmission dynamics and control of SARS: The 2002–2003 epidemic. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2004, 359, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. FDA Approves First COVID-19 Vaccine. FDA News Release 2021. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-covid-19-vaccine (accessed on 13 February 2025).
- U.S. Food and Drug Administration. FDA Takes Additional Action in Fight Against COVID-19 by Issuing Emergency Use Authorization for Second COVID-19 Vaccine. Available online: https://www.prnewswire.com/news-releases/fda-takes-additional-action-in-fight-against-covid-19-by-issuing-emergency-use-authorization-for-second-covid-19-vaccine-301196303.html (accessed on 24 January 2025).
- Mallapaty, S. India’s DNA Covid vaccine is a first—more are coming. Nature 2021, 597, 161–162. [Google Scholar] [CrossRef]
- Yu, D. China grants emergency use of new vaccines as it eases COVID-19 policy. BioWorld 2022, 13. [Google Scholar]
- European Medicines Agency. Vaccine AstraZeneca. 2021. Available online: https://www.bioworld.com/articles/692398-china-grants-emergency-use-of-new-vaccines-as-it-eases-covid-19-policy?v=preview (accessed on 13 February 2025).
- European Medicines Agency. Fact Sheet for Healthcare Providers Administering Vaccine (Vaccination Providers). Available online: https://healthpolicy-watch.news/european-medicines-agency-astrazeneca/ (accessed on 13 February 2025).
- Taylor, A. WHO grants emergency use authorization for Chinese-made Sinopharm coronavirus vaccine. The Washington Post, 2021; p. 1. [Google Scholar]
- World Health Organization. Coronavirus disease (COVID-19) Epidemiological Updates and Monthly Operational Updates. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 13 February 2025).
- Gnimadi, T.A.C.; Kadio, K.J.-J.O.; Mathew, M.J.; Diallo, H.; Soumah, A.K.; Keita, A.K.; Hounmenou, C.G.; Fernandez-Nuñez, N.; Vidal, N.; Guichet, E. Genetic Diversity and Spatiotemporal Distribution of SARS-CoV-2 Variants in Guinea: A Meta-Analysis of Sequence Data (2020–2023). Viruses 2025, 17, 204. [Google Scholar] [CrossRef]
- Perez-Gomez, R. The development of SARS-CoV-2 variants: The gene makes the disease. J. Dev. Biol. 2021, 9, 58. [Google Scholar] [CrossRef]
- O’Toole, Á.; Pybus, O.G.; Abram, M.E.; Kelly, E.J.; Rambaut, A. Pango lineage designation and assignment using SARS-CoV-2 spike gene nucleotide sequences. BMC Genom. 2022, 23, 121. [Google Scholar] [CrossRef]
- Lundstrom, K. Role of Nucleic Acid Vaccines for the Management of Emerging Variants of SARS-CoV-2. In SARS-CoV-2 Variants and Global Population Vulnerability; Apple Academic Press: Palm Bay, FL, USA, 2023; pp. 285–316. [Google Scholar]
- Callaway, E. Making sense of coronavirus mutations. Nature 2020, 585, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, T.; Nemudryi, A.; Nemudraia, A.; McVey, A.; Little, A.; Taylor, D.N.; Walk, S.T.; Wiedenheft, B. The rise and fall of SARS-CoV-2 variants and ongoing diversification of omicron. Viruses 2022, 14, 2009. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Smith, D.M. SARS-CoV-2 variants of concern. Yonsei Med. J. 2021, 62, 961. [Google Scholar] [CrossRef]
- Sun, Q.; Zeng, J.; Tang, K.; Long, H.; Zhang, C.; Zhang, J.; Tang, J.; Xin, Y.; Zheng, J.; Sun, L. Variation in synonymous evolutionary rates in the SARS-CoV-2 genome. Front. Microbiol. 2023, 14, 1136386. [Google Scholar] [CrossRef] [PubMed]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Kaku, Y.; Uriu, K.; Okumura, K.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 KP. 3.1. 1 variant. Lancet Infect. Dis. 2024, 24, e609. [Google Scholar] [CrossRef]
