Abstract
Phage display has advanced the discovery of peptides that selectively bind to a wide variety of cell surface molecules, offering new modalities to modulate disease-related protein–protein interactions (PPIs). These cell-binding peptides occupy a unique pharmaceutical space between small molecules and large biologics, and their growing popularity has opened up new avenues for targeting cell surface proteins that were previously considered undruggable. This work provides an overview of methods for identifying cell-selective peptides using phage display combinatorial libraries, covering in vitro, ex vivo, and in vivo biopanning approaches. It addresses key considerations in library design, including the peptide conformation (linear vs. cyclic) and length, and highlights examples of clinically approved peptides developed through phage display. It also discusses the on-phage chemical cyclization of peptides to overcome the limitations of genetically encoded disulfide bridges and emphasizes advances in combining next-generation sequencing (NGS) with phage display to improve peptide selection and analysis workflows. Furthermore, due to the often suboptimal binding affinity of peptides identified in phage display selections, this article discusses affinity maturation techniques, including random mutagenesis and rational design through structure–activity relationship (SAR) studies to optimize initial peptide candidates. By integrating these developments, this review outlines practical strategies and future directions for harnessing phage display in targeting challenging cell surface proteins.
1. Introduction
Forty years ago, in the spring of 1985, a paper was published in Science that promised the birth of a new technology [1]. This novel technical achievement, which was later called phage display, laid the foundation for numerous scientific discoveries in the years that followed. Phage display involves expressing foreign peptides or proteins on the surface of bacteriophages. In its four-decade-long existence, this molecular display technique has made a significant contribution to the field of peptide discovery, providing high-diversity, genetically encoded libraries [2,3] that can be searched to discover binders to a variety of biologically relevant targets. Cell surface proteins are one of the most important biological targets widely used in phage display studies. In this context, phage display enables the identification of peptide ligands capable of binding to proteins involved in critical biological processes and disease pathways. The selective enrichment of high-affinity peptides is achieved through the affinity selection of phage display libraries on cell surface proteins. Cell surface protein-binding peptides, as short amino acid sequences selected from phage display combinatorial libraries, allow scientists to probe, map, and ultimately manipulate signal transduction pathways. By performing the library selection on cell surface proteins and receptors that are either recombinantly produced or are present on intact cells and tissues, phage display offers a highly adaptable platform for isolating peptides that selectively bind to biomolecules associated with a broad range of diseases. Applications of this technology span different scientific areas, such as oncology, immunology, metabolomics, and neurology. In addition to direct biological functions on cells, cell-binding peptides identified by phage display also hold great potential for developing targeted drug delivery [4,5] and imaging [6,7] modalities. Today, phage display has matured into a powerful approach for developing peptides with potential diagnostic and therapeutic applications [8].
The capacity of phage display to identify selective binders to diverse cell surface proteins has not only advanced our understanding of cell biology and disease mechanisms but has also had a profound impact on the identification of therapeutic peptides [9,10,11]. This interface between peptide selection and clinical translation introduces phage display as a powerful discovery engine that drives the development of next-generation peptide-based therapeutics, constituting a dynamic and rapidly evolving area of biomedical research. Therapeutic peptides represent a distinct class of pharmacological agents with molecular weights typically ranging from 500 to 5000 Daltons and occupy a unique pharmaceutical space between small molecules and large biologics [12]. The research on therapeutic peptides began with early studies on natural human hormones—such as insulin, oxytocin, vasopressin, somatostatin, and the gonadotropin-releasing hormone—all of which are short peptides, and the investigation of their specific physiological functions in the body [13]. Insulin was the first commercialized peptide drug, developed by Eli Lilly and Company, and received FDA approval in 1923. The groundbreaking success of insulin as a drug sparked widespread public interest in peptide therapeutics, paving the way for other animal-derived peptide drugs to enter clinical applications. Notable examples include the adrenocorticotropic hormone and calcitonin, which became important early contributors to the peptide pharmacopeia. The second half of the 20th century (from the 1950s to the 1990s) witnessed the discovery and characterization of an increasing number of peptide hormones and their receptors with therapeutic potential. The onset of the 21st century marked a new era in peptide drug development, driven by substantial breakthroughs in structural biology, omics technologies, recombinant methodologies, and novel synthetic and analytical methods. Since 2000, over 30 non-insulin peptide drugs have received regulatory approval globally [14]. In recent decades, peptides have gained increasing recognition as biologically active and therapeutically valuable molecules. Their favorable pharmacological characteristics and intrinsic features have positioned them as attractive candidates in modern drug development pipelines. Peptides present multiple advantages for clinical applications, such as high specificity, favorable cell and tissue penetration, easy synthesis, low production costs, reduced immunogenicity, and amenability to modifications [12,15,16,17]. Over time, the therapeutic applications of peptides have progressed steadily and continue to evolve with ongoing advancements in drug design and treatment strategies. The development of peptide drugs remains a hot topic in pharmaceutical research. The soaring popularity of peptide therapeutics has opened up new avenues for creating novel platforms that can modulate undruggable targets associated with a wide range of diseases [18,19]. The current review aims to provide an overview of the current knowledge on phage display and its application for the discovery and refinement of peptides binding to cell surface proteins. We begin by discussing the foundational principles of phage display technology and the molecular architecture of peptide libraries, with a particular focus on the different conformations (linear vs. cyclic) and lengths of displayed peptides during library design. We then explore the various modes of target presentation in the affinity selection of phage display peptide libraries and investigate how the biological context of the target impacts the selection outcome. Finally, we discuss in detail the various approaches that are employed to mature the binding affinity of peptides isolated from phage display libraries, making them optimal for the potential translation into the clinic.
2. Phage Display and Combinatorial Peptide Libraries
Bacteriophages, also known as phages, are viruses that specifically infect bacteria. Phage display was born in the mid-1980s, when George Smith introduced the concept that filamentous bacteriophage can serve as an expression vector capable of displaying antigens cloned into the phage genome on the virion surface [1]. He inserted foreign DNA fragments into gene III of the f1 phage and demonstrated that, as the phage assembles inside the host bacterium and is extruded from the cell, the foreign sequences are displayed on the phage surface as fusions to the pIII coat protein without disrupting the infectivity of the phage particle. The peptides encoded by the inserted sequences were in an accessible form and could directly interact with a target of interest. Since then, various phages, such as T4 [20], T7 [21], λ (lambda) [22], MS2 [23], and Qβ [24], have been used for display purposes. However, filamentous phages of the Ff family, including M13, fd, and f1, remain the most widely adopted platforms for the display of biomolecules, with M13 as the most predominant in the field [25,26]. The dominance of Ff bacteriophages over other phages for ligand display is attributed to their ability to achieve titers higher than those of other known phages, an efficient transformation that enables the generation of diverse libraries, and the robust stability of their virions under high temperatures, detergents, and extreme pH conditions. These properties collectively facilitate a broad range of biopanning conditions for the affinity selection of phage display libraries [26]. Additionally, these phages exhibit great genetic flexibility, making them well-suited for genetic engineering. This flexibility allows the enlargement of the genome size to be accompanied by a proportional increase in both the length of the phage particle and the number of pVIII molecules. As a result, the encapsulated DNA does not impose a limitation on the size of the phage particle [27]. These filamentous phages infect F plasmid-containing Gram-negative bacteria, like Escherichia coli, and follow a non-lytic life cycle. After the replication of the phage DNA, the translation of phage proteins, and the assembly of phage particles, new virions are released from infected bacteria without causing the lysis of the host cell. Although all five coat proteins of the M13 phage have been used for the surface presentation of foreign amino acids, pIII (one of the minor coat proteins with 406 residues) and pVIII (the major coat protein with 50 residues) are the most important coat proteins that have been broadly employed for peptide display [28]. The integration of the idea of combinatorial chemistry [29] with phage display led to the generation of phage display peptide libraries [2,3]. These combinatorial phage libraries house a large repertoire of peptide sequences that can be searched to identify ligands with desired binding characteristics. In phage display libraries, the physical linkage between the displayed peptide and the nucleotide sequence that encodes it, known as the genotype–phenotype link, is a foundational pillar of the technique that enables the sequence identification of displayed peptides with sought-after binding features [26,30]. Figure 1 illustrates the general structure of the M13 phage with its coat proteins and the physical linkage between the genotype and phenotype in phage display.
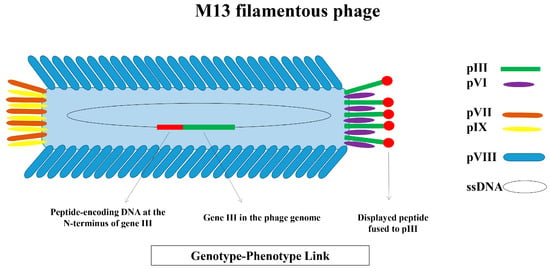
Figure 1.
The structure of the M13 filamentous phage, and the schematic illustration of the genotype–phenotype link M13 is a rod-shaped bacteriophage with a single-stranded DNA (ssDNA) genome encapsulated by five coat proteins. Four minor coat proteins (pIII and pVI at one end; pVII and pIX at the other) are present in 3 to 5 copies per virion, while the major coat protein (pVIII) is present in approximately 2700 copies and covers the phage body. M13 is the most widely used bacteriophage for the surface display of ligands, including peptides. In phage display, foreign peptide-encoding DNA is typically inserted at the N-terminus of gene III, resulting in the expression of the peptide on the phage surface while fused to the pIII coat protein. This physical linkage between the encoding DNA (genotype) and the displayed peptide (phenotype) enables the sequence identification of peptides displayed on phages isolated during biopanning.
Peptides can adopt different structural conformations on the virion surface, which must be carefully considered in the design of phage display libraries. Linear libraries consist of peptides of varying lengths, typically ranging from fewer than 10 to over 40 amino acids. The linear form offers structural simplicity, allowing an easier direct interaction with target molecules. In contrast, cyclic libraries are generated by introducing cysteine residues into the peptide sequences, resulting in the formation of intramolecular disulfide bonds [31]. These covalent linkages create looped structures and diverse folded conformations in the displayed peptides. Peptides with such secondary structures provide advantages for target binding. The conformational constraint imparted by disulfide bridges often enhances the binding affinity and selectivity of displayed peptides [32,33], which is particularly beneficial when targeting complex binding sites or increasing the peptide half-life. Disulfide bridges are a key structural feature in many natural miniproteins, such as scorpion venoms and plant toxins [34]. To date, dozens of cyclic peptides have received FDA approval. A comprehensive list of FDA-approved cyclic peptide drugs in the past two decades and their respective indications can be found in [35]. More specifically, several clinically approved peptide drugs with cyclic structures have been developed employing phage display technology. Notably, pegcetacoplan, used for the treatment of paroxysmal nocturnal hemoglobinuria (PNH), is a pegylated cyclic peptide targeting and inhibiting complement protein C3, which was discovered and optimized through phage display [36]. Ecallantide, approved for the treatment of acute hereditary angioedema (HAE), is a human plasma kallikrein inhibitor containing three intramolecular disulfide bonds identified through selection of phage display peptide libraries [37]. In addition to the cyclic peptide drugs already approved, several promising candidates are in late-stage clinical development and may receive regulatory approval in the near future. These examples illustrate the expanding role of cyclic peptides in modern drug discovery, which offer robust and effective therapeutic options across a variety of medical areas. The advent and further advancements in display technologies, such as phage display, along with the development of sophisticated synthetic chemistry approaches (e.g., automated synthesis platforms) have played a significant role in discovering and optimizing cyclic peptides. While disulfide bridges offer clear structural and functional advantages by constraining peptide conformation and enhancing binding affinity and specificity, their incorporation into phage-displayed peptide libraries is not without challenges. A key limitation is the susceptibility of disulfide bonds to reducing intracellular environments [38,39], which can lead to bond cleavage and conformational instability. The reducing conditions in the cytoplasm of host bacteria can disrupt the structural integrity of cyclic peptides. Additionally, the sensitivity of disulfide bridges to reducing environments poses a challenge for evaluating hits from such libraries in cell-based assays [39]. Cyclic conformation can also impair the propagation efficiency of phage particles [40], potentially due to rigid or bulky structures that are less compatible with the phage coat proteins. This incompatibility may compromise the proper phage assembly during the amplification of phage display libraries. Collectively, these effects can reduce the library diversity and ultimately impact the outcome of the selection in phage display experiments. Thus, while cyclic peptides offer structural and functional benefits, a careful design is required to balance the stability and library diversity in phage display. Peptides displayed on phages can also be cyclized by applying various chemical strategies. The development of methods for chemically modifying peptides displayed on phages has significantly expanded the range of accessible cyclic peptide architectures, thereby enhancing the structural diversity and pharmacological potential of phage display libraries [41]. Chemical cyclization on phages was first applied to generate bicyclic peptides [42]. These molecules are composed of two macrocyclic rings, formed by chemically linking specific residues within the peptide sequence. Compared to monocyclic peptides of a similar molecular weight, bicyclic peptides exhibit greater conformational rigidity, which often results in an enhanced binding affinity for their targets and an increased resistance to proteolytic degradation [43]. Phage-displayed peptides can be chemically converted into bicyclic structures by reacting three cysteine residues in the peptide with a reagent containing three thiol-reactive groups. Reagents were selected to ensure efficient cyclization through cysteine residues while preserving the structural integrity of phage coat proteins, thereby maintaining the phage infectivity. This strategy was used to induce bicyclization by treating a phage library, displaying the peptide with the ACX6CX6CG format fused to the p3 coat protein, with 1,3,5-tris(bromomethyl)benzene (TBMB). The subsequent biopanning of this library against the proteases plasma kallikrein and cathepsin G led to the isolation of bicyclic peptides with nanomolar-range inhibitory constants [42]. Beyond TBMB, other chemical reagents have been employed to generate cyclic peptides on phages. For instance, azobenzene linkers have been used to generate and identify light-responsive cyclic peptide ligands [44,45], while ortho-phthalaldehyde (OPA) has enabled the formation of isoindole-bridged cyclic peptides, which have been successfully applied to isolate ligands targeting several therapeutically relevant proteins [46].