- Scarpa, F.; Branda, F.; Ceccarelli, G.; Romano, C.; Locci, C.; Pascale, N.; Azzena, I.; Fiori, P.L.; Casu, M.; Pascarella, S. SARS-CoV-2 XEC: A Genome-Based Survey. Microorganisms 2025, 13, 253. [Google Scholar] [CrossRef]
- Li, P.; Faraone, J.N.; Hsu, C.C.; Chamblee, M.; Liu, Y.; Zheng, Y.-M.; Xu, Y.; Carlin, C.; Horowitz, J.C.; Mallampalli, R.K. Immune Evasion, Cell-Cell Fusion, and Spike Stability of the SARS-CoV-2 XEC Variant: Role of Glycosylation Mutations at the N-terminal Domain. bioRxiv 2024. [Google Scholar] [CrossRef]
- Kaku, Y.; Okumura, K.; Kawakubo, S.; Uriu, K.; Chen, L.; Kosugi, Y.; Uwamino, Y.; Begum, M.S.T.M.; Leong, S.; Ikeda, T.; et al. Virological characteristics of the SARS-CoV-2 XEC variant. Lancet Infect. Dis. 2024, 24, e736. [Google Scholar] [CrossRef]
- Branda, F.; Ciccozzi, M.; Scarpa, F. Genetic variability of the recombinant SARS-CoV-2 XEC: Is it a new evolutionary dead-end lineage? New Microbes New Infect. 2024, 62, 101520. [Google Scholar] [CrossRef] [PubMed]
- Gangavarapu, K.; Latif, A.A.; Mullen, J.L.; Alkuzweny, M.; Hufbauer, E.; Tsueng, G.; Haag, E.; Zeller, M.; Aceves, C.M.; Zaiets, K. Outbreak. info genomic reports: Scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat. Methods 2023, 20, 512–522. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Risk Evaluation of for SARS-CoV-2 Variant Under Monitoring: XEC. Available online: https://www.who.int/publications/m/item/risk-evaluation-of-for-sars-cov-2-variant-under-monitoring-xec (accessed on 12 February 2025).
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky Pond, S.L.; Scheffler, K. FUBAR: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Uriu, K.; Kosugi, Y.; Okumura, K.; Yamasoba, D.; Uwamino, Y.; Kuramochi, J.; Sadamasu, K.; Yoshimura, K.; Asakura, H. Virological characteristics of the SARS-CoV-2 KP. 2 variant. Lancet Infect. Dis. 2024, 24, e416. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.; Mellis, I.A.; Wu, M.; Mohri, H.; Gherasim, C.; Valdez, R.; Purpura, L.J.; Yin, M.T.; Gordon, A. Antibody evasiveness of SARS-CoV-2 subvariants KP. 3.1. 1 and XEC. Cell Rep. 2025, 44, 115543. [Google Scholar] [CrossRef]
- Aljabali, A.A.A.; Lundstrom, K.; Hromić-Jahjefendić, A.; Abd El-Baky, N.; Nawn, D.; Hassan, S.S.; Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications. Preprints 2025. [Google Scholar] [CrossRef]
- World Health Organization. COVID-19 Epidemiological Update—12 March 2025. Available online: https://www.who.int/publications/m/item/covid-19-epidemiological-update-edition-177 (accessed on 3 May 2025).
- World Health Organization. Executive Summary Initial Risk Evaluation of XEC. Available online: https://www.who.int/docs/default-source/coronaviruse/09122024_xec_ire.pdf?sfvrsn=13695ab6_2 (accessed on 3 May 2025).
- Centers for Disease Control and Prevention. SARS-CoV-2 Variant XEC Increases as KP.3.1.1 Slows. Available online: https://www.cdc.gov/ncird/whats-new/sars-cov-2-variant-xec-increases-as-kp-3-1-1-slows.html (accessed on 12 February 2025).
- Souza, U.J.B.d.; Spilki, F.R.; Tanuri, A.; Roehe, P.M.; Campos, F.S. Two Years of SARS-CoV-2 Omicron Genomic Evolution in Brazil (2022–2024): Subvariant Tracking and Assessment of Regional Sequencing Efforts. Viruses 2025, 17, 64. [Google Scholar] [CrossRef]
- COVID Data Tracker. Center for Disease Control and Prevention [Online]. 2022. Available online: https://covid.cdc.gov/covid-data-tracker/#datatracker-home (accessed on 12 June 2022).