The length of displayed peptides is another critical factor in the design of phage display libraries [47]. Even short peptides, when fully randomized, yield a library size that far exceeds the capacity of phage display systems. The theoretical sequence space increases exponentially with the peptide length. Given the 20 naturally occurring amino acids, the total number of possible sequences for a peptide of length N is 20N. However, the real sequence space that can be covered by a phage display library is typically around 108–109 unique variants, due to limitations in the transformation efficiency during the library construction [48]. This restriction means that the complete coverage of the sequence space is only achievable for peptides with up to six fully randomized positions (hexamer libraries). For peptides of seven or more residues, only a small fraction of the total sequence space is represented by the library. The fraction of the theoretical sequence space that can be represented by the library is dramatically reduced with the increased length of displayed peptides (Table 1). For example, a 20-mer library encompasses such a vast theoretical sequence space that only a minuscule portion (9.54 × 10−18) of the sequence space can be covered in a peptide library displayed on the phage.

Table 1.
The relationship between the number of fully randomized positions and the percentage of the whole sequence space that can be represented by a phage display random peptide library (known as the coverage percentage). The total number of possible sequences (theoretical sequence space) is calculated as 20N, where N is the number of fully randomized positions in the peptide library. The coverage percentage is determined by dividing 109, which is the maximum achievable diversity in phage display peptide libraries, by the theoretical sequence space (the 2nd column). In practice, the maximum diversity of 109 is difficult to reach, and thus, the coverage percentage is very likely to be lower than what is shown in the table. The full randomization is unbiased, meaning that each position of the library is allowed to vary completely, using all 20 possible amino acids. However, in the case of a partial randomization in which the variation in each position of the library is biased toward a subset of, not all, amino acids, the possible sequence space becomes smaller, and the coverage percentage represented by the library is significantly increased. Partial randomization is applied to build secondary libraries (described in Section 6).
Both the length and conformation of displayed peptides have also been demonstrated to significantly affect the propagation capacity of phage clones, potentially leading to unbalanced changes in the composition and diversity of the library and consequently a biased selection of target-binding peptides during biopanning [40]. Therefore, the careful balancing of the peptide length and conformational complexity is crucial to maximize the functional diversity represented in the library and to minimize the selection bias during biopanning.
3. Affinity Selection of Phage Display Peptide Libraries Through Biopanning
Biopanning is the core selection process used to isolate target-specific peptides from phage display libraries. It relies on affinity-based interactions between the target molecule and a vast phage-displayed peptide repertoire, often comprising millions of unique variants [3,49]. The process begins with the incubation of the phage library with the target of interest. Phage particles that do not bind, or bind weakly, are removed through a series of stringent washing steps. In contrast, strongly bound phages are retained and subsequently eluted. The eluted phage pool is then amplified by infecting the host bacteria, enabling the replication and secretion of new virions into the culture medium. This process is typically repeated over multiple rounds, enriching the population of phages displaying peptides with a high affinity and specificity for the target. After the enrichment, the DNA is extracted from the phage pool and sequenced to identify the peptide-coding regions. Conventionally, the identification of isolated peptide sequences in the biopanning output has been performed by plating the recovered phage pool and sequencing a limited number of viral plaques that appear on the plate. However, in recent years, a growing number of phage display studies are employing high-throughput sequencing (HTS), also known as next-generation sequencing (NGS), which enables the parallel sequencing of the DNA extracted from thousands to millions of isolated phage clones [50,51,52,53,54,55,56]. NGS-based phage display brings significant advantages over the conventional low-throughput sequencing of the biopanning output, yielding quantitative insights into the peptide composition of phage pools [57,58]. The incorporation of NGS into biopanning has challenged some of the core assumptions underlying phage display selection. For example, analyzing the sequence content of the naïve library by NGS has found a significant bias in the nucleotide and amino acid composition, highlighting that the unselected library is not truly random. Consequently, the peptide enrichment during biopanning is not solely driven by the binding affinity for the target. These deviations from randomness can skew the selection outcomes, leading to the erroneous isolation of false-positive hits or the failure to identify genuine binders [48]. The NGS-based sequencing of millions of phage clones from the biopanning output has also shown that a single round of biopanning can be sufficient to identify target-specific binders, thereby accelerating ligand discovery. Omitting further rounds significantly reduces amplification-associated biases, helping to preserve potentially high-affinity binders that might otherwise be lost [59]. Additionally, NGS enables the identification of rare target-binding peptide motifs and allows for a more accurate determination of consensus sequences and sub-groups of consensus sequences within the selected peptide pool [60]. Another important application of NGS is to enable us to uncover the corruption of the selection output, a phenomenon not captured by conventional Sanger sequencing [58]. Corruption can occur when certain clones become overrepresented during biopanning, not because of the true binding affinity for the target, but due to propagation advantages. These clones dominate the phage pool during biopanning rounds, resulting in the undesirable enrichment and isolation of nonspecific binders. The NGS-assisted detection of such corruption in earlier rounds of selection can inform the decision to terminate the biopanning or halt the downstream validation of isolated peptides. By overcoming some of the major bottlenecks of Sanger sequencing and providing deeper insights into the peptide composition of the selection output, NGS helps avoid the misidentification of nonspecifically enriched target-unrelated peptides (TUPs) and improves the reliability of biopanning findings [58]. The comprehensive profiling of peptide populations by NGS generates large sequence datasets that are ideal for training machine learning (ML) models. These models hold great potential for the computational deconvolution of selection noise (nonspecific binders) vs. true target-specific peptides, as well as the identification of sequence motifs or clusters associated with target binding. Before the biopanning on the target, a target-independent capture and elimination can also be incorporated into the selection protocol, intended to remove binders that interact with off-target molecules present in the selection system. The main objective of this modification, which is called negative or subtractive selection, is to deplete the library of binders to the components of the selection apparatus, thereby minimizing background noise caused by cross-reactivity [61,62,63]. This can be achieved by pre-incubating the library with the solid support containing all the elements of the selection apparatus except the target. Afterward, the subtracted phage supernatant retrieved from the negative selection is collected and used as the input for the positive selection, where the target is exposed to the subtracted library. To introduce a higher level of specificity to the ligand selection, a structurally similar control protein molecule that lacks the desired epitope(s) can be employed during the negative selection to discard library members binding nonspecifically to common features between the target and control molecules. When whole cells expressing the target receptor on their surface are used in library selection, control cells either not expressing or expressing very low levels of the target molecule are included in the negative selection [64]. Despite its benefits, the negative selection can bring down the diversity of the library, potentially eliminating useful candidates if performed in a highly stringent manner.
Once the amino acid sequences of identified peptides are known, the peptides are chemically synthesized, and their target binding properties are initially evaluated using a variety of in vitro assays. Peptides are selected against the target in biopanning while they are displayed on the phage. However, we should make sure that the free (synthetic) peptide retains the same binding capacity toward the target. Therefore, it is necessary to confirm the binding for the target while peptides are chemically synthesized and removed from the phage scaffold. After the initial characterization, peptides that show promising results proceed to ex vivo and in vivo assays. Ex vivo assays assess the peptide binding for the target in biological samples using cells or tissue extracts from animals or humans, providing a more physiologically relevant environment for the binding characterization. The final confirmation of the target-binding capacity before entering clinical trials is conducted in vivo by evaluating the peptide behavior and its target binding in living organisms using animal models. Different assays are employed at each stage—in vitro, ex vivo, and in vivo—of the binding characterization. This stepwise workflow for downstream characterization ensures that only the most promising candidates identified through biopanning advance to further characterization, thereby reducing costs and minimizing unnecessary testing. Figure 2 depicts the phage display biopanning workflow, including the iterative rounds of the library selection on the target, the sequence analysis of the biopanning output to identify candidate peptides with a potential target-binding capacity, and ultimately the downstream characterization to validate the target-binding specificity and affinity of candidate peptides through a variety of in vitro, ex vivo, and in vivo assays.
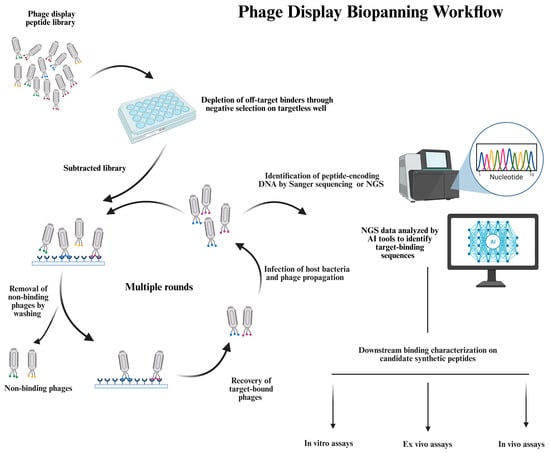
Figure 2.
A schematic overview of the phage display biopanning workflow and downstream validation. This figure illustrates the stepwise process of identifying target-specific peptides from a combinatorial phage display library. Initially, the library undergoes negative selection against a targetless solid support to remove phages that bind nonspecifically to the components of the selection system. The resulting subtracted library, depleted of off-target binders and obtained from the negative selection, is employed as input for the positive selection on the target of interest. After incubation, unbound or loosely bound phages are removed by stringent washing steps, and target-bound phages are eluted and then amplified in host bacterial cells to generate an enriched phage pool for subsequent rounds of selection. After multiple rounds of enrichment, the phage pool is subjected to DNA sequencing using either the Sanger approach or NGS. NGS offers a quantitative analysis of the peptide representation within the pool and enables data-driven interpretations using computational tools, such as artificial intelligence (AI) algorithms, to distinguish true binders from selection noise. One or more candidate peptides enriched in biopanning and identified through sequencing are chemically synthesized, and their target binding is investigated out of the phage scaffold through a stepwise workflow, starting with in vitro assays, proceeding to ex vivo analyses, and further advancing to in vivo testing in relevant animal models. This sequential validation ensures functional relevance and minimizes failures in subsequent validation phases, ultimately facilitating the discovery of target-selective peptide ligands.
4. Target Presentation in Biopanning
The choice of the target type is one of the most critical considerations in designing an effective biopanning strategy, as it directly influences peptide selection outcomes and the physiological relevance. The target employed in biopanning can be presented to the library in different modes (Figure 3). In this section, these modes are discussed critically, highlighting the advantages and disadvantages of each presentation format.
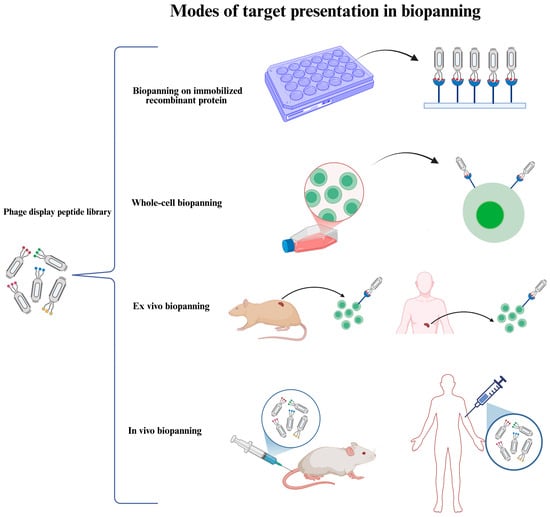
Figure 3.
Different modes of target presentation in biopanning The target employed in the selection of phage display peptide libraries might be presented in various modes: (1) directly adsorbed or indirectly immobilized purified recombinant protein (e.g., the extracellular domain of a cell surface protein), (2) present on cultured cells (whole-cell biopanning), (3) present on cells derived from animal or human tissues (ex vivo biopanning), and (4) present within the animal or human body (in vivo biopanning).
4.1. Biopanning on Recombinant Proteins
Due to its convenience, the use of the recombinantly produced cell surface protein is the most widely used approach for the target presentation in biopanning. However, the method utilized to immobilize the recombinant protein has a substantial impact on the success of the selection process [65,66]. The target cell surface protein can be tethered to a variety of solid supports, including multi-well polystyrene plates, polystyrene tubes, Petri dishes, magnetic beads, nitrocellulose surfaces, and monolithic columns [49].
A common strategy involves the direct or passive adsorption of the recombinant protein onto a solid support [66]. Although this method does not require any modification of the protein, it can induce some conformational changes or partial denaturation, resulting in occluded or inaccessible binding sites for ligands [67]. Consequently, some binders selected using this approach may fail to recognize the target cell surface protein in its native, soluble form. An alternative to direct adsorption is solution-phase biopanning, in which the target is first incubated with the phage library in a homogeneous solution, and the resulting phage–target complexes are subsequently captured on plates or magnetic beads. This two-step mode of capture allows for better control over binding kinetics without the complications associated with surface reactions [49]. The affinity capture in this method typically requires the target to carry an affinity tag. One common approach involves biotinylating the target and capturing the phage–target complex on a streptavidin-coated polystyrene plate. Biotin is a widely used affinity tag that captures target molecules based on a high-affinity interaction between biotinylated protein and streptavidin molecules coated on the solid support [3]. The phage–target complex can also be captured using magnetic beads specific for the target. The specificity of beads for the target is created by various affinity tags. For example, if the target protein is fused to affinity tags such as glutathione S-transferase (GST), the maltose-binding protein (MBP), or a polyhistidine tag, the phage–target complex can be captured using glutathione-, amylose-, or nickel-coated beads, respectively. To minimize the nonspecific selection of bead-binding peptides, a negative selection step can be included in the biopanning protocol by pre-incubating the phage library with the bead matrix in the absence of the target protein. Compared to the direct coating method, solution biopanning provides several advantages, including the enhanced accessibility of binding sites on the target, directional presentation of the target to the phage-displayed peptide with minimal disruption to its native conformation, reduced risk of the partial denaturation of the target due to the adsorption to the plastic surfaces, and significantly lower quantities of the target protein required per experiment [30].
4.2. Biopanning on Whole Cells
Whole cells can also be employed in the phage display selection. This type of biopanning can be performed on both suspension and adherent cells. In whole-cell biopanning, the target cell surface protein is present on live cells and thus retains its natural conformation, including proper folding, a quaternary structure (for multi-domain proteins), correct post-translational modifications, and interactions with neighboring molecules [68]. In addition, this method eliminates the need for the purification of the target protein. This is particularly advantageous when dealing with transmembrane cell surface receptors, as the purification of these proteins is difficult due to a significant number of hydrophobic residues [69,70]. Whole-cell biopanning is a potent strategy for isolating peptides that bind to cell surface proteins. However, the protocol of whole-cell biopanning can also be adapted to identify internalizing peptides [63,71,72,73]. This can be achieved by increasing the temperature of the library incubation with cells to 37 °C (instead of 4 °C for cell surface-bound phages) to allow for the receptor-mediated endocytosis of phages and their corresponding displayed peptides. It also needs a differential phage recovery that is performed through the surface stripping of target cells to remove extracellularly bound phages and then lysing the cells to release internalized ones. Cell-internalizing peptides facilitate the intracellular transport of molecules and therefore hold great potential for the targeted delivery of therapeutic cargoes into cells. As noted, whole-cell biopanning enables peptide selection in a more relevant cellular context with a preserved cell surface topography. Nevertheless, it deserves to be mentioned that cultured cells used in library selection have undergone a long-term culture along with repeated passaging, which can result in a wide variety of genetic mutations and chromosomal abnormalities. These cell proliferation-associated genotypic changes lead cultured cells to deviate from the original phenotype (a phenomenon known as phenotypic drift) [74,75], which might change the conformation, expression level, and other features of the target protein on the cell surface. Thus, cell-selective peptides identified through biopanning on cultured cells might demonstrate a low reproducibility and poor translational potential.
4.3. Ex Vivo Biopanning
Ex vivo target presentation represents another valuable approach in phage display selection. Ex vivo biopanning performed on animal- or patient-derived tissues offers several advantages over whole-cell biopanning [76,77,78]. This approach of library selection avoids the drawbacks associated with the long-term culture and continuous proliferations, providing a more physiologically relevant context due to the presence of a natural tissue architecture, including the extracellular matrix (ECM), cell–cell interactions, and vascular structures that are lost in whole-cell biopanning. These features increase the translational relevance of peptides identified through ex vivo phage display. In spite of these benefits, the access to fresh tissue samples is not easy for all research groups, tissue processing requirements yield a lower throughput compared to whole-cell biopanning, and the presence of multiple cell types in tissue samples causes heterogeneity versus more homogeneous cultured cells in whole-cell biopanning. These challenges can affect the reproducibility and consistency of the results of ex vivo phage display.
4.4. In Vivo Biopanning
Despite the promise of in vitro whole-cell and ex vivo biopanning, these selection schemes fail to fully capture the complex and varied environment of a living body, where factors like blood circulation, immune system activity, communications between organs, the diverse cellular populations, dynamic microenvironments, and in vivo physiological barriers all play important roles. In vivo phage display provides a more physiological platform for peptide selection by allowing phages to interact with the target within the full biological context of a living system [79]. In this selection modality, the phage library is injected into the tail vein of an animal and allowed to circulate for a short period, during which phage-displayed peptides can directly bind to specific targets on cells within tissues or organs. The animal then undergoes the perfusion of the left ventricle with saline to wash away unbound phages, followed by the euthanasia of the animal and the collection of the target tissues (e.g., xenograft tumors) or organs. These tissues are homogenized to recover bound phages. Phages retrieved from these homogenates are used to infect host bacteria for amplification. In the case of planning more than one round, the amplified phage pool is subsequently injected into another animal with the same genetic background. In vivo phage display is largely dependent on the more readily accessible vascular system of the target tissue or organ, and thus, the capillary vessels can pose a significant barrier for phage particles to traverse. Therefore, many peptides selected by using this approach tend to bind to endothelial cells of the vasculature of the target tissue or organ rather than the tissue or organ itself. Given the high degree of vascular heterogeneity among tissues, it is possible to isolate phages that bind to the differentially expressed cell surface proteins of endothelial cells in different tissues or organs [80]. By using in vivo phage display, peptides are selected in the complex physiological environment of an animal’s body with desired pharmacokinetic and pharmacodynamic characteristics. There are numerous studies in the literature reporting in vivo phage display in animal models, identifying a variety of vascular-targeting as well as tissue- or organ-targeting peptides [7,54,81,82,83,84,85,86]. However, in vivo phage display has rarely been reported in humans [81,87,88,89].
Each mode of the target presentation in phage display selection—recombinant protein, whole-cell, ex vivo, and in vivo systems—offers unique advantages and presents specific limitations. While recombinant proteins allow for high-throughput screening, they may lack physiological relevance. Whole-cell and ex vivo methods better preserve natural conformations but suffer from limitations in reproducibility or accessibility. In vivo biopanning, though powerful, is technically demanding and skewed toward vascular targets. Therefore, a combinatorial approach that integrates multiple presentation formats can significantly improve the identification of physiologically relevant, cell-selective peptides. Figure 3 provides a visual summary of the various modes of target presentation in the selection of phage display peptide libraries.
5. The Application of Phage Display for the Discovery of Cell-Selective Peptides
Targeting cell surface proteins is a powerful strategy for drug development [90,91]. Peptides are particularly promising tools for modulating these proteins. Here, we explore mechanisms by which peptides can interact with cell surface protein targets and then highlight the application of phage display technology for discovering cell-selective peptides. We aim to provide an overview of peptides identified over the past decade through the affinity selection of phage display libraries, highlighting the potential of this technology for developing therapeutic peptides for various diseases.
5.1. Peptides and Their Interactions with Cell Surface Proteins
Cell surface proteins play a crucial role in mediating biochemical and electrical communications between cells, enabling vital biological processes. Many transmembrane proteins and receptors have binding sites that are accessible to drugs, including peptides. The significance of these proteins and receptors in disease-associated cellular and molecular processes has created a huge interest in developing drugs targeting them [92,93,94]. Cell surface proteins do not act alone. They rely heavily on protein–protein interactions (PPIs) to transmit signals across the cell membrane. Advancements in omics technologies and structural biology have led to the discovery of many PPIs. PPIs play essential roles in regulating the mechanisms behind many cellular signaling pathways, and thus, their impaired regulation is a characteristic of many human diseases [95]. Cell surface-binding peptides hold potential as incredibly valuable tools in the development of therapeutics for a variety of diseases. Peptides might compete with the natural endogenous ligands to bind to the target cell surface protein and then interact with biologically relevant sites on the surface of the target molecule. Although this interaction involves a large contact surface, the majority of the binding energy is typically mediated by only a few key amino acid residues on both the ligand and the receptor. Upon the peptide binding, the cell surface protein or receptor undergoes conformational changes in the extracellular domain that enable or disable interactions with other molecules, such as proteins, triggering intracellular signal transduction pathways. Sometimes, the peptide interaction with the cell surface protein or receptor does not interfere with the binding of the native ligand, like peptides that bind to a monomeric receptor and prevent receptor oligomerization. For example, peptides derived from the transmembrane domains (TMDs) of G protein-coupled receptors (GPCRs) have been shown to disrupt receptor dimerization without affecting the binding of the natural ligand [96]. TMD peptides are short stretches of amino acids that can be discovered through screening approaches or designed rationally based on the structure or sequence of their target receptors, providing unique insights into receptor function and regulation [97]. Notably, a peptide can exhibit biological activity even without directly interacting with the receptor’s ligand binding site. For instance, it may act allosterically to modulate the receptor function, thereby affecting the binding of the endogenous ligand or the activity of the downstream signal transduction pathways [98].
Some membrane receptors are challenging targets for peptide drug discovery. In this context, GPCRs, which form one of the largest families of drug targets [99], have a complex structure that restricts their use as the target of biopanning. These receptors possess multiple transmembrane domains, and their extracellular ligand-binding region is composed of several distinct parts. Their structure is therefore highly dependent on preserving the integrity of the entire molecule, which is embedded within the cell membrane [98].
5.2. Cell Surface Protein-Binding Peptides Identified by Phage Display
Most of the approved peptide drugs are agonists, and peptide antagonists historically have faced a slower market entry. This is explained by the fact that agonists can achieve therapeutic effects by occupying only 5% to 20% of target receptor molecules, whereas antagonists must compete with the endogenous ligand and typically require a > 50% receptor occupancy for efficacy. In addition, peptide antagonists often function through allosteric receptor interactions, where their large surface area may offer minimal advantages over competing small-molecule drugs. Nevertheless, peptide antagonists demonstrate an obvious superiority over small molecules when the target requires an inhibitor with a large surface area and structural complexity (e.g., ion channels) to ensure subtype selectivity and minimize off-target effects [100]. Peptides targeting the erythropoietin receptor (EpoR) [101] and thrombopoietin receptor (TpoR) [102] are important examples of agonists of membrane receptors identified by phage display. In subsequent years, the TpoR-binding peptide underwent further modifications and was later developed into an FDA-approved drug. This drug, called romiplostim, is being used routinely in the clinic for the treatment of chronic immune thrombocytopenic purpura (ITP). Romiplostim is composed of two identical single-chain subunits, each containing a peptide with two TPOR-binding domains covalently linked to a human IgG1 Fc domain, forming a peptibody. The attachment to the Fc fragment increases the stability and half-life in the bloodstream [103,104].
Although only a small number of peptides identified through phage display have progressed to approved drugs, numerous candidates remain under investigation at various stages of research and development, holding significant potential as future therapeutics. Furthermore, the field itself continues to expand, with the scientific community actively identifying new peptides that exhibit promising biological activities. In particular, the increasing number of peptide candidates with known binding to cell surface proteins provides new opportunities for developing peptide-based therapeutics. Table 2 presents examples of cell surface-binding peptides identified by phage display over the past decade. These peptides have been reported to bind selectively to either cells or cell surface-associated proteins and receptors. Some of these phage-display-derived peptides might offer potential applications as therapeutic agents in the future.

Table 2.
A list of peptide sequences binding to cells or cell surface proteins identified in the recent decade from phage display libraries. The table indicates the sequence of isolated peptide(s), the target of the library selection, the involved disease, information about peptide properties and their biological function, and the publication reporting the peptide(s). In these reports, the target is a cell surface biomolecule that is presented to the library in different modes: a purified recombinant protein as well as a protein on the surface of either cultured cell lines (in vitro biopanning) or xenograft animal models of the relevant cell (in vivo biopanning). The presentation mode of the targets is noted in the table for each study. All targets (recombinant proteins and cells) are of human origin, unless otherwise noted. The references are arranged in the table in reverse chronological order (from newest to oldest).
6. Affinity Maturation of Peptides Derived from Phage Display by Building Secondary Libraries
Affinity maturation is a crucial step in peptide engineering that aims to improve the binding affinity and overall functionality of peptides identified through phage display. While the primary selection can yield candidate binders, these peptides often exhibit suboptimal affinities for their targets. To address this limitation, affinity maturation employs iterative cycles of mutation and selection to fine-tune peptide sequences for enhanced binding affinity. A central element of this process is the construction of secondary (and higher-order) libraries, which use initial peptide hits from a primary selection as the input for further diversification and selection [143]. In this section, we explore key strategies used in peptide affinity maturation, including the random approach and rational design. We then discuss how the number of peptide sequences selected as the input for constructing secondary and higher-order libraries influences affinity selection outcomes, comparing greedy and non-greedy strategies that can be employed to optimize the binding properties of peptides.