- Rubin, R. What to Know About XEC, the New SARS-CoV-2 Variant Expected to Dominate Winter’s COVID-19 Wave. JAMA 2024, 332, 1961–1962. [Google Scholar] [CrossRef]
- Rizzo-Valente, V.S.; Oliveira, J.S.; Vizzoni, V.F.; Rizzo-Valente, V.S.; do Brasil, M.; Oliveira, J.S. XEC: International spread of a new sublineage of Omicron SARS-CoV-2. Authorea 2024. [Google Scholar] [CrossRef]
- Seemann, T.; Lane, C.R.; Sherry, N.L.; Duchene, S.; Gonçalves da Silva, A.; Caly, L.; Sait, M.; Ballard, S.A.; Horan, K.; Schultz, M.B. Tracking the COVID-19 pandemic in Australia using genomics. Nat. Commun. 2020, 11, 4376. [Google Scholar] [CrossRef]
- Li, J.; Lai, S.; Gao, G.F.; Shi, W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature 2021, 600, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.K. Genesis of Recombinant XEC Variant and Comparable SWISS-Modelling of Spike of LB. 1.7 and KP. 3.1. 1 Subvariants Coronaviruses. SunText Rev. Virol. 2024, 5, 152. [Google Scholar]
- Tegally, H.; San, J.E.; Cotten, M.; Moir, M.; Tegomoh, B.; Mboowa, G.; Martin, D.P.; Baxter, C.; Lambisia, A.W.; Diallo, A. The evolving SARS-CoV-2 epidemic in Africa: Insights from rapidly expanding genomic surveillance. Science 2022, 378, eabq5358. [Google Scholar] [CrossRef]
- Ozer, E.A.; Simons, L.M.; Adewumi, O.M.; Fowotade, A.A.; Omoruyi, E.C.; Adeniji, J.A.; Olayinka, O.A.; Dean, T.J.; Zayas, J.; Bhimalli, P.P. Multiple expansions of globally uncommon SARS-CoV-2 lineages in Nigeria. Nat. Commun. 2022, 13, 688. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.C. Genomic surveillance for SARS-CoV-2 variants: Circulation of omicron lineages—United States, January 2022–May 2023. MMWR. Morb. Mortal. Wkly. Rep. 2023, 72, 651–656. [Google Scholar] [CrossRef]
- Hussain, A.; Hussain, A.; Eldaif, W.A.H.; Rashid, M. The XEC COVID-19 Variant: A Global Threat Demanding Immediate Action. Coronaviruses 2024, in press. [Google Scholar] [CrossRef]
- Branda, F.; Ciccozzi, M.; Scarpa, F. On the new SARS-CoV-2 variant KP. 3.1. 1: Focus on its genetic potential. Infect. Dis. 2024, 56, 903–906. [Google Scholar] [CrossRef]
- Branda, F.; Ciccozzi, M.; Scarpa, F. Features of the SARS-CoV-2 KP. 3 variant mutations. Infect. Dis. 2024, 56, 894–896. [Google Scholar] [CrossRef]
- Fossum, E.; Vikse, E.L.; Robertson, A.H.; Wolf, A.-S.; Rohringer, A.; Trogstad, L.; Mjaaland, S.; Hungnes, O.; Bragstad, K. Low levels of neutralizing antibodies against SARS-CoV-2 KP. 3.1. 1 and XEC in serum from seniors in May 2024. Influenza Other Respir. Viruses 2024, 19, e70102. [Google Scholar] [CrossRef]
- GISAID. Tracking of hCoV-19 Variants. Available online: https://gisaid.org/hcov19-variants/ (accessed on 13 February 2025).
- Waafira, A.; Subbaram, K.; Faiz, R.; Naher, Z.U.; Manandhar, P.L.; Ali, S. A new and more contagious XEC subvariant of SARS-CoV-2 may lead to massive increase in COVID-19 cases. New Microbes New Infect. 2024, 62, 101517. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, F.; Byrd, J.B.; Yu, H.; Ye, X.; He, Y. Differential COVID-19 Symptoms Given Pandemic Locations, Time, and Comorbidities During the Early Pandemic. Front. Med. 2022, 9, 770031. [Google Scholar] [CrossRef] [PubMed]