6.1. Definition and Scope of Affinity Maturation
A fundamental paradigm to improve the biological activity of peptides is affinity maturation. This strategy is mainly dependent on directed evolution and is classically used to enhance the binding affinity of a peptide. However, the underlying principles of affinity maturation—namely, iterative mutagenesis and selection—can also be employed to improve traits beyond binding affinity. These include pharmacokinetic and pharmacodynamic properties such as the peptide stability and solubility. In such cases, the maturation strategy focuses on optimizing these alternative traits, either within the phage display framework or during post-display peptide engineering. Due to the classical and predominant use of this strategy for the maturation of the binding affinity, here we focus on how this powerful approach can serve to refine the binding affinity of peptides toward a desired target. In a broad sense, affinity maturation is defined as a process in which iterative rounds of mutagenesis and selection are applied to enhance the binding affinity of a peptide that is specific for a given target. This concept is inspired by the natural immune response in antibody-encoding genes in B cells, where the somatic hypermutation and selection generate antibodies with a progressively increasing affinity for an antigen over time [144]. In the phage display context, the highest-affinity binders might be strongly underrepresented or entirely absent in the naïve library. This is of particular importance for longer peptides, as the coverage of their complete theoretical sequence spaces is beyond the capacities of the library construction methods. Additionally, biases that occur during the selection (e.g., inefficient recovery of tight binders) and the amplification of the selected pool (e.g., outcompeting high-affinity binders displayed on slow-propagating phages by low-affinity binders displayed on fast-propagating phages) can skew the selection toward the enrichment of weak binders. The net outcome is that initial hits identified in phage display selection may have suboptimal affinity. To address this challenge and enhance the probability of the discovery of the strongest binders, a secondary library can be constructed based on the sequence(s) identified in the primary selection [143]. At this stage, the peptides remain genetically encoded and displayed on the surface of phages, allowing the process of mutagenesis and selection to continue within the phage display system. It is only after high-affinity binders have been identified that peptide sequences are synthesized and tested independently for functional characterization or therapeutic development. In affinity maturation, randomization is performed on the most promising peptide hit(s) from a previous biopanning, followed by a secondary biopanning under a higher selection stringency. By doing so, the affinity of peptides that bind to the target in the micromolar range can be optimized to reach the nanomolar or even picomolar range [145].
6.2. Strategies for Peptide Affinity Maturation: Random Approach and Rational Design
Various strategies are employed for the affinity maturation of phage display-derived peptides, which can be classified into two broad categories of the random approach and rational design (Figure 4). The random approach involves the unbiased exploration of the vast sequence space of peptides through stochastic methods and introduces diversity into peptide sequences without prior knowledge of the structure–function relationship. The random approach includes techniques such as random mutagenesis [146] and DNA shuffling [147,148]. In random mutagenesis, random mutations are introduced into the peptide sequence (e.g., by error-prone PCR), whereas in DNA shuffling, sequences from different peptide variants are recombined to generate a library of chimeric peptides that might contain variants with enhanced binding characteristics.
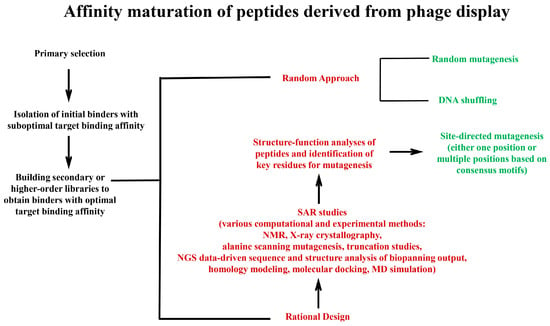
Figure 4.
A schematic overview of the main strategies used for the affinity maturation of peptides derived from phage display selection. Peptides identified from the selection of phage display libraries exhibiting suboptimal affinities are used as the input for affinity maturation by building secondary or higher-order libraries. The diagram illustrates the two principal strategies for affinity maturation: the random approach and rational design. In the random approach, mutations are introduced into peptide sequences using stochastic methods such as random mutagenesis (e.g., error-prone PCR) and DNA shuffling, which generates secondary libraries that are then subjected to selection for enhanced binding affinity. In the rational design, structure–activity relationship (SAR) data, obtained from computational (e.g., molecular docking, molecular dynamics) and experimental methods (e.g., alanine scanning, truncation studies), provide insights into residues predicted to improve binding affinity upon modification, guiding the site-directed mutagenesis of these residues. The directed mutagenesis of multiple positions is enabled by analyzing the peptide pool of the biopanning output and identifying consensus sequence motifs in the peptide repertoire. In both strategies, the iterative cycles of mutagenesis and selection progressively improve binding affinity, with a potential extension to tertiary (and higher-order) libraries for further optimization.
In contrast, the rational design uses a combination of in silico and experimental methods to obtain peptides with an improved binding affinity in a targeted manner. In this context, structure–activity relationship (SAR) studies are highly important in systematically evaluating how structural changes affect the biological function as well as in identifying key residues involved in binding [149,150,151,152,153]. While SAR studies are often applied to peptides outside the phage display platform, they can inform rational design methodologies for constructing secondary libraries within the phage display system. In SAR studies, data obtained through nuclear magnetic resonance (NMR), X-ray crystallography, homology modeling, and computational tools play a pivotal role in the structure- and sequence-guided rational design of peptides to enhance the binding affinity without compromising their structural integrity or biological function. Computational methods like molecular docking and molecular dynamics (MD) simulations can be utilized to predict the best binding poses of a peptide to its target and identify residues with strong binding interactions between the peptide and the target, respectively [154]. Additionally, large sequence datasets generated by the NGS analysis of the biopanning output can also serve as a rich source to gain quantitative information about the abundance and enrichment of peptide variants during library selection, which can be effectively employed for data-driven peptide engineering. Experimental methods such as alanine scanning mutagenesis and truncation studies also play important roles in SAR studies. In alanine scanning mutagenesis, each amino acid of a peptide is replaced with alanine [155]. Residues that significantly reduce binding upon mutation are considered critical. Another type of SAR analysis is truncation study that involves removing a segment of a peptide to identify the essential amino acids or regions for binding [156]. The diverse information acquired from sequence- and structure-based analyses in SAR analyses provides valuable insights into key residues involved in binding. Based on these insights, residues predicted to enhance binding affinity when modified can be selectively targeted for mutagenesis. An important technique for the rational design of peptides for this purpose is site-directed mutagenesis [157]. The candidate residues are targeted by this method, either to introduce a specific amino acid substitution or to explore multiple alternatives, sometimes even all 19 other amino acids. In some cases, sequence and structural analyses suggest that multiple positions should be selected for mutagenesis to achieve optimal results. In this regard, the consensus motifs identified by the analysis of the results of biopanning data can be used to design secondary focused libraries for the affinity maturation of phage display-derived peptides [158,159,160]. A consensus motif is a recurring sequence pattern observed among a group of peptides that have been identified in biopanning against a specific target. The highly conserved residues of a consensus motif are used as a scaffold and thus kept intact (fixed positions), whereas semi-conserved or variable positions are mutated for further optimization. The motifs can help in identifying binding hotspots, regions on the peptide that interact most strongly with the target. Secondary focused libraries constructed based on consensus motifs preserve the core structure of the parental binder while permitting the fine-tuning of specific residues to improve binding affinity. In some cases, the combinatorial optimization of motifs can occur by merging multiple consensus motifs identified in the same or different selections. Figure 4 is a schematic diagram of the strategies used for the affinity maturation of phage display-derived peptides.
The process of the evolution of the binding affinity maturation of peptides can be further extended beyond primary (first-generation) and secondary (second-generation) libraries by constructing tertiary (third-generation) libraries. Tertiary libraries are generated based on the input from secondary libraries. This iterative process holds potential for the gradual improvement of the binding affinity of peptides. Such a stepwise evolutionary selection strategy has been used to construct phage display secondary and tertiary sub-libraries to identify peptides that bind to and inhibit hemagglutinin (HA) (the membrane protein of type A influenza viruses). The selection from secondary and tertiary sub-libraries led to an enhanced binding affinity of peptides and narrowed down the minimum inhibitory sequence to a 5-residue motif critical for inhibition. These HA inhibitors are considered promising candidates for the development of antiviral drugs. It is of interest to note that the selection and optimization process, including the construction of secondary and tertiary sub-libraries, was carried out within the phage display system. However, the most promising candidates were later synthesized and tested as free peptides to confirm their inhibitory activity and therapeutic relevance [161].
6.3. Greedy vs. Non-Greedy Strategies for Optimization of Peptide Binding Affinity
Regarding the number of peptide sequences selected as the input for building secondary and higher-order libraries, two main strategies might be employed (Figure 5). The first one, which is known as the greedy strategy, involves selecting the best clone (the strongest binder), called the initial champion, and then subjecting this clone to either random or rational mutagenesis. This generates a clan of closely related variants. Again, the best binder from this clan is selected and mutagenized. This alternating process of selection and mutation is repeated until a peptide with optimal binding for the target is obtained. In this optimization program, each iteration selects “greedily” for the very best binder available within the population [162]. A significant limitation of this optimization scheme is that only close relatives of the initial champion are explored. Therefore, a minuscule region within the vast space of all possible sequences is searched to find the most fit sequence(s). However, it is entirely possible this localized search in the neighborhood of only the first-best sequence overlooks superior sequences that reside in distant, unexplored regions of the sequence space, including the neighborhoods of the second-best sequence, third-best sequence, fourth-best sequence, and so on. As a consequence, a more thoughtful alternative to the greedy plan is to relax the stringency of earlier selection round(s) to search in a broader part of the possible sequence space for finding more fit sequences. Based on this, rather than selecting the very best clone in the current population, a broader subpopulation of clones is selected that exhibits affinities above a certain moderate threshold. This baseline threshold is sufficiently permissive to retain “dark horses”, variants in the output of the primary selection that are (somewhat) inferior to the initial champion but possess the potential to be mutated and reach fitness levels even higher than the initial champion. In this non-greedy binding optimization strategy, a cocktail of sequences with diverse fitness levels (above the defined baseline level), containing thousands or millions of different variants, is selected and subjected to mutagenesis en masse, generating many clans of mutants. In the next round, a mixture of sequences with broad fitness levels (above a slightly higher threshold than the previous round) is selected from different clans of mutants. This process is repeated while increasing the selection stringency with each successive round. At the end of the final round, a stringent selection is employed to select the very best sequence in the existing population. By applying this scheme, dark horse sequences that reside in a different neighborhood than the initial champion can be discovered [162,163]. When sequences are similar and lie in the same neighborhood in the sequence space, they often converge on the same local optimum through mutational trajectories. However, the non-greedy strategy provides the opportunity to explore dark horse sequences from other neighborhoods of the sequence space, increasing the probability of finding the very best sequence that could exist in the possible sequence space. It is important to note that even by using the non-greedy strategy, it is not possible to explore the entire sequence space, and there is always the chance that a significantly better binder exists somewhere in the vast expanse of the sequence space. However, considering the impracticality of conducting a truly global search, the close neighborhoods of the best binder in the sequence space remain the most promising areas to become the focus of ligand optimization efforts [162]. Figure 5 illustrates a schematic comparison of the greedy and non-greedy strategies for optimizing the binding affinity of phage display-derived peptides.
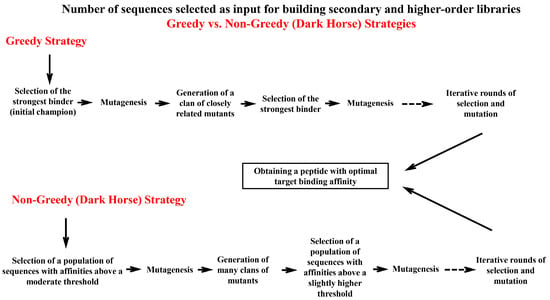
Figure 5.
A comparison of greedy and non-greedy strategies for the selection of input sequence(s) when building secondary and higher-order libraries. The diagram compares the greedy (top) and non-greedy (bottom) strategies for the optimization of binding affinity. In the greedy strategy, the strongest binder (initial champion) from the primary selection is selected and subjected to mutagenesis. This process is repeated for further cycles of mutagenesis and selection, leading to a localized exploration of the sequence space (in the neighborhood of only the first-best sequence). This localized search risks missing superior binders in distant, unexplored regions of the sequence space (e.g., the neighborhoods of the second-best sequence, third-best sequence, and so on). In contrast, the non-greedy strategy retains a broader selection window by including a subpopulation of binders above a moderate affinity threshold during each cycle of selection and mutagenesis. This strategy is capable of capturing dark horse sequences that may be inferior to the strongest binder in terms of target binding affinity but hold the potential to surpass the affinity of the strongest binder through further rounds of mutagenesis and selection. By iteratively increasing the selection stringency during cycles of in vitro evolution and including a population of sequences with affinities slightly higher than the previous round, the non-greedy strategy enables the simultaneous exploration of multiple sequence neighborhoods. Broadening the exploration of the sequence landscape enhances the probability of discovering peptides with optimal target binding affinity.
7. Concluding Remarks and Future Perspectives
Over many years, phage display has proven to be a reliable and powerful method for finding peptides that bind to a wide variety of clinically relevant targets [164,165]. One of the most transformative contributions of phage display has been its role in revolutionizing the discovery of cell surface-binding peptides. By enabling the selection of peptides that recognize specific molecular signatures on the surface of cells—either through purified targets or complex biological systems such as whole cells, tissues, and living organisms—phage display has fundamentally reshaped the way targeting ligands are developed. This has had wide-reaching implications for drug delivery, molecular imaging, and the design of precision therapeutics. In addition, the expanding use of phage display in complex biological environments, such as ex vivo and in vivo settings, represents the promise of selecting peptides with a high physiological relevance. These applications have evolved phage display from a pioneering display technology into a cornerstone of modern pharmaceutical research.
The success of phage display depends not only on the construction of diverse and well-designed peptide libraries but also on the strategic presentation of targets and the optimization of affinity selection protocols. Importantly, the transition from peptide discovery to therapeutic application necessitates improvements in binding affinity and pharmacological properties. The development of secondary libraries, constructed by either random approach or rational design through SAR studies, serves as a crucial step in maturing peptides toward clinical relevance. Several technical innovations have made phage display a more powerful strategy for peptide discovery. In this context, advancements in phage display technology have enabled the construction of sophisticated peptide libraries with topologically constrained formats. Among these, bicyclic and multicyclic peptide libraries represent a major innovation, offering an enhanced conformational rigidity, optimized resistance to proteolytic degradation, and improved receptor-binding affinity compared to their linear or even monocyclic counterparts [166,167,168]. The structural constraints imposed by multiple cycles allow these peptides to mimic protein-like interfaces more effectively, facilitating high-affinity and selective interactions with challenging biological targets, such as protein–protein interaction surfaces or cryptic binding pockets in cell surface proteins. Moreover, expanding the chemical diversity of displayed peptides through genetic code expansion by introducing noncanonical amino acids with novel reactivities and chemistries has opened new avenues for improving the stability and specificity of peptides identified by phage display [169,170]. The incorporation of NGS into phage display in recent years has further enhanced the precision and throughput of this technology in peptide discovery [56,57], allowing for a deeper exploration of peptide repertoires and more informed decision-making during lead optimization.
Looking ahead, several areas offer exciting potential for further innovation in phage display. The integration of AI and ML into phage display workflows is poised to radically change the discovery of target-binding peptides [52,171]. The rapid expansion of sequencing data generated by NGS-based phage display has created a fertile ground for the application of AI and ML tools. By analyzing large-scale sequencing data from NGS, AI and ML algorithms can uncover hidden sequence–function relationships, predict the peptide-target binding affinity, cluster functionally relevant motifs, and prioritize candidates for further testing—far beyond what traditional methods can achieve. Additionally, these tools can guide the design of secondary libraries by predicting beneficial mutations or identifying underrepresented yet functionally promising “dark horse” candidates. As these computational approaches continue to mature and integrate with experimental workflows, they will increasingly complement wet-lab methods, enabling the faster, smarter, and more efficient identification of cell-selective peptide ligands with diagnostic and therapeutic potential. AI-assisted computational approaches are transforming how we interpret, navigate, and exploit the vast sequence space explored during biopanning.
Despite the promise of phage display in identifying cell-selective peptides, using this technology is not without restrictions. One of the most significant and technically challenging limitations of phage display, particularly when applied to complex biological targets such as the cell surface, is the undesirable enrichment of nonspecific binders, which can significantly compromise the efficiency and specificity of the selection process [172]. These nonspecific binders can broadly be categorized into two types: selection-related nonspecific binders, which interact with off-target components present in the selection system (e.g., plastic surfaces, blocking buffer), and propagation-related nonspecific binders, which are enriched not because of target selectivity but due to intrinsic propagation advantages that favor their amplification during biopanning rounds [173]. This phenomenon distorts the apparent outcome of selection, allowing peptides that do not specifically bind to the intended target to dominate the biopanning output. These issues are particularly challenging in cell-based biopanning, where the heterogeneity of the cell surface presents a significant technical barrier. Mammalian cell surfaces are highly complex and dynamic mosaics composed of a wide variety of biomolecules. This heterogeneity makes it difficult to identify peptides that bind selectively and reproducibly to a unique molecular marker, especially given the short length of peptides, which limits their interaction surface area and structural complexity. Further compounding the issue is that the molecular identity of the peptide’s target is often unknown or undefined in cell-based selections [174], which can severely hinder the downstream validation, mechanistic elucidation, and functional interpretation. Peptides isolated through cell-based biopanning may bind to a wide range of possible targets. Additionally, cell culture artifacts pose a substantial challenge. Cells grown in vitro often exhibit gene expression patterns, membrane compositions, and glycosylation profiles that diverge significantly from their native tissue context, leading to the enrichment of peptide binders that lack physiological relevance. Addressing these challenges requires the rigorous incorporation of counter-selection strategies and the utilization of robust bioinformatics pipelines capable of filtering out selection-related nonspecific peptides and propagation-biased sequences during data analysis. Nevertheless, the isolation of nonspecific binders remains a critical limitation that must be carefully managed to fully realize the promise of phage display in discovering functionally relevant, high-affinity cell-selective peptides.
Using the full potential of phage display for the discovery of cell-selective peptides will depend on addressing its major limitations. However, this molecular display framework remains not only a foundational technology in peptide discovery but also a continually evolving platform that adapts to the needs of modern biomedical research. Its synergy with increasingly sophisticated selection methodologies, chemical innovation, and computational tools will further enhance our capacity to discover and optimize high-affinity cell surface-binding peptides, bringing us closer to a future of more precise, effective, and personalized therapies.
Author Contributions
Conceptualization: B.B. and S.G.; Data curation: B.B. and S.G.; Visualization: B.B.; Writing—original draft: B.B. and S.G.; Writing—review and editing: B.B. and S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
Correction Statement
This article has been republished with a minor correction to the existing affiliation information. This change does not affect the scientific content of the article.
Abbreviations
| AgNPs | Silver nanoparticles |
| AI | Artificial intelligence |
| BBB | Blood–brain barrier |
| BTIC | Brain tumor initiating cell |
| BSA | Bovine serum albumin |
| CCR7 | C-C chemokine receptor type 7 |
| CPT | Camptothecin |
| DAS | Dorsal air sac |
| ECM | Extracellular matrix |
| EGFRvIII | Epidermal growth factor tyrosine kinase receptor variant III |
| EPC | Endothelial progenitor cell |
| EpCAM | Epithelial cell adhesion molecule |
| EpoR | Erythropoietin receptor |
| ESCC | Esophageal squamous cell carcinoma |
| FGFR2 | Fibroblast growth factor receptor 2 |
| FLAP | 5-lipoxygenase-activating protein |
| FRα | Folate receptor α |
| GPCR | G-protein coupled receptor |
| GPC-3 | Glypican-3 |
| GST | Glutathione S-transferase |
| HA | Hemagglutinin |
| HAE | Hereditary angioedema |
| hCMEC | Human cerebral microvascular endothelial cell |
| HCC | Hepatocellular carcinoma |
| HESC | Human immortalized epithelial-like eutopic endometrial stromal cell |
| \ | |
| HTS | High-throughput sequencing |
| HuCCT-1 | Human cholangiocarcinoma cell line T1 |
| IGF2BP2 | Insulin-like growth factor 2 mRNA-binding protein 2 |
| ITP | Immune thrombocytopenic purpura |
| LCC | Large carcinoma cell |
| LGR5 | Leucine-rich repeat-containing G-protein coupled receptor 5 |
| LMP1 | Latent membrane protein 1 |
| MBP | Maltose-binding protein |
| MD simulation | Molecular dynamics simulation |
| ML | Machine learning |
| MMP | Matrix metalloproteinase |
| NGS | Next-generation sequencing |
| NMR | Nuclear magnetic resonance |
| NPC | Nasopharyngeal carcinoma |
| NSCLC | Non-small cell lung cancer |
| OPA | Ortho-phthalaldehyde |
| PAI1 | Plasminogen activator inhibitor 1 |
| PBI | Phage binding index |
| PD-L1 | Programmed death-ligand 1 |
| PDX | Patient-derived xenograft |
| pIII, pVI, pVII, pVIII, pIX | Coat proteins (3, 6, 7, 8, 9) of the filamentous M13 phage |
| PNH | Paroxysmal nocturnal hemoglobinuria |
| PPI | Protein–protein interaction |
| PSMA | Prostate-specific membrane antigen |
| SAR | Structure–activity relationship |
| SPIONs | Super-paramagnetic iron oxide nanoparticles |
| ssDNA | Single-stranded DNA |
| TBMB | 1,3,5-tris(bromomethyl)benzene |
| TIMP-1 | Tissue inhibitor of metalloproteinases 1 (metalloproteinase inhibitor 1) |
| TMDs | Transmembrane domains |
| TpoR | Thrombopoietin receptor |
| TUPs | Target-unrelated peptides |
References
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Scott, J.K.; Smith, G.P. Searching for peptide ligands with an epitope library. Science 1990, 249, 386–390. [Google Scholar] [CrossRef]
- Parmley, S.F.; Smith, G.P. Antibody-selectable filamentous fd phage vectors: Affinity purification of target genes. Gene 1988, 73, 305–318. [Google Scholar] [CrossRef]
- Gray, B.P.; Li, S.; Brown, K.C. From phage display to nanoparticle delivery: Functionalizing liposomes with multivalent peptides improves targeting to a cancer biomarker. Bioconjug. Chem. 2013, 24, 85–96. [Google Scholar] [CrossRef]
- Dókus, L.E.; Lajkó, E.; Ranđelović, I.; Mező, D.; Schlosser, G.; Kőhidai, L.; Tóvári, J.; Mező, G. Phage display-based homing peptide-daunomycin conjugates for selective drug targeting to PANC-1 pancreatic cancer. Pharmaceutics 2020, 12, 576. [Google Scholar] [CrossRef]
- Qin, Y.; Cheng, S.; Li, Y.; Zou, S.; Chen, M.; Zhu, D.; Gao, S.; Wu, H.; Zhu, L.; Zhu, X. The development of a Glypican-3-specific binding peptide using in vivo and in vitro two-step phage display screening for the PET imaging of hepatocellular carcinoma. Biomater. Sci. 2020, 8, 5656–5665. [Google Scholar] [CrossRef] [PubMed]
- Soendergaard, M.; Newton-Northup, J.R.; Deutscher, S.L. In vivo phage display selection of an ovarian cancer targeting peptide for SPECT/CT imaging. Am. J. Nucl. Med. Mol. Imaging 2014, 4, 561–570. [Google Scholar] [PubMed]
- Pierzynowska, K.; Morcinek-Orłowska, J.; Gaffke, L.; Jaroszewicz, W.; Skowron, P.M.; Węgrzyn, G. Applications of the phage display technology in molecular biology, biotechnology and medicine. Crit. Rev. Microbiol. 2024, 50, 450–490. [Google Scholar] [CrossRef]
- Saw, P.E.; Song, E.-W. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell 2019, 10, 787–807. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Tang, Y.; Chen, Q.; Liu, Y. The screening of therapeutic peptides for anti-inflammation through phage display technology. Int. J. Mol. Sci. 2022, 23, 8554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Gao, H.; Qing, G. Phage display derived peptides for Alzheimer’s disease therapy and diagnosis. Theranostics 2022, 12, 2041–2062. [Google Scholar] [CrossRef] [PubMed]
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013, 81, 136–147. [Google Scholar] [CrossRef]
- Henninot, A.; Collins, J.C.; Nuss, J.M. The current state of peptide drug discovery: Back to the future? J. Med. Chem. 2018, 61, 1382–1414. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef]