- Padilla, S.; Ledesma, C.; García-Abellán, J.; García, J.A.; Fernández-González, M.; de la Rica, A.; Galiana, A.; Gutiérrez, F.; Masiá, M. Long COVID across SARS-CoV-2 variants, lineages, and sublineages. iScience 2024, 27, 109536. [Google Scholar] [CrossRef]
- Saltnes-Lillegård, C.; Rustøen, T.; Beitland, S.; Puntillo, K.; Hagen, M.; Lerdal, A.; Hofsø, K. Self-reported symptoms experienced by intensive care unit patients: A prospective observational multicenter study. Intensive Care Med. 2023, 49, 1370–1382. [Google Scholar] [CrossRef] [PubMed]
- Alhosni, F.; Al Qadire, M.; Omari, O.A.; Al Raqaishi, H.; Khalaf, A. Symptom prevalence, severity, distress and management among patients with chronic diseases. BMC Nurs. 2023, 22, 155. [Google Scholar] [CrossRef]
- Omori, T.; Hanafusa, M.; Kondo, N.; Miyazaki, Y.; Okada, S.; Fujiwara, T.; Kuramochi, J. Specific sequelae symptoms of COVID-19 of Omicron variant in comparison with non-COVID-19 patients: A retrospective cohort study in Japan. J. Thorac. Dis. 2024, 16, 3170–3180. [Google Scholar] [CrossRef]
- Kaku, Y.; Okumura, K.; Padilla-Blanco, M.; Kosugi, Y.; Uriu, K.; Hinay, A.A.; Chen, L.; Plianchaisuk, A.; Kobiyama, K.; Ishii, K.J. Virological characteristics of the SARS-CoV-2 JN. 1 variant. Lancet Infect. Dis. 2024, 24, e82. [Google Scholar] [CrossRef]
- Arora, P.; Happle, C.; Kempf, A.; Nehlmeier, I.; Stankov, M.V.; Dopfer-Jablonka, A.; Behrens, G.M.N.; Pöhlmann, S.; Hoffmann, M. Impact of JN.1 booster vaccination on neutralisation of SARS-CoV-2 variants KP.3.1.1 and XEC. Lancet Infect. Dis. 2024, 24, e732–e733. [Google Scholar] [CrossRef] [PubMed]
- Dadonaite, B.; Brown, J.; McMahon, T.E.; Farrell, A.G.; Figgins, M.D.; Asarnow, D.; Stewart, C.; Lee, J.; Logue, J.; Bedford, T.; et al. Spike deep mutational scanning helps predict success of SARS-CoV-2 clades. Nature 2024, 631, 617–626. [Google Scholar] [CrossRef]
- Liu, J.; Yu, Y.; Jian, F.; Yang, S.; Song, W.; Wang, P.; Yu, L.; Shao, F.; Cao, Y. Enhanced immune evasion of SARS-CoV-2 variants KP.3.1.1 and XEC through N-terminal domain mutations. Lancet Infect. Dis. 2025, 25, e6–e7. [Google Scholar] [CrossRef]
- Liu, J.; Yu, Y.; Yang, S.; Jian, F.; Song, W.; Yu, L.; Shao, F.; Cao, Y. Virological and antigenic characteristics of SARS-CoV-2 variants LF. 7.2. 1, NP. 1, and LP. 8.1. Lancet Infect. Dis. 2025, 25, e128–e130. [Google Scholar] [CrossRef]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; Robertson, D.L. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chuang, C.H.; Shen, T.F.; Lin, C.S.; Yang, H.P.; Li, H.C.; Chen, C.L.; Lin, I.F.; Chiu, C.H. Risk reduction analysis of mix-and-match vaccination strategy in healthcare workers during SARS-CoV-2 Omicron variant predominant period: A multi-center cohort study in Taiwan. Hum. Vaccin. Immunother 2023, 19, 2237387. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Kaku, Y.; Okumura, K.; Uriu, K.; Zhu, Y.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 LP. 8.1 variant. Lancet Infect. Dis. 2025, 25, e193. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, S.C.; Ando, N. X-rays in the Cryo-Electron Microscopy Era: Structural Biology’s Dynamic Future. Biochemistry 2018, 57, 277–285. [Google Scholar] [CrossRef]