- Kaspar, A.A.; Reichert, J.M. Future directions for peptide therapeutics development. Drug Discov. Today 2013, 18, 807–817. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Tsomaia, N. Peptide therapeutics: Targeting the undruggable space. Eur. J. Med. Chem. 2015, 94, 459–470. [Google Scholar] [CrossRef]
- Atangcho, L.; Navaratna, T.; Thurber, G.M. Hitting undruggable targets: Viewing stabilized peptide development through the lens of quantitative systems pharmacology. Trends Biochem. Sci. 2019, 44, 241–257. [Google Scholar] [CrossRef]
- Jiang, J.; Abu-Shilbayeh, L.; Rao, V.B. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect. Immun. 1997, 65, 4770–4777. [Google Scholar] [CrossRef]
- Sharma, S.C.; Memic, A.; Rupasinghe, C.N.; Duc, A.C.E.; Spaller, M.R. T7 phage display as a method of peptide ligand discovery for PDZ domain proteins. Pept. Sci. Orig. Res. Biomol. 2009, 92, 183–193. [Google Scholar] [CrossRef]
- González-Cano, P.; Gamage, L.N.; Marciniuk, K.; Hayes, C.; Napper, S.; Hayes, S.; Griebel, P.J. Lambda display phage as a mucosal vaccine delivery vehicle for peptide antigens. Vaccine 2017, 35, 7256–7263. [Google Scholar] [CrossRef]
- Chackerian, B.; do Carmo Caldeira, J.; Peabody, J.; Peabody, D.S. Peptide epitope identification by affinity selection on bacteriophage MS2 virus-like particles. J. Mol. Biol. 2011, 409, 225–237. [Google Scholar] [CrossRef]
- Skamel, C.; Aller, S.G.; Bopda Waffo, A. In vitro evolution and affinity-maturation with coliphage Qβ display. PLoS ONE 2014, 9, e113069. [Google Scholar] [CrossRef]
- Pande, J.; Szewczyk, M.M.; Grover, A.K. Phage display: Concept, innovations, applications and future. Biotechnol. Adv. 2010, 28, 849–858. [Google Scholar] [CrossRef]
- Rakonjac, J.; Russel, M.; Khanum, S.; Brooke, S.J.; Rajič, M. Filamentous phage: Structure and biology. In Recombinant Antibodies for Infectious Diseases; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–20. [Google Scholar]
- Ebrahimizadeh, W.; Rajabibazl, M. Bacteriophage vehicles for phage display: Biology, mechanism, and application. Curr. Microbiol. 2014, 69, 109–120. [Google Scholar] [CrossRef]
- Hamzeh-Mivehroud, M.; Alizadeh, A.A.; Morris, M.B.; Church, W.B.; Dastmalchi, S. Phage display as a technology delivering on the promise of peptide drug discovery. Drug Discov. Today 2013, 18, 1144–1157. [Google Scholar] [CrossRef]
- Geysen, H.M.; Meloen, R.H.; Barteling, S.J. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc. Natl. Acad. Sci. USA 1984, 81, 3998–4002. [Google Scholar] [CrossRef]
- Aghebati-Maleki, L.; Bakhshinejad, B.; Baradaran, B.; Motallebnezhad, M.; Aghebati-Maleki, A.; Nickho, H.; Yousefi, M.; Majidi, J. Phage display as a promising approach for vaccine development. J. Biomed. Sci. 2016, 23, 66. [Google Scholar] [CrossRef]
- O’Neil, K.T.; Hoess, R.H.; Jackson, S.A.; Ramachandran, N.S.; Mousa, S.A.; DeGrado, W.F. Identification of novel peptide antagonists for GPIIb/IIIa from a conformationally constrained phage peptide library. Proteins Struct. Funct. Bioinform. 1992, 14, 509–515. [Google Scholar] [CrossRef]
- Ladner, R.C. Constrained peptides as binding entities. Trends Biotechnol. 1995, 13, 426–430. [Google Scholar] [CrossRef]
- Ho, K.L.; Yusoff, K.; Seow, H.F.; Tan, W.S. Selection of high affinity ligands to hepatitis B core antigen from a phage-displayed cyclic peptide library. J. Med. Virol. 2003, 69, 27–32. [Google Scholar] [CrossRef]
- Kolmar, H. Biological diversity and therapeutic potential of natural and engineered cystine knot miniproteins. Curr. Opin. Pharmacol. 2009, 9, 608–614. [Google Scholar] [CrossRef]
- Li, X.; Craven, T.W.; Levine, P.M. Cyclic peptide screening methods for preclinical drug discovery: Miniperspective. J. Med. Chem. 2022, 65, 11913–11926. [Google Scholar] [CrossRef]
- Heo, Y.-A. Pegcetacoplan: A review in paroxysmal nocturnal haemoglobinuria. Drugs 2022, 82, 1727–1735. [Google Scholar] [CrossRef]
- Cicardi, M.; Levy, R.J.; McNeil, D.L.; Li, H.H.; Sheffer, A.L.; Campion, M.; Horn, P.T.; Pullman, W.E. Ecallantide for the treatment of acute attacks in hereditary angioedema. N. Engl. J. Med. 2010, 363, 523–531. [Google Scholar] [CrossRef]
- Wang, X.S.; Chen, P.H.C.; Hampton, J.T.; Tharp, J.M.; Reed, C.A.; Das, S.K.; Wang, D.S.; Hayatshahi, H.S.; Shen, Y.; Liu, J. A genetically encoded, phage-displayed cyclic-peptide library. Angew. Chem. 2019, 131, 16051–16056. [Google Scholar] [CrossRef]
- Sohrabi, C.; Foster, A.; Tavassoli, A. Methods for generating and screening libraries of genetically encoded cyclic peptides in drug discovery. Nat. Rev. Chem. 2020, 4, 90–101. [Google Scholar] [CrossRef]
- Kamstrup Sell, D.; Sinkjaer, A.W.; Bakhshinejad, B.; Kjaer, A. Propagation capacity of phage display peptide libraries is affected by the length and conformation of displayed peptide. Molecules 2023, 28, 5318. [Google Scholar] [CrossRef]
- Deyle, K.; Kong, X.-D.; Heinis, C. Phage selection of cyclic peptides for application in research and drug development. Acc. Chem. Res. 2017, 50, 1866–1874. [Google Scholar] [CrossRef]
- Heinis, C.; Rutherford, T.; Freund, S.; Winter, G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol. 2009, 5, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, L.; Ivarsson, Y. Genetically encoded cyclic peptide phage display libraries. ACS Cent. Sci. 2020, 6, 336–338. [Google Scholar] [CrossRef] [PubMed]
- Bellotto, S.; Chen, S.; Rentero Rebollo, I.; Wegner, H.A.; Heinis, C. Phage selection of photoswitchable peptide ligands. J. Am. Chem. Soc. 2014, 136, 5880–5883. [Google Scholar] [CrossRef]
- Jafari, M.R.; Deng, L.; Kitov, P.I.; Ng, S.; Matochko, W.L.; Tjhung, K.F.; Zeberoff, A.; Elias, A.; Klassen, J.S.; Derda, R. Discovery of light-responsive ligands through screening of a light-responsive genetically encoded library. ACS Chem. Biol. 2014, 9, 443–450. [Google Scholar] [CrossRef]
- Xiang, H.; Bai, L.; Zhang, X.; Dan, T.; Cheng, P.; Yang, X.; Ai, H.; Li, K.; Lei, X. A facile strategy for the construction of a phage display cyclic peptide library for the selection of functional macrocycles. Chem. Sci. 2024, 15, 11847–11855. [Google Scholar] [CrossRef]
- Kay, B.K.; Adey, N.B.; Yun-Sheng, H.; Manfredi, J.P.; Mataragnon, A.H.; Fowlkes, D.M. An M13 phage library displaying random 38-amino-acid peptides as a source of novel sequences with affinity to selected targets. Gene 1993, 128, 59–65. [Google Scholar] [CrossRef]
- Sloth, A.B.; Bakhshinejad, B.; Jensen, M.; Stavnsbjerg, C.; Liisberg, M.B.; Rossing, M.; Kjaer, A. Analysis of compositional bias in a commercial phage display peptide library by next-generation sequencing. Viruses 2022, 14, 2402. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef]
- Bibi, N.; Niaz, H.; Hupp, T.; Kamal, M.A.; Rashid, S. Screening and identification of PLK1-Polo box binding peptides by high-throughput sequencing of phage-selected libraries. Protein Pept. Lett. 2019, 26, 620–633. [Google Scholar] [CrossRef]
- Juds, C.; Schmidt, J.; Weller, M.G.; Lange, T.; Beck, U.; Conrad, T.; Börner, H.G. Combining phage display and next-generation sequencing for materials sciences: A case study on probing polypropylene surfaces. J. Am. Chem. Soc. 2020, 142, 10624–10628. [Google Scholar] [CrossRef]
- He, B.; Li, B.; Chen, X.; Zhang, Q.; Lu, C.; Yang, S.; Long, J.; Ning, L.; Chen, H.; Huang, J. PDL1Binder: Identifying programmed cell death ligand 1 binding peptides by incorporating next-generation phage display data and different peptide descriptors. Front. Microbiol. 2022, 13, 928774. [Google Scholar] [CrossRef]
- Mansour, S.; Adhya, I.; Lebleu, C.; Dumpati, R.; Rehan, A.; Chall, S.; Dai, J.; Errasti, G.; Delacroix, T.; Chakrabarti, R. Identification of a novel peptide ligand for the cancer-specific receptor mutation EGFRvIII using high-throughput sequencing of phage-selected peptides. Sci. Rep. 2022, 12, 20725. [Google Scholar] [CrossRef] [PubMed]
- Pleiko, K.; Põšnograjeva, K.; Haugas, M.; Paiste, P.; Tobi, A.; Kurm, K.; Riekstina, U.; Teesalu, T. In vivo phage display: Identification of organ-specific peptides using deep sequencing and differential profiling across tissues. Nucleic Acids Res. 2021, 49, e38. [Google Scholar] [CrossRef] [PubMed]
- Spiliotopoulos, A.; Maurer, S.K.; Tsoumpeli, M.T.; Bonfante, J.A.; Owen, J.P.; Gough, K.C.; Dreveny, I. Next-generation phage display to identify peptide ligands of deubiquitinases. In Deubiquitinases: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2022; pp. 189–218. [Google Scholar]
- Kohl, F.; Laufkötter, O.; Firth, M.; Krimpenfort, L.; Mangla, P.; Ansarizadeh, M.; Geylan, G.; Eklund, L.; De Maria, L.; Jakobsson, L. Identification of cell type-specific cell-penetrating peptides through in vivo phage display leveraged by next generation sequencing. Biomed. Pharmacother. 2025, 182, 117740. [Google Scholar] [CrossRef] [PubMed]
- Sloth, A.B.; Bakhshinejad, B.; Stavnsbjerg, C.; Rossing, M.; Kjaer, A. Depth of sequencing plays a determining role in the characterization of phage display peptide libraries by NGS. Int. J. Mol. Sci. 2023, 24, 5396. [Google Scholar] [CrossRef]
- Sell, D.K.; Bakhshinejad, B.; Sinkjaer, A.W.; Dawoodi, I.M.; Wiinholt, M.N.; Sloth, A.B.; Stavnsbjerg, C.; Kjaer, A. Using NGS to Uncover the Corruption of a Peptide Phage Display Selection. Curr. Issues Mol. Biol. 2024, 46, 10590–10605. [Google Scholar] [CrossRef]
- AC’t Hoen, P.; Jirka, S.M.; Ten Broeke, B.R.; Schultes, E.A.; Aguilera, B.; Pang, K.H.; Heemskerk, H.; Aartsma-Rus, A.; van Ommen, G.J.; den Dunnen, J.T. Phage display screening without repetitious selection rounds. Anal. Biochem. 2012, 421, 622–631. [Google Scholar] [CrossRef]
- Rentero Rebollo, I.; Sabisz, M.; Baeriswyl, V.; Heinis, C. Identification of target-binding peptide motifs by high-throughput sequencing of phage-selected peptides. Nucleic Acids Res. 2014, 42, e169. [Google Scholar] [CrossRef]
- Munisso, M.C.; Yamaoka, T. Novel peptides for small-caliber graft functionalization selected by a phage display of endothelial-positive/platelet-negative combined selection. J. Mater. Chem. B 2017, 5, 9354–9364. [Google Scholar] [CrossRef]
- Tan, Y.; Liu, W.; Zhu, Z.; Lang, L.; Wang, J.; Huang, M.; Zhang, M.; Yang, C. Selection and identification of transferrin receptor-specific peptides as recognition probes for cancer cells. Anal. Bioanal. Chem. 2018, 410, 1071–1077. [Google Scholar] [CrossRef]
- Abbineni, G.; Modali, S.; Safiejko-Mroczka, B.; Petrenko, V.A.; Mao, C. Evolutionary selection of new breast cancer cell-targeting peptides and phages with the cell-targeting peptides fully displayed on the major coat and their effects on actin dynamics during cell internalization. Mol. Pharm. 2010, 7, 1629–1642. [Google Scholar] [CrossRef]
- Bakhshinejad, B.; Sadeghizadeh, M. Identification of a novel colon adenocarcinoma cell targeting peptide using phage display library biopanning. Biotechnol. Appl. Biochem. 2022, 69, 2753–2765. [Google Scholar] [CrossRef] [PubMed]
- Koide, A.; Wojcik, J.; Gilbreth, R.N.; Reichel, A.; Piehler, J.; Koide, S. Accelerating phage-display library selection by reversible and site-specific biotinylation. Protein Eng. Des. Sel. 2009, 22, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Colazet, M.; Chames, P. Phage display and selections on purified antigens. In Antibody Engineering: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2018; pp. 165–178. [Google Scholar]
- Butler, J.; Ni, L.; Nessler, R.; Joshi, K.; Suter, M.; Rosenberg, B.; Chang, J.; Brown, W.; Cantarero, L. The physical and functional behavior of capture antibodies adsorbed on polystyrene. J. Immunol. Methods 1992, 150, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Liu, I.-J.; Lu, R.-M.; Wu, H.-C. Advancement and applications of peptide phage display technology in biomedical science. J. Biomed. Sci. 2016, 23, 8. [Google Scholar] [CrossRef]
- Kubicek, J.; Block, H.; Maertens, B.; Spriestersbach, A.; Labahn, J. Expression and purification of membrane proteins. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 541, pp. 117–140. [Google Scholar]
- Even-Desrumeaux, K.; Chames, P. Phage display and selections on cells. In Antibody Engineering: Methods and Protocols, Second Edition; Springer: Berlin/Heidelberg, Germany, 2012; pp. 225–235. [Google Scholar]
- Zhu, X.; Wu, H.; Luo, S.; Xianyu, Z.; Zhu, D. Screening and identification of a novel hepatocellular carcinoma cell binding peptide by using a phage display library. J. Huazhong Univ. Sci. Technol. [Med. Sci.] 2008, 28, 299–303. [Google Scholar] [CrossRef]
- Gross, A.L.; Gillespie, J.W.; Petrenko, V.A. Promiscuous tumor targeting phage proteins. Protein Eng. Des. Sel. 2016, 29, 93–103. [Google Scholar] [CrossRef]
- Caprini, A.; Silva, D.; Zanoni, I.; Cunha, C.; Volontè, C.; Vescovi, A.; Gelain, F. A novel bioactive peptide: Assessing its activity over murine neural stem cells and its potential for neural tissue engineering. New Biotechnol. 2013, 30, 552–562. [Google Scholar] [CrossRef]
- Kaur, R.; Jain, R.; Budholiya, N.; Rathore, A.S. Long term culturing of CHO cells: Phenotypic drift and quality attributes of the expressed monoclonal antibody. Biotechnol. Lett. 2023, 45, 357–370. [Google Scholar] [CrossRef]
- Yao, L.; Bestwick, C.; Bestwick, L.A.; Maffulli, N.; Aspden, R.M. Phenotypic drift in human tenocyte culture. Tissue Eng. 2006, 12, 1843–1849. [Google Scholar] [CrossRef]
- Hoffman, J.A.; Laakkonen, P.; Porkka, K.; Bernasconi, M.; Ruoslahti, E. In vivo and ex vivo selections using phage-displayed libraries. In Phage Display: A Practical Approach; Lowman, H.B., Clackson, T., Eds.; Oxford University Press: New York, NY, USA, 2004; pp. 171–192. [Google Scholar] [CrossRef]
- Koivistoinen, A.; Ilonen, I.; Punakivi, K.; Räsänen, J.V.; Helin, H.; Sihvo, E.; Bergman, M.; Salo, J. A novel peptide (Thx) homing to non-small cell lung cancer identified by ex vivo phage display. Clin. Transl. Oncol. 2013, 15, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Odermatt, A.; Audigé, A.; Frick, C.; Vogt, B.; Frey, B.M.; Frey, F.J.; Mazzucchelli, L. Identification of receptor ligands by screening phage-display peptide libraries ex vivo on microdissected kidney tubules. J. Am. Soc. Nephrol. 2001, 12, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Pasqualini, R.; Ruoslahti, E. Organ targeting in vivo using phage display peptide libraries. Nature 1996, 380, 364–366. [Google Scholar] [CrossRef]
- Bábíčková, J.; Tóthová, Ľ.; Boor, P.; Celec, P. In vivo phage display—A discovery tool in molecular biomedicine. Biotechnol. Adv. 2013, 31, 1247–1259. [Google Scholar] [CrossRef]
- Arap, W.; Kolonin, M.G.; Trepel, M.; Lahdenranta, J.; Cardó-Vila, M.; Giordano, R.J.; Mintz, P.J.; Ardelt, P.U.; Yao, V.J.; Vidal, C.I. Steps toward mapping the human vasculature by phage display. Nat. Med. 2002, 8, 121–127. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Q.; Pang, Z.; Wang, Y.; Liu, Q.; Guo, L.; Jiang, X. Identification of peptide sequences that target to the brain using in vivo phage display. Amino Acids 2012, 42, 2373–2381. [Google Scholar] [CrossRef]
- Li, J.; Feng, L.; Jiang, X. In vivo phage display screen for peptide sequences that cross the blood–cerebrospinal-fluid barrier. Amino Acids 2015, 47, 401–405. [Google Scholar] [CrossRef]
- Larimer, B.M.; Thomas, W.D.; Smith, G.P.; Deutscher, S.L. Affinity maturation of an ERBB2-targeted SPECT imaging peptide by in vivo phage display. Mol. Imaging Biol. 2014, 16, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Hyvönen, M.; Laakkonen, P. Identification and characterization of homing peptides using in vivo peptide phage display. In Cell-Penetrating Peptides: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2015; pp. 205–222. [Google Scholar]
- Du, B.; Han, H.; Wang, Z.; Kuang, L.; Wang, L.; Yu, L.; Wu, M.; Zhou, Z.; Qian, M. Targeted drug delivery to hepatocarcinoma in vivo by phage-displayed specific binding peptide. Mol. Cancer Res. 2010, 8, 135–144. [Google Scholar] [CrossRef]
- Arap, W.; Pasqualini, R. The human vascular mapping project. Selection and utilization of molecules for tumor endothelial targeting. Haemostasis 2001, 31, 30–31. [Google Scholar] [PubMed]
- Staquicini, F.I.; Cardó-Vila, M.; Kolonin, M.G.; Trepel, M.; Edwards, J.K.; Nunes, D.N.; Sergeeva, A.; Efstathiou, E.; Sun, J.; Almeida, N.F. Vascular ligand-receptor mapping by direct combinatorial selection in cancer patients. Proc. Natl. Acad. Sci. USA 2011, 108, 18637–18642. [Google Scholar] [CrossRef] [PubMed]
- Krag, D.N.; Shukla, G.S.; Shen, G.-P.; Pero, S.; Ashikaga, T.; Fuller, S.; Weaver, D.L.; Burdette-Radoux, S.; Thomas, C. Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res. 2006, 66, 7724–7733. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Zhu, Y.; Chen, L. Advances in targeting cell surface signalling molecules for immune modulation. Nat. Rev. Drug Discov. 2013, 12, 130–146. [Google Scholar] [CrossRef]
- Kuhlmann, L.; Cummins, E.; Samudio, I.; Kislinger, T. Cell-surface proteomics for the identification of novel therapeutic targets in cancer. Expert Rev. Proteom. 2018, 15, 259–275. [Google Scholar] [CrossRef]
- Herz, J.M.; Thomsen, W.J.; Yarbrough, G.G. Molecular approaches to receptors as targets for drug discovery. J. Recept. Signal Transduct. 1997, 17, 671–776. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, M.I.; Clemmensen, L.S.; Bredt, D.S.; Bettler, B.; Strømgaard, K. Targeting receptor complexes: A new dimension in drug discovery. Nat. Rev. Drug Discov. 2020, 19, 884–901. [Google Scholar] [CrossRef]
- Arinaminpathy, Y.; Khurana, E.; Engelman, D.M.; Gerstein, M.B. Computational analysis of membrane proteins: The largest class of drug targets. Drug Discov. Today 2009, 14, 1130–1135. [Google Scholar] [CrossRef]
- Safari-Alighiarloo, N.; Taghizadeh, M.; Rezaei-Tavirani, M.; Goliaei, B.; Peyvandi, A.A. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol. Hepatol. Bed Bench 2014, 7, 17–31. [Google Scholar] [PubMed]
- Harikumar, K.G.; Dong, M.; Cheng, Z.; Pinon, D.I.; Lybrand, T.P.; Miller, L.J. Transmembrane segment peptides can disrupt cholecystokinin receptor oligomerization without affecting receptor function. Biochemistry 2006, 45, 14706–14716. [Google Scholar] [CrossRef]
- Westerfield, J.M.; Barrera, F.N. Membrane receptor activation mechanisms and transmembrane peptide tools to elucidate them. J. Biol. Chem. 2020, 295, 1792–1814. [Google Scholar] [CrossRef]
- Molek, P.; Strukelj, B.; Bratkovic, T. Peptide phage display as a tool for drug discovery: Targeting membrane receptors. Molecules 2011, 16, 857–887. [Google Scholar] [CrossRef] [PubMed]
- Lorente, J.S.; Sokolov, A.V.; Ferguson, G.; Schiöth, H.B.; Hauser, A.S.; Gloriam, D.E. GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2025, 24, 458–479. [Google Scholar] [CrossRef] [PubMed]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef]
- Wrighton, N.C.; Farrell, F.X.; Chang, R.; Kashyap, A.K.; Barbone, F.P.; Mulcahy, L.S.; Johnson, D.L.; Barrett, R.W.; Jolliffe, L.K.; Dower, W.J. Small peptides as potent mimetics of the protein hormone erythropoietin. Science 1996, 273, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Cwirla, S.E.; Balasubramanian, P.; Duffin, D.J.; Wagstrom, C.R.; Gates, C.M.; Singer, S.C.; Davis, A.M.; Tansik, R.L.; Mattheakis, L.C.; Boytos, C.M. Peptide agonist of the thrombopoietin receptor as potent as the natural cytokine. Science 1997, 276, 1696–1699. [Google Scholar] [CrossRef]
- Cines, D.B.; Yasothan, U.; Kirkpatrick, P. Romiplostim. Nat. Rev. Drug Discov. 2008, 7, 887–889. [Google Scholar] [CrossRef]
- Frampton, J.E.; Lyseng-Williamson, K.A. Romiplostim. Drugs 2009, 69, 307–317. [Google Scholar] [CrossRef]
- Sun, Y.; Qian, Y.; Qiu, L.; Zhu, X.; Ning, H.; Pang, L.; Niu, X.; Liu, Y.; Zhou, X.; Chen, G. A novel peptide targeting CCR7 inhibits tumor cell lymph node metastasis. Cancer Immunol. Immunother. 2025, 74, 153. [Google Scholar] [CrossRef]
- Ammous-Boukhris, N.; Rached, N.; Mosbah, A.; Ayadi, W.; Gargouri, A.; Mokdad-Gargouri, R. Selection of Peptides Targeting the LMP1 Oncoprotein Via Phage Display Differential Screening on Cancer Cells. Int. J. Pept. Res. Ther. 2025, 31, 40. [Google Scholar] [CrossRef]
- Quilumba-Dutan, V.; Carreón-Álvarez, C.; Sanabria-Ayala, V.; Hidalgo-Figueroa, S.; Chakraborty, S.; Valsami-Jones, E.; López-Revilla, R.; Rodríguez-López, J.L. Assessment of Phage-Displayed Peptides Targeting Cancer Cell Surface Proteins: A Comprehensive Molecular Docking Study. J. Pept. Sci. 2025, 31, e70004. [Google Scholar] [CrossRef]
- Jirwankar, Y.; Nair, A.; Marathe, S.; Dighe, V. Phage Display Identified Novel Leydig Cell Homing Peptides for Testicular Targeting. ACS Pharmacol. Transl. Sci. 2024, 7, 809–822. [Google Scholar] [CrossRef]
- Yun, S.K.; Yang, S.M.; Kwak, M.H.; Park, J.M. Development and validation of cyclic peptide probe for gastric cancer based on phage display technique. Pept. Sci. 2024, 116, e24339. [Google Scholar] [CrossRef]
- Wu, X.; Feng, S.; Chang, T.-S.; Zhang, R.; Jaiswal, S.; Choi, E.-Y.K.; Duan, Y.; Jiang, H.; Wang, T.D. Detection of Hepatocellular Carcinoma in an Orthotopic Patient-Derived Xenograft with an Epithelial Cell Adhesion Molecule-Specific Peptide. Cancers 2024, 16, 2818. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Wang, S.H.; Zhao, L.; Wang, H.M.; Wang, L.; Shi, R.R.; Liang, S.C.; Li, B.F.; Chen, B. Screening and identification of vascular endothelial cell targeting peptide in gastric cancer through novel integrated in vitro and in vivo strategy. BMC Cancer 2024, 24, 1595. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zheng, L.; Wu, K.; Zhang, B. Identification and validation of a new peptide targeting pancreatic beta cells. Molecules 2022, 27, 2286. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.; Xiao, Y.; Wang, L.; Liang, M.; Li, Z.; Wu, X.; Cao, Q. Identification of an IGF2BP2-Targeted Peptide for Near-Infrared Imaging of Esophageal Squamous Cell Carcinoma. Molecules 2022, 27, 7609. [Google Scholar] [CrossRef] [PubMed]
- Furman, O.; Zaporozhets, A.; Tobi, D.; Bazylevich, A.; Firer, M.A.; Patsenker, L.; Gellerman, G.; Lubin, B.C.R. Novel cyclic peptides for targeting EGFR and EGRvIII mutation for drug delivery. Pharmaceutics 2022, 14, 1505. [Google Scholar] [CrossRef]
- Jin, H.; Gao, X.; Xiao, L.; He, H.; Cheng, S.; Zhang, C.; Hou, Y.; Song, F.; Su, X.; Gao, Q. Screening and identification of a specific peptide binding to breast cancer cells from a phage-displayed peptide library. Biotechnol. Lett. 2021, 43, 153–164. [Google Scholar] [CrossRef]
- Tooyserkani, R.; Rasaee, M.J.; Bandehpour, M.; WPM Löwik, D. Novel anti-PD-L1 peptide selected from combinatorial phage library inhibits tumor cell growth and restores T-cell activity. J. Drug Target. 2021, 29, 771–782. [Google Scholar] [CrossRef]
- Yan, J.; Yu, X.; Chen, X.; Liu, F.; Chen, F.; Ding, N.; Yu, L.; Meng, F.; Shen, J.; Wei, J. Identification of a glypican-3 binding peptide from a phage-displayed peptide library for PET imaging of hepatocellular carcinoma. Front. Oncol. 2021, 11, 679336. [Google Scholar] [CrossRef]
- Simón-Gracia, L.; Kiisholts, K.; Petrikaitė, V.; Tobi, A.; Saare, M.; Lingasamy, P.; Peters, M.; Salumets, A.; Teesalu, T. Homing peptide-based targeting of tenascin-C and fibronectin in endometriosis. Nanomaterials 2021, 11, 3257. [Google Scholar] [CrossRef]
- Kwak, M.H.; Yi, G.; Yang, S.M.; Choe, Y.; Choi, S.; Lee, H.S.; Kim, E.; Lim, Y.B.; Na, K.; Choi, M.G.; et al. A dodecapeptide selected by phage display as a potential theranostic probe for colon cancers. Transl. Oncol. 2020, 13, 100798. [Google Scholar] [CrossRef]
- Asar, M.C.; Franco, A.; Soendergaard, M. Phage display selection, identification, and characterization of novel pancreatic cancer targeting peptides. Biomolecules 2020, 10, 714. [Google Scholar] [CrossRef] [PubMed]