- Link-Gelles, R. Early estimates of updated 2023–2024 (monovalent XBB. 1.5) COVID-19 vaccine effectiveness against symptomatic SARS-CoV-2 infection attributable to co-circulating Omicron variants among immunocompetent adults—Increasing Community Access to Testing Program, United States, September 2023–January 2024. MMWR. Morb. Mortal. Wkly. Rep. 2024, 73, 77–83. [Google Scholar]
- Wang, X.; Jiang, S.; Ma, W.; Li, X.; Wei, K.; Xie, F.; Zhao, C.; Zhao, X.; Wang, S.; Li, C. Enhanced neutralization of SARS-CoV-2 variant BA. 2.86 and XBB sub-lineages by a tetravalent COVID-19 vaccine booster. Cell Host Microbe 2024, 32, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Elneima, O.; Hurst, J.R.; Echevarria, C.; Quint, J.K.; Walker, S.; Siddiqui, S.; Novotny, P.; Pfeffer, P.E.; Brown, J.S.; Shankar-Hari, M. Long-term impact of COVID-19 hospitalisation among individuals with pre-existing airway diseases in the UK: A multicentre, longitudinal cohort study–PHOSP-COVID. ERJ Open Res. 2024, 10, 00982–2023. [Google Scholar] [CrossRef]
- Chatterjee, S.; Bhattacharya, M.; Nag, S.; Dhama, K.; Chakraborty, C. A Detailed Overview of SARS-CoV-2 Omicron: Its Sub-Variants, Mutations and Pathophysiology, Clinical Characteristics, Immunological Landscape, Immune Escape, and Therapies. Viruses 2023, 15, 167. [Google Scholar] [CrossRef]
- St. Louis, M.E.; Walke, H.; Perry, H.; Nsubuga, P.; White, M.E.; Dowell, S. Surveillance in Low-Resource Settings: Challenges and Opportunities in the Current Context of Global Health. In Principles and Practice of Public Health Surveillance, 3rd ed.; Oxford Academic: Oxford, UK, 2010. [Google Scholar] [CrossRef]
- Mack, A.; Choffnes, E.R.; Sparling, P.F.; Hamburg, M.A.; Lemon, S.M. Global Infectious Disease Surveillance and Detection: Assessing the Challenges" Finding Solutions: Workshop Summary; National Academies Press: Washington, DC, USA, 2007. [Google Scholar]
- Adesola, R.O.; Idris, I. Global health alert on the emergence of SARS-CoV-2 variants. Bull. Natl. Res. Cent. 2024, 48, 131. [Google Scholar] [CrossRef]
- Esonova, G.; Abdurakhimov, A.; Ibragimova, S.; Kurmaeva, D.; Gulomov, J.; Mirazimov, D.; Sohibnazarova, K.; Abdullaev, A.; Turdikulova, S.; Dalimova, D. Complete genome sequencing of SARS-CoV-2 strains that were circulating in Uzbekistan over the course of four pandemic waves. PLoS ONE 2024, 19, e0298940. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering, and Medicine. Wastewater-Based Disease Surveillance for Public Health Action; The National Academies Press: Washington, DC, USA, 2023. [Google Scholar]
- World Health Organization. COVID-19 Weekly Epidemiological Update, Edition 155. 10 August 2023. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---10-august-2023 (accessed on 13 February 2025).
- Cobar, O.; Cobar, S. Omicron Variants World Prevalence, WHO COVID-19 Dashboard, ECDC Communicable Disease Threat Report, and CDC COVID Data Tracker Review. 2024. Available online: https://www.researchgate.net/profile/Oscar-Cobar/publication/381638924_Omicron_Variants_World_Prevalence_168_WHO_COVID-19_Dashboard_ECDC_Communicable_Disease_Threat_Report_and_CDC_COVID_Data_Tracker_Review/links/6677cdd2d21e220d89c92086/Omicron-Variants-World-Prevalence-168-WHO-COVID-19-Dashboard-ECDC-Communicable-Disease-Threat-Report-and-CDC-COVID-Data-Tracker-Review.pdf (accessed on 13 February 2025).
- Bocharov, G.; Volpert, V.; Ludewig, B.; Meyerhans, A. Modelling of Experimental Infections. In Mathematical Immunology of Virus Infections; Springer: Cham, Germany, 2018; pp. 97–152. [Google Scholar] [CrossRef]
- Herzog, S.A.; Blaizot, S.; Hens, N. Mathematical models used to inform study design or surveillance systems in infectious diseases: A systematic review. BMC Infect. Dis. 2017, 17, 775. [Google Scholar] [CrossRef]
- Khoury, D.S.; Schlub, T.E.; Cromer, D.; Steain, M.; Fong, Y.; Gilbert, P.B.; Subbarao, K.; Triccas, J.A.; Kent, S.J.; Davenport, M.P. Correlates of Protection, Thresholds of Protection, and Immunobridging among Persons with SARS-CoV-2 Infection. Emerg. Infect. Dis. 2023, 29, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Cai, R.; Zhang, L.; Zhang, J.; Zhang, Z.; Zhu, A.; Li, H.; Zhuang, Z.; Chen, L.; Chen, J.; et al. In vivo determination of protective antibody thresholds for SARS-CoV-2 variants using mouse models. Emerg. Microbes Infect. 2025, 14, 2459140. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Y.; Casner, R.G.; Yu, J.; Nair, M.S.; Ho, J.; Reddem, E.R.; Tzang, C.C.; Huang, Y.; Shapiro, L. Optimizing a Human Monoclonal Antibody for Better Neutralization of SARS-CoV-2. bioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Hashemian, S.M.R.; Sheida, A.; Taghizadieh, M.; Memar, M.Y.; Hamblin, M.R.; Bannazadeh Baghi, H.; Sadri Nahand, J.; Asemi, Z.; Mirzaei, H. Paxlovid (Nirmatrelvir/Ritonavir): A new approach to COVID-19 therapy? Biomed. Pharmacother. 2023, 162, 114367. [Google Scholar] [CrossRef]
- Shahzad, M.; Upshur, R.; Donnelly, P.; Bharmal, A.; Wei, X.; Feng, P.; Brown, A.D. A population-based approach to integrated healthcare delivery: A scoping review of clinical care and public health collaboration. BMC Public Health 2019, 19, 708. [Google Scholar] [CrossRef]
- Wang, X.; Kattan, M.W. Cohort Studies: Design, Analysis, and Reporting. Chest 2020, 158, S72–S78. [Google Scholar] [CrossRef]
- Domingo-Fernández, D.; Baksi, S.; Schultz, B.; Gadiya, Y.; Karki, R.; Raschka, T.; Ebeling, C.; Hofmann-Apitius, M.; Kodamullil, A.T. COVID-19 Knowledge Graph: A computable, multi-modal, cause-and-effect knowledge model of COVID-19 pathophysiology. Bioinformatics 2021, 37, 1332–1334. [Google Scholar] [CrossRef]
- Vasireddy, D.; Vanaparthy, R.; Mohan, G.; Malayala, S.V.; Atluri, P. Review of COVID-19 Variants and COVID-19 Vaccine Efficacy: What the Clinician Should Know? J. Clin. Med. Res. 2021, 13, 317–325. [Google Scholar] [CrossRef]
- Forchette, L.; Sebastian, W.; Liu, T. A Comprehensive Review of COVID-19 Virology, Vaccines, Variants, and Therapeutics. Curr. Med. Sci. 2021, 41, 1037–1051. [Google Scholar] [CrossRef]
- Pérez-Vargas, J.; Worrall, L.J.; Olmstead, A.D.; Ton, A.-T.; Lee, J.; Villanueva, I.; Thompson, C.A.H.; Dudek, S.; Ennis, S.; Smith, J.R.; et al. A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants. Emerg. Microbes Infect. 2023, 12, 2246594. [Google Scholar] [CrossRef]
- Büyükköroğlu, G.; Şenel, B. Chapter 16-Engineering Monoclonal Antibodies: Production and Applications. In Omics Technologies and Bio-Engineering; Barh, D., Azevedo, V., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 353–389. [Google Scholar]