- Rahn, J.J.; Lun, X.; Jorch, S.K.; Hao, X.; Venugopal, C.; Vora, P.; Ahn, B.Y.; Babes, L.; Alshehri, M.M.; Cairncross, J.G. Development of a peptide-based delivery platform for targeting malignant brain tumors. Biomaterials 2020, 252, 120105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Huang, J.; Li, W.; Zhang, Z.; Zhu, M.; Feng, Y.; Zhao, Y.; Li, Y.; Lu, S.; He, S. Screening and identification of a CD44v6 specific peptide using improved phage display for gastric cancer targeting. Ann. Transl. Med. 2020, 8, 1442. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Ito, S.; Masuda, T.; Couraud, P.-O.; Ohtsuki, S. Novel cyclic peptides facilitating transcellular blood-brain barrier transport of macromolecules in vitro and in vivo. J. Control. Release 2020, 321, 744–755. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Q.; Hong, A.; Chen, X. Screening and identification of small peptides targeting fibroblast growth factor receptor2 using a phage display peptide library. J. Vis. Exp. (JoVE) 2019, 30, e60189. [Google Scholar] [CrossRef] [PubMed]
- Munisso, M.C.; Yamaoka, T. Evolution of phage display approaches to select highly specific hemocompatible peptides. Tissue Eng. Part C Methods 2019, 25, 288–295. [Google Scholar] [CrossRef]
- Bakhshinejad, B.; Nasiri, H. Identification of a novel tumor-binding peptide for lung Cancer through in-vitro panning. Iran. J. Pharm. Res. IJPR 2018, 17, 396–407. [Google Scholar] [PubMed]
- Hou, L.; Zhu, D.; Liang, Y.; Tian, X.; Li, L.; Wang, P.; Zhu, L.; Weng, X.; Wang, Y.; Li, Y. Identification of a specific peptide binding to colon cancer cells from a phage-displayed peptide library. Br. J. Cancer 2018, 118, 79–87. [Google Scholar] [CrossRef]
- Li, G.; Yin, Q.; Ji, H.; Wang, Y.; Liu, H.; Jiang, L.; Zhu, F.; Li, B. A study on screening and antitumor effect of CD55-specific ligand peptide in cervical cancer cells. Drug Des. Dev. Ther. 2018, 12, 3899–3912. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Peng, J.; He, J.; Li, Q.; Zhou, J.; Liang, X.; Tang, S. Selection and identification of novel peptides specifically targeting human cervical cancer. Amino Acids 2018, 50, 577–592. [Google Scholar] [CrossRef]
- Xing, L.; Xu, Y.; Sun, K.; Wang, H.; Zhang, F.; Zhou, Z.; Zhang, J.; Zhang, F.; Caliskan, B.; Qiu, Z. Identification of a peptide for folate receptor alpha by phage display and its tumor targeting activity in ovary cancer xenograft. Sci. Rep. 2018, 8, 8426. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Ito, S.; Kurogi-Hirayama, M.; Ohtsuki, S. Identification of cyclic peptides for facilitation of transcellular transport of phages across intestinal epithelium in vitro and in vivo. J. Control. Release 2017, 262, 232–238. [Google Scholar] [CrossRef]
- Fukuta, T.; Asai, T.; Kiyokawa, Y.; Nakada, T.; Bessyo-Hirashima, K.; Fukaya, N.; Hyodo, K.; Takase, K.; Kikuchi, H.; Oku, N. Targeted delivery of anticancer drugs to tumor vessels by use of liposomes modified with a peptide identified by phage biopanning with human endothelial progenitor cells. Int. J. Pharm. 2017, 524, 364–372. [Google Scholar] [CrossRef]
- Yu, C.-W.; Fu, C.-Y.; Hung, L.-Y.; Wang, C.-H.; Chiang, N.-J.; Wang, Y.-C.; Shan, Y.-S.; Lee, G.-B. Screening of peptide specific to cholangiocarcinoma cancer cells using an integrated microfluidic system and phage display technology. Microfluid. Nanofluid 2017, 21, 145. [Google Scholar] [CrossRef]
- Li, C.; Gao, N.; Xue, Q.; Ma, N.; Hu, Y.; Zhang, J.; Chen, B.; Hou, Y. Screening and identification of a specific peptide binding to cervical cancer cells from a phage-displayed peptide library. Biotechnol. Lett. 2017, 39, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.-H.; Hsiao, J.-K.; Lin, M.-H.; Chang, C.; Lan, C.-H.; Wu, H.-C. Lung cancer-targeting peptides with multi-subtype indication for combinational drug delivery and molecular imaging. Theranostics 2017, 7, 1612–1632. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, F.L.; Ferreira, D.; Martins, I.M.; Suarez-Diez, M.; Azeredo, J.; Kluskens, L.D.; Rodrigues, L.R. Screening and characterization of novel specific peptides targeting MDA-MB-231 claudin-low breast carcinoma by computer-aided phage display methodologies. BMC Cancer 2016, 16, 881. [Google Scholar] [CrossRef]
- Liu, Y.; Xia, X.; Wang, Y.; Li, X.; Zhou, G.; Liang, H.; Feng, G.; Zheng, C. Screening and identification of a specific peptide for targeting hypoxic hepatoma cells. Mol. Cell. Probes 2016, 30, 246–253. [Google Scholar] [CrossRef]
- Wang, L.; Hu, Y.; Li, W.; Wang, F.; Lu, X.; Han, X.; Lv, J.; Chen, J. Identification of a peptide specifically targeting ovarian cancer by the screening of a phage display peptide library. Oncol. Lett. 2016, 11, 4022–4026. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Qi, C.L.; Kong, M.; Liu, T.T.; Li, L.; Li, B.J. Screening specific polypeptides of breast cancer stem cells from a phage display random peptide library. Oncol. Lett. 2016, 12, 4727–4731. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Jia, H.; Li, W.; Hou, Y.; Lu, S.; He, S. Screening and identification of a phage display derived peptide that specifically binds to the CD44 protein region encoded by variable exons. J. Biomol. Screen. 2016, 21, 44–53. [Google Scholar] [CrossRef][Green Version]
- Jin, W.; Qin, B.; Chen, Z.; Liu, H.; Barve, A.; Cheng, K. Discovery of PSMA-specific peptide ligands for targeted drug delivery. Int. J. Pharm. 2016, 513, 138–147. [Google Scholar] [CrossRef]
- Cornelison, G.L.; Pflanz, N.C.; Tipps, M.E.; Mihic, S.J. Identification and characterization of heptapeptide modulators of the glycine receptor. Eur. J. Pharmacol. 2016, 780, 252–259. [Google Scholar] [CrossRef]
- Bozovičar, K.; Jenko Bizjan, B.; Meden, A.; Kovač, J.; Bratkovič, T. Focused peptide library screening as a route to a superior affinity ligand for antibody purification. Sci. Rep. 2021, 11, 11650. [Google Scholar] [CrossRef] [PubMed]
- Kepler, T.B.; Munshaw, S.; Wiehe, K.; Zhang, R.; Yu, J.-S.; Woods, C.W.; Denny, T.N.; Tomaras, G.D.; Alam, S.M.; Moody, M.A. Reconstructing a B-cell clonal lineage. II. Mutation, selection, and affinity maturation. Front. Immunol. 2014, 5, 170. [Google Scholar] [CrossRef]
- Bhat, A.; Roberts, L.R.; Dwyer, J.J. Lead discovery and optimization strategies for peptide macrocycles. Eur. J. Med. Chem. 2015, 94, 471–479. [Google Scholar] [CrossRef]
- Labrou, N.E. Random mutagenesis methods for in vitro directed enzyme evolution. Curr. Protein Pept. Sci. 2010, 11, 91–100. [Google Scholar] [CrossRef]
- Harayama, S. Artificial evolution by DNA shuffling. Trends Biotechnol. 1998, 16, 76–82. [Google Scholar] [CrossRef]
- Stemmer, W.P. Rapid evolution of a protein in vitro by DNA shuffling. Nature 1994, 370, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, J. Research progress in structure-activity relationship of bioactive peptides. J. Med. Food 2015, 18, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Ciulla, M.G.; Gelain, F. Structure–activity relationships of antibacterial peptides. Microb. Biotechnol. 2023, 16, 757–777. [Google Scholar] [CrossRef]
- Guha, R. On exploring structure–activity relationships. In Silico Models for Drug Discovery; Springer: Berlin/Heidelberg, Germany, 2013; pp. 81–94. [Google Scholar]
- Jamieson, A.G.; Boutard, N.; Sabatino, D.; Lubell, W.D. Peptide scanning for studying structure-activity relationships in drug discovery. Chem. Biol. Drug Des. 2013, 81, 148–165. [Google Scholar] [CrossRef]
- Zhou, P.; Wang, C.; Ren, Y.; Yang, C.; Tian, F. Computational peptidology: A new and promising approach to therapeutic peptide design. Curr. Med. Chem. 2013, 20, 1985–1996. [Google Scholar] [CrossRef]
- Vidal-Limon, A.; Aguilar-Toalá, J.E.; Liceaga, A.M. Integration of molecular docking analysis and molecular dynamics simulations for studying food proteins and bioactive peptides. J. Agric. Food Chem. 2022, 70, 934–943. [Google Scholar] [CrossRef]
- Raguine, L.; Ali, M.; Bender, V.; Diefenbach, E.; Doddareddy, M.R.; Hibbs, D.; Manolios, N. Alanine scan of an immunosuppressive peptide (CP): Analysis of structure–function relationships. Chem. Biol. Drug Des. 2013, 81, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Haskell-Luevano, C.; Sawyer, T.K.; Hendrata, S.; North, C.; Panahinia, L.; Stum, M.; Staples, D.J.; Castrucci, A.M.D.L.; Hadley, M.E.; Hruby, V.J. Truncation studies of α-melanotropin peptides identify tripeptide analogues exhibiting prolonged agonist bioactivity. Peptides 1996, 17, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Carrigan, P.E.; Ballar, P.; Tuzmen, S. Site-directed mutagenesis. In Disease Gene Identification: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2010; pp. 107–124. [Google Scholar]
- Ladner, R.C.; Sato, A.K.; Gorzelany, J.; de Souza, M. Phage display-derived peptides as therapeutic alternatives to antibodies. Drug Discov. Today 2004, 9, 525–529. [Google Scholar] [CrossRef]
- Deshayes, K.; Schaffer, M.L.; Skelton, N.J.; Nakamura, G.R.; Kadkhodayan, S.; Sidhu, S.S. Rapid identification of small binding motifs with high-throughput phage display: Discovery of peptidic antagonists of IGF-1 function. Chem. Biol. 2002, 9, 495–505. [Google Scholar] [CrossRef]
- Alteen, M.G.; Meek, R.W.; Kolappan, S.; Busmann, J.A.; Cao, J.; O’Gara, Z.; Chou, Y.; Derda, R.; Davies, G.J.; Vocadlo, D.J. Phage display uncovers a sequence motif that drives polypeptide binding to a conserved regulatory exosite of O-GlcNAc transferase. Proc. Natl. Acad. Sci. USA 2023, 120, e2303690120. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Onishi, A.; Saito, T.; Shimada, A.; Inoue, H.; Taki, T.; Nagata, K.; Okahata, Y.; Sato, T. Sialic acid-mimic peptides as hemagglutinin inhibitors for anti-influenza therapy. J. Med. Chem. 2010, 53, 4441–4449. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Smith, G.P. Affinity maturation of phage-displayed peptide ligands. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1996; Volume 267, pp. 3–27. [Google Scholar]
- Smith, G.P.; Yu, J. In search of dark horses: Affinity maturation of phage-displayed ligands. Mol. Divers. 1996, 2, 2–4. [Google Scholar] [CrossRef]
- Zambrano-Mila, M.S.; Blacio, K.E.S.; Vispo, N.S. Peptide phage display: Molecular principles and biomedical applications. Ther. Innov. Regul. Sci. 2020, 54, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Cimen, A.N.; Porsuk, M.H.; Kutlu, O.; Cetinel, S. Phage Display–Selected Peptides: Research and Clinical Applications in Cancer Imaging. J. Pept. Sci. 2025, 31, e70034. [Google Scholar] [CrossRef]
- Angelini, A.; Cendron, L.; Chen, S.; Touati, J.; Winter, G.; Zanotti, G.; Heinis, C. Bicyclic peptide inhibitor reveals large contact interface with a protease target. ACS Chem. Biol. 2012, 7, 817–821. [Google Scholar] [CrossRef]
- Rebollo, I.R.; Heinis, C. Phage selection of bicyclic peptides. Methods 2013, 60, 46–54. [Google Scholar] [CrossRef]
- Chen, F.J.; Pinnette, N.; Gao, J. Strategies for the construction of multicyclic phage display libraries. ChemBioChem 2024, 25, e202400072. [Google Scholar] [CrossRef]
- Allen, G.L.; Grahn, A.K.; Kourentzi, K.; Willson, R.C.; Waldrop, S.; Guo, J.; Kay, B.K. Expanding the chemical diversity of M13 bacteriophage. Front. Microbiol. 2022, 13, 961093. [Google Scholar] [CrossRef]
- Hampton, J.T.; Liu, W.R. Diversification of phage-displayed peptide libraries with noncanonical amino acid mutagenesis and chemical modification. Chem. Rev. 2024, 124, 6051–6077. [Google Scholar] [CrossRef]
- Croce, G.; Lani, R.; Tardivon, D.; Bobisse, S.; de Tiani, M.; Bragina, M.; Perez, M.A.; Michaux, J.; Pak, H.S.; Michel, A. Phage display enables machine learning discovery of cancer antigen–specific TCRs. Sci. Adv. 2025, 11, eads5589. [Google Scholar] [CrossRef] [PubMed]
- Bakhshinejad, B.; Kjaer, A. On the origin of non-specific binders isolated in the selection of phage display peptide libraries. Front. Microbiol. 2025, 16, 1571679. [Google Scholar] [CrossRef] [PubMed]
- Bakhshinejad, B.; Zade, H.M.; Shekarabi, H.S.Z.; Neman, S. Phage display biopanning and isolation of target-unrelated peptides: In search of nonspecific binders hidden in a combinatorial library. Amino Acids 2016, 48, 2699–2716. [Google Scholar] [CrossRef] [PubMed]
- Gray, B.P.; Brown, K.C. Combinatorial peptide libraries: Mining for cell-binding peptides. Chem. Rev. 2014, 114, 1020–1081. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).