- Stone, C.A.; Spiller, B.W.; Smith, S.A. Engineering therapeutic monoclonal antibodies. J. Allergy Clin. Immunol. 2024, 153, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.; Akl, E.A.; Duda, S.; Solo, K.; Yaacoub, S.; Schünemann, H.J.; El-Harakeh, A.; Bognanni, A.; Lotfi, T.; Loeb, M. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: A systematic review and meta-analysis. Lancet 2020, 395, 1973–1987. [Google Scholar] [CrossRef] [PubMed]
- Um, S.; Lee, R.S. Effects of KF94 Face Mask on Cardiopulmonary Function and Subjective Sensation During Graded Exercise: A Comparison of KF94 2D and 3D Face Masks. In Proceedings of the Sports Analytics: First International Conference, ISACE 2024, Paris, France, 12–13 July 2024; pp. 302–312. [Google Scholar]
- Collins, A.P.; Service, B.C.; Gupta, S.; Mubarak, N.; Zeini, I.M.; Osbahr, D.C.; Romeo, A.A. N95 respirator and surgical mask effectiveness against respiratory viral illnesses in the healthcare setting: A systematic review and meta-analysis. J. Am. Coll. Emerg. Physicians Open 2021, 2, e12582. [Google Scholar] [CrossRef]
- Bhatia, B.; Sonar, S.; Khan, S.; Bhattacharya, J. Pandemic-Proofing: Intercepting Zoonotic Spillover Events. Pathogens 2024, 13, 1067. [Google Scholar] [CrossRef]
- Pauciullo, S.; Zulian, V.; La Frazia, S.; Paci, P.; Garbuglia, A.R. Spillover: Mechanisms, Genetic Barriers, and the Role of Reservoirs in Emerging Pathogens. Microorganisms 2024, 12, 2191. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.T.; Larson, J.C.; Buckingham, S.L.; Maton, K.I.; Crowley, D.M. Bridging the research-policy divide: Pathways to engagement and skill development. Am. J. Orthopsychiatry 2019, 89, 434–441. [Google Scholar] [CrossRef]
- Françoise, M.; Frambourt, C.; Goodwin, P.; Haggerty, F.; Jacques, M.; Lama, M.-L.; Leroy, C.; Martin, A.; Calderon, R.M.; Robert, J.; et al. Evidence based policy making during times of uncertainty through the lens of future policy makers: Four recommendations to harmonise and guide health policy making in the future. Arch. Public Health 2022, 80, 140. [Google Scholar] [CrossRef]
| Mutation | Location | Source Lineage | Functional Role | Reference |
|---|---|---|---|---|
| T22N | Spike (NTD) | KS.1.1 | Creates glycosylation site; enhances immune evasion | [29] |
| F59S | Spike (NTD) | De novo | Enhances infectivity via structural changes | [35] |
| Q493E | Spike (RBD) | KP.3.3 | Increases ACE2 affinity; immune escape | [36] |
| VuMs | No. of Nations | No. of Submitted Sequences | Prevalence (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Epiweek 37 | Epiweek 47 | Epiweek 34 | Epiweek 35 | Epiweek 30 | Epiweek 40 | Epiweek 37 | Epiweek 44 | Epiweek 46 | Epiweek 47 | ||
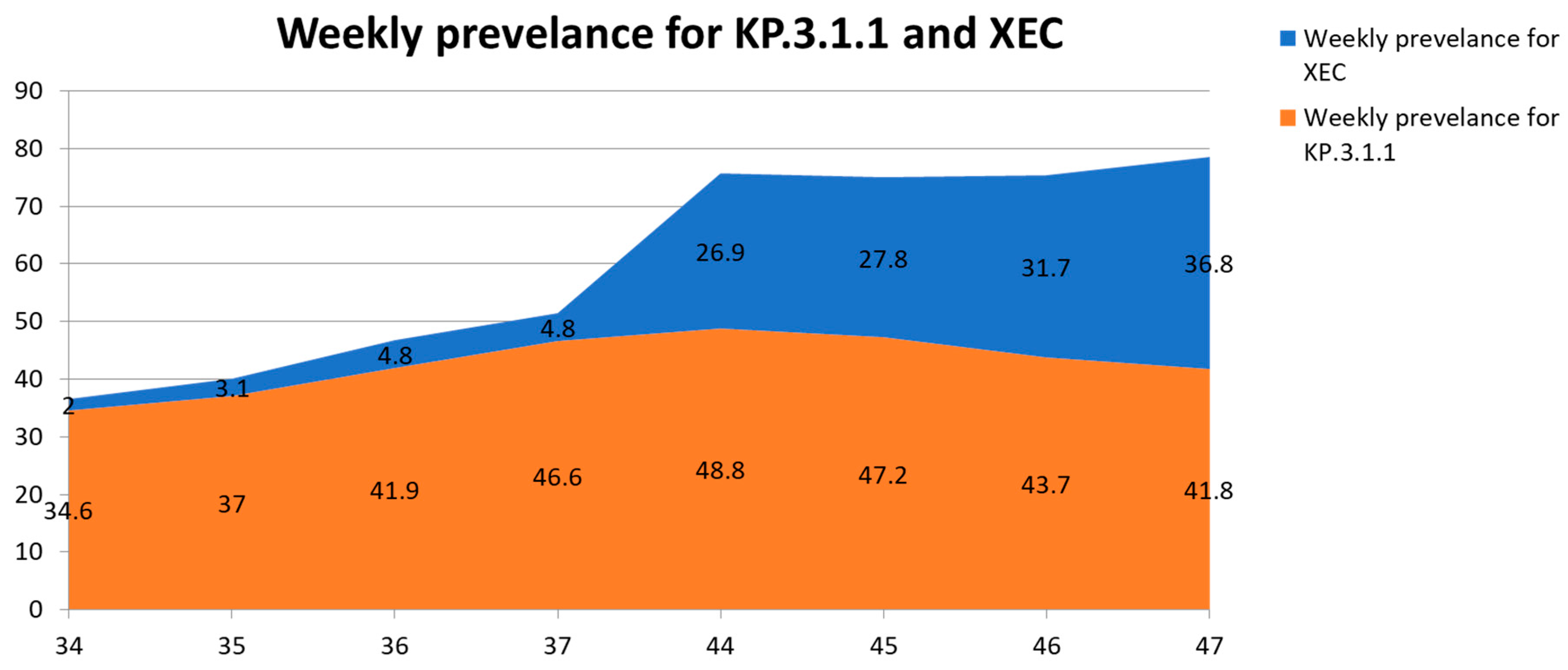
| XEC | 29 | 1263 | 13,331 | 2 | 3.1 | 4.8 | 4.8 | 26.9 | 27.8 | 31.7 | 36.8 |
| KP.3.1.1 | 54 | 29,557 | 65,234 | 34.6 | 37 | 41.9 | 46.6 | 48.8 | 47.2 | 43.7 | 41.8 |
| KP.3 | 70 | 45,327 | 56,177 | 18.8 | 18.4 | 16.9 | 14.4 | 8.4 | 7.7 | 8.2 | 6.1 |
| KP.2 | 82 | 27,976 | 33,287 | 12 | 10.7 | 8.9 | 8.1 | 1.3 | 1.3 | 1.2 | 0.9 |
| LB.1 | 73 | 12,675 | 16,166 | 6.9 | 7.3 | 6.3 | 6.3 | 0.9 | 0.9 | 0.6 | 0.6 |
| JN.1.18 | 89 | 6318 | 7962 | 2.3 | 2 | 1.7 | 1.2 | 1.1 | 1 | 1.2 | 0.9 |
| VUMs | Region | Epiweek 34 * | Epiweek 37 * |
|---|---|---|---|
| XEC | Europe | 5.3 | 12.0 |
| The Western Pacific | 0.2 | 2.0 | |
| The Americas | 0.9% | 2.8% | |
| Eastern Mediterranean region, Africa, Southeast Asia | Not reported | Not reported | |
| KP.3.1.1 | Europe | 48.2 | 50.4 |
| The Western Pacific | 13.5 | 24.2 | |
| The Americas | 34.1 | 49.2 | |
| Southeast Asia | A single reported sequence | A single reported sequence |
| Country | No. of All Sequenced SARS-CoV-2 Cases | No. of XEC Cases |
|---|---|---|
| Netherlands | 10,440 | 69 |
| Denmark | 11,425 | 92 |
| Slovenia | 1483 | 19 |
| Germany | 6891 | 99 |
| Czech Republic | 542 | 8 |
| Italy | 7065 | 35 |
| France | 21,277 | 110 |
| Sweden | 12,910 | 72 |
| Austria | 2238 | 13 |
| Ireland | 5475 | 36 |
| UK | 43,625 | 122 |
| Portugal | 1749 | 5 |
| Luxembourg | 1483 | 5 |
| Poland | 2377 | 9 |
| Croatia | 970 | 4 |
| Israel | 4370 | 5 |
| Belgium | 3367 | 4 |
| Australia | 16,260 | 22 |
| Finland | 2699 | 4 |
| Canada | 64,554 | 140 |
| Spain | 26,388 | 61 |
| Norway | 768 | 2 |
| Hong Kong | 972 | 1 |
| USA | 186,795 | 168 |
| Taiwan | 1399 | 1 |
| South Korea | 20,753 | 4 |
| Japan | 28,004 | 3 |
| Brazil | 10,547 | 1 |
| China | 17,640 | 1 |
| Variants | Ages 0–4 | Ages 5–11 | Ages 12–19 | Ages 20–39 | Ages 40–59 | Ages 60–79 | Ages 80 and Over | Total |
|---|---|---|---|---|---|---|---|---|
| XEC | 32.3% | 25.0% | 26.7% | 27.8% | 32.7% | 32.4% | 31.9% | 31.9% |
| KP.3.1.1 | 30.1% | 25.0% | 13.3% | 38.3% | 30.7% | 33.1% | 31.9% | 32.3% |
| Other Omicron | 18.3% | 25.0% | 20.0% | 13.6% | 16.5% | 11.0% | 11.2% | 11.9% |
| Other recombinant | 1.1% | 0.0% | 0.0% | 1.2% | 1.2% | 1.1% | 1.0% | 1.0% |
| Variants | Age 0–4 | Age 5–11 | Age 12–19 | Age 20–39 | Age 40–59 | Age 60–79 | Age 80 and over | Total |
|---|---|---|---|---|---|---|---|---|
| XEC | 32.3% | 25.0% | 26.7% | 27.8% | 32.7% | 32.4% | 31.9% | 31.9% |
| KP.3.1.1 | 30.1% | 25.0% | 13.3% | 38.3% | 30.7% | 33.1% | 31.9% | 32.3% |
| Other Omicron | 18.3% | 25.0% | 20.0% | 13.6% | 16.5% | 11.0% | 11.2% | 11.9% |
| Other recombinant | 1.1% | 0.0% | 0.0% | 1.2% | 1.2% | 1.1% | 1.0% | 1.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aljabali, A.A.A.; Lundstrom, K.; Hromić-Jahjefendić, A.; El-Baky, N.A.; Nawn, D.; Hassan, S.S.; Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications. Viruses 2025, 17, 985. https://doi.org/10.3390/v17070985
Aljabali AAA, Lundstrom K, Hromić-Jahjefendić A, El-Baky NA, Nawn D, Hassan SS, Rubio-Casillas A, Redwan EM, Uversky VN. The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications. Viruses. 2025; 17(7):985. https://doi.org/10.3390/v17070985
Chicago/Turabian StyleAljabali, Alaa A. A., Kenneth Lundstrom, Altijana Hromić-Jahjefendić, Nawal Abd El-Baky, Debaleena Nawn, Sk. Sarif Hassan, Alberto Rubio-Casillas, Elrashdy M. Redwan, and Vladimir N. Uversky. 2025. "The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications" Viruses 17, no. 7: 985. https://doi.org/10.3390/v17070985
APA StyleAljabali, A. A. A., Lundstrom, K., Hromić-Jahjefendić, A., El-Baky, N. A., Nawn, D., Hassan, S. S., Rubio-Casillas, A., Redwan, E. M., & Uversky, V. N. (2025). The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications. Viruses, 17(7), 985. https://doi.org/10.3390/v17070985